Abstract
The RecA protein plays a principal role in the bacterial SOS response to DNA damage. The induction of the SOS response is well understood and involves the cleavage of the LexA repressor catalyzed by the RecA nucleoprotein filament. In contrast, our understanding of the regulation and termination of the SOS response is much more limited. RecX and DinI are two major regulators of RecA’s ability to promote LexA cleavage and a strand exchange reaction and are believed to modulate its activity in ongoing SOS events. DinI’s function in the SOS response remains controversial since its interaction with the RecA filament is concentration-dependent and may result in either stabilization or depolymerization of the filament. The 17 C-terminal residues of RecA modulate the interaction between DinI and RecA. We demonstrate that DinI binds to the active RecA filament in two distinct structural modes. In the first mode DinI binds to the C-terminus of a RecA protomer. In the second mode DinI resides deeply in the groove of the RecA filament with its negatively charged C-terminal helix proximal to the L2 loop of RecA. The deletion of the 17 C-terminal residues of RecA favors the second mode of binding. We suggest that the negatively charged C-terminus of RecA prevents DinI from entering the groove and protects the RecA filament from depolymerization. Polymorphic binding of DinI to RecA filaments implies an even more complex role of DinI in the bacterial SOS response.
Introduction
RecA is a key player in maintaining the integrity of the bacterial genome. It is crucial for DNA repair via homologous recombination and required for the induction of the SOS response in bacteria1–3. The RecA protein of E. coli contains 352 amino acid residues and consists of a large core domain and a smaller C-terminal domain (residues 271-352). The last 24 residues were found to be disordered in all crystal structures4–8, except the M. smegmatis RecA complexed with dATP9. Most bacterial RecA proteins contain a high concentration of negatively charged residues at their C-terminus10.
Double-stranded DNA (dsDNA) breakages or stalled replication forks produce regions of single-stranded DNA (ssDNA). Polymerization of RecA monomers on ssDNA initiates transcription of more than 40 proteins involved in the SOS response in bacteria11. Synthesis of these proteins is normally inhibited by the LexA repressor which can cleave itself12 upon binding within the helical groove of the RecA-DNA filament13. Also, the RecA-DNA filament acts as an ATP-dependent recombinase and is capable of promoting DNA pairing and strand exchange14. The repair of the damaged dsDNA results in degradation of ssDNA and depolymerization of the RecA nucleoprotein filaments which in turn restores the pool of LexA repressor and shuts down the SOS system.
To prevent undesired DNA rearrangements, bacteria developed a sophisticated system to control the formation of the RecA-ssDNA filament. RecA activity is modulated by its C-terminus in a Mg2+-dependent fashion15. Deletion of the 25 C-terminal residues results in faster RecA nucleation16. The 17 C-terminal RecA residues prevent binding of RecA to dsDNA, favoring RecA polymerization on ssDNA, and modulate RecA’s ability to displace SSB protein from ssDNA17. Recently crystal structures of the RecA-ssDNA/dsDNA filaments have been solved18, but these did not provide any structural information about the RecA C-terminus because this region was used as linker to construct a RecA polypeptide.
The other two well known modulators of RecA filaments are RecX and DinI. RecX protein is an intrinsic inhibitor of RecA activities, and overexpression of RecA in the absence of the RecX gene is toxic for bacteria19,20. RecX inhibits the RecA-dependent strand exchange reaction and co-protease activity by means of slow depolymerization of RecA-DNA filaments21,22. Similarly to LexA, RecX binds deep in the helical groove of RecA filaments13. The competition between LexA and RecX for binding within the RecA helical groove may contribute to inhibition of LexA cleavage by RecX. Removal of the 17 C-terminal RecA residues (RecAΔC17) significantly alters the ability of RecX to inhibit the strand exchange reaction and DNA-dependent ATPase activity23.
Another member of the SOS regulon is the DinI gene which encodes a small protein containing 81 amino acid residues24,25. Originally it was shown that the over-expression of DinI protein conferred severe UV sensitivity on wild-type cells and resulted in the inhibition of LexA and UmuD cleavage26. Later it was suggested that under normal expression levels DinI would inhibit UmuD cleavage so as to limit SOS mutagenesis, while having little effect on LexA processing26. The role of DinI as a down-regulator of the SOS response was also supported by the observation that the maximum binding of DinI to RecA filaments occurred at later stages of the SOS response27. It was proposed that a negatively charged C-terminal helix in DinI could compete with ssDNA for binding to the L2 loop of RecA, which in turn would displace the ssDNA from the RecA nucleoprotein filament and inhibit the strand exchange reaction27. At a stoichiometric ratio of DinI to RecA, DinI has a substantial stabilizing effect on the RecA filament28. It also allows LexA cleavage and does not interfere with an on-going strand exchange reaction. The deletion of 17 C-terminal residues of RecA has a dramatic effect on the RecA-DinI interaction28 – it alters DinI affinity for the RecA filament. In contrast to the wild type RecA, RecAΔC17 has reduced strand exchange activity when DinI is present. We used electron microscopy and image analysis to locate the binding site of DinI on the RecA filament and to explore the structural consequences of the deletion of the 17 C-terminal residues of RecA on its ability to interact with DinI.
Results and discussion
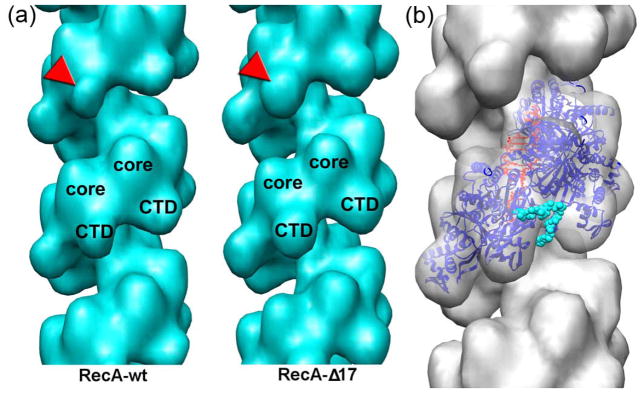
The formation of the RecA filament requires ATP-binding, but ATP hydrolysis is not required for the co-protease activity29 or for the catalysis of the strand exchange reaction30. In the presence of ATP RecA forms disordered filaments due to protomer dissociation upon ATP hydrolysis. Such filaments are inappropriate for image analysis. We have shown previously that the long and ordered RecA filaments formed in the presence of non-hydrolysable ATP analogs are in an active extended conformation31. In this study we used AMP-PNP to form RecA-DNA or RecAΔC17-DNA filaments alone (Fig. 1a and b), or in the presence of DinI (Fig. 1c and d). We extracted 6,381 segments of RecA and 5,198 of RecAΔC17 to generate 3D-reconstructions of those filaments (Fig 2a). Both sets converged to a pitch of ~92 Å consistent with the extended “active” form of the filament. Paradoxically, wild type RecA reconstruction possessed a weaker electron density in the C-terminal domain (CTD) region than the RecAΔC17 reconstruction (Fig 2a, red arrowheads), but otherwise the two reconstructions were similar. This was consistent with our previous observations that the deletion of 18 C-terminal residues resulted in conformational changes in the CTD32. Recently crystal structures of RecA-ssDNA/dsDNA filaments have been solved18. Since our filaments were formed on dsDNA, we used the high resolution structure of the RecA-dsDNA filament (PDB 3CMT) to evaluate the reconstruction of the RecAΔC17 filament. At a resolution of ~23 Å no perturbations of the original crystal structure were required to dock it into the electron density map (Fig 2b). This suggests that no large scale conformational changes to the RecA protomers within the extended RecA filament take place upon the deletion of the 17 terminal residues. The alignment of the RecA protomer from the RecA-dsDNA heteroduplex filament with the crystal structure of the M. smegmatis RecA9 places the missing 17 residues in the region spanning from the outer portion of the CTD to the channel in between two adjacent RecA protomers (Fig 2b, cyan spheres).
Fig. 1.
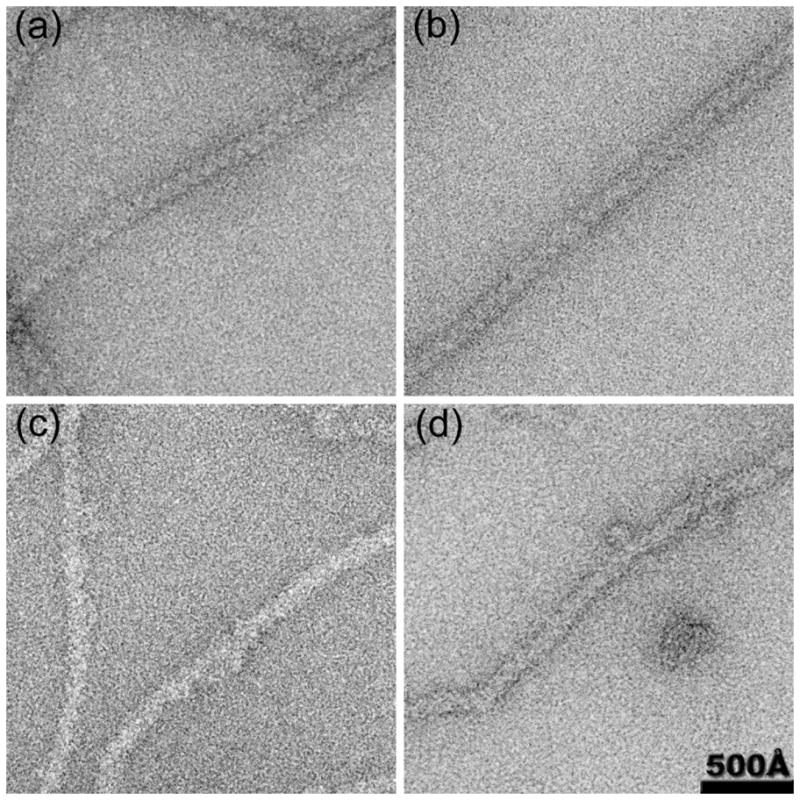
Electron micrographs of RecA-DNA filaments (a) and RecAΔC17-DNA filaments (b) show strong periodic striations that arise from the one-start right handed RecA filament helix. These striations disappear when DinI is added to wt-RecA (c) or RecAΔC17 filaments (d).
Fig. 2.
3D-reconstructions of RecA and the RecAΔC17 filaments (a) are similar in the core region, but differ in the C-terminal domain (CTD) region (red arrowheads). The crystal structure of the RecA-dsDNA filament18 fits without perturbation into the RecAΔC17 reconstruction (b). The RecA protomers are shown as blue ribbons, while the dsDNA molecule is in red. The position of the 24 C-terminal residues found ordered in the crystal structures of the M. smegmatis RecA9 are shown as cyan spheres.
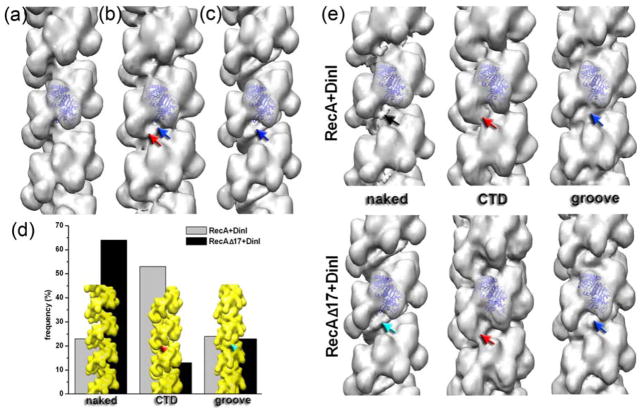
Incubation of the RecA or RecAΔC17 filaments with DinI resulted in the disappearance of the helical pattern (Fig 1c,d) suggesting that DinI was bound to these filaments. We collected 5,891 segments of wt-RecA and 5,773 segments of RecAΔC17 filaments decorated with DinI. The 3D-reconstructions of the two sets are shown in Fig 3b and c, respectively. The wt-RecA-DinI reconstruction (Fig 3b) showed two regions of density absent in the map of the pure RecAΔC17 (Fig 3a) – the first site was an attachment to the CTD (Fig 3b, red arrow), while the second site was a continuous density located in the helical groove (Fig 3b, blue arrow). A continuous density located in the helical groove was observed in the RecAΔC17-DinI reconstruction (Fig 3c, blue arrow), though no binding of DinI to RecA’s CTD was detected in the overall reconstruction. It was unclear whether the two regions of binding of DinI found in the RecA-DinI reconstruction (Fig 3b, red and blue arrows) reflected a single mode of DinI-RecA interaction, or represented a mixture of two distinct modes.
Fig. 3.
Comparisonof the 3D-reconstructions of pure RecA ΔC17 filament (a), RecA-DinI (b) and RecAΔC17-DinI (c) complexes reveals additional density bound to the C-terminal domain of RecA (red arrow) along with the continuous density residing in the helical groove (blue arrows). Cross-correlation sorting using model volumes (d, yellow surfaces) revealed differences in the frequency of the two modes of DinI binding to RecA (d, grey and black bars). 3D-reconstructions of each of the six classes shown in (d) are shown in (e) as solid surfaces. While the reconstructions of the CTD (red arrows) and the groove (blue arrows) modes from the RecA-DinI and RecAΔC17-DinI complexes are similar, the reconstructions of the naked classes differ. Poorly decorated segments from RecA-DinI filaments have few traces of the DinI bound (black arrow), while such filaments extracted from the RecAΔC17-DinI complex possess continuous density in the helical groove (cyan arrow).
The single particles approach to helical reconstruction allows us to sort segments of filaments by the occupancy and the mode of binding. We generated three model volumes (Fig 3d) – naked RecA filament, a RecA filament having additional density at its CTD (Fig 3d, red arrowhead), and a RecA filament with a continuous density in the helical groove (Fig 3d, cyan arrowhead). The three model volumes were projected and cross-correlated with the images of RecA-DinI and RecAΔC17-DinI complexes. The frequency distribution for such a sorting is shown in Fig 3d as grey and black bars, respectively. The majority of the RecA-DinI images (~54%) had the highest correlation with the model having additional density attached to the CTD, while a quarter of segments were assigned to the “groove” class (Fig 3d, grey bars). The deletion of the 17 C-terminal residues did not alter the frequency of the groove mode, but significantly reduced the population of the “CTD” class (Fig 3d, black bars). Each class was iterated in the IHRSR procedure starting from a solid featureless cylinder until it converged to a stable solution. The resultant reconstructions are shown in Fig 3e. The poorly decorated RecA-DinI filaments yielded a reconstruction possessing weak traces of DinI (Fig 3e, black arrow), while the same class from the RecAΔC17-DinI complex resulted in a map having continuous additional density in the helical groove (Fig 3e, cyan arrow). This suggested that the cooperativity of DinI binding to RecA was higher than that to RecAΔC17. The CTD class reconstructions had a substantial additional mass attached to the CTD (Fig 3e, red arrows). The groove classes from both sets yielded maps having substantial continuous density in the helical groove of the RecA filament (Fig 3e, blue arrows). Despite similar frequencies of the groove classes (Fig 3d), and because of the substantial additional density in the helical groove of the naked RecAΔC17-DinI class reconstruction, our sorting suggested that the groove of the RecAΔC17 filaments was more attractive to DinI than that of unmodified RecA filaments.
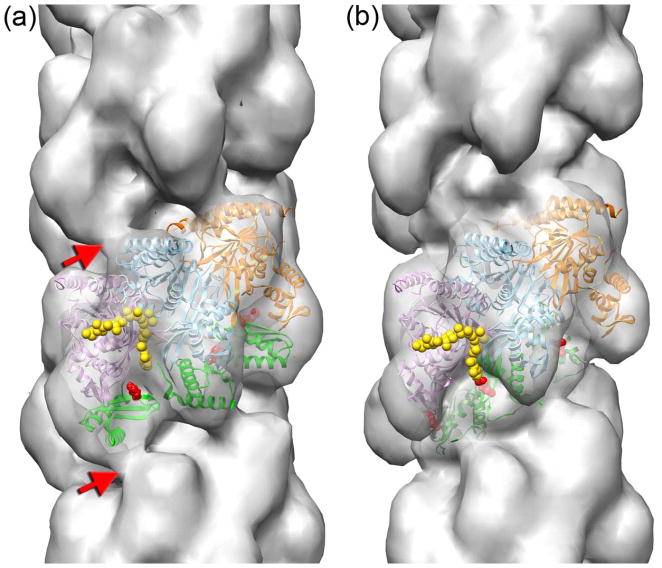
Since each class was reconstructed starting from a featureless solid cylinder, the details of the reconstructions reflected the real modes of interaction of DinI with the RecA filament rather than being model-biased solutions. We therefore used the reconstructions of the CTD and the groove classes of the RecAΔC17-DinI complex to build atomic models of the two structural modes. The RecA protomer from the crystal structure of the RecA-dsDNA filament18 was used to model the position of RecA protomers in the RecA-DinI complex, while keeping the interface between the adjacent RecA protomers similar to that in the original crystal. Three consecutive RecA protomers docked into the density map are shown in Fig 4 as magenta, cyan, and orange ribbons. Docking of RecA into the two decorated maps allowed us to determine the density attributed to the DinI in each reconstruction. In both modes of binding the additional density perfectly matched the size and the shape of the DinI molecule (Fig 4, green ribbons). The models suggested that the two modes were mutually exclusive. That is, a DinI molecule could not be bound to the same RecA protomer in both modes without extensive steric clashes. If DinI were randomly bound to RecA filaments at either of the two sites, we would never be able to sort out the two discrete modes that we observe. Instead, our results are consistent with a cooperativity of binding so that within the lengths of the segments we are analyzing (~ 22 RecA subunits) there is predominantly one mode present. What could be responsible for such cooperativity? In the groove mode (Fig 4b) there appears to be an interaction between adjacent DinI molecules bound to RecA actin subunits, and such a DinI–DinI interaction could contribute to the cooperative nature of the binding. Since in the CTD-mode the two DinI molecules do not interact with each other, it is likely that in that particular mode the cooperativity in the interaction must reside mainly within RecA filament. That is, when DinI binds to one RecA protomer in CTD mode, structural changes in RecA filament greatly increase the likelihood that DinI will bind in this same manner to adjacent RecA subunits. This is consistent with many, many papers showing such internal cooperativity within RecA filaments33,34.
Fig. 4.
3D-reconstructions of the CTD (a) and the groove (b) modes are shown with three consecutive RecA protomers docked into the electron density maps (magenta, cyan, and orange ribbons). Three DinI molecules in each mode are shown as green ribbons. The 17 C-terminal residues present in the crystal structure of the M. smegmatis RecA9 are shown as yellow spheres. In the CTD mode the DinI protein bridges adjacent RecA protomers across the helical strand (red arrows). Mutation in R43 (red spheres) significantly diminishes the ability of DinI to stimulate ATP hydrolysis by RecA.
In general, the docking of a high resolution structure of a protein into a 23 Å resolution map is quite ambiguous35. Due to a substantial asymmetry of the RecA and DinI molecules when filtered to 23 Å, the orientation of both molecules in the resultant atomic models was unambiguous. In addition, the two models are supported by DinI mutagenesis studies which show that substitution of Arg 43 of DinI significantly reduces its ability to stimulate the RecA ATPase activity (Supplementary data). In both atomic models that residue is located in proximity to the RecA CTD (Fig 4a,b, red spheres).
We used the recent crystal structure of the M. smegmatis RecA9 to reveal the position of the 17 C-terminal residues in our atomic models (Fig 4, yellow spheres). Interestingly, in both modes the C-terminus was located far from the DinI molecule (Fig 4a,b). The removal of the 17 C-terminal residues of RecA did not alter the mode of binding of DinI to RecA; rather it only changed the frequency of the existing modes (Fig 3d), suggesting that the C-terminus is not involved in direct interaction with DinI. Why then does the deletion of these C-terminal residues alter the interaction between RecA and DinI? Most of the bacterial RecA proteins contain a high concentration of negatively charged residues at their C-terminus10. In all but one RecA crystal structure this region of RecA has been found to be disordered. The DinI molecule has an extended negatively charged ridge36 in its C-terminal helix. If the C-terminus of RecA is located in the groove of the filament, electrostatic forces may preclude extensive binding of DinI in the helical groove and favor the CTD mode of binding. Alternatively, the deletion of the C-terminus may cause conformational changes in RecA, and this could alter the frequency of the two modes. This is consistent with our observations that the deletion of the 17 C-terminal residues stabilizes the CTD of RecA (Fig 2a). In addition, the extreme C-terminal residues could be making a direct favorable contact with DinI increasing the frequency of this mode. A high resolution structure of the RecA-DinI complex is required to distinguish among these possibilities.
It has been shown that at low concentration DinI can stabilize the RecA filament without interfering with LexA cleavage and an ongoing strand exchange reaction28. When DinI binds to RecA in the CTD mode, it bridges adjacent RecA protomers across the helical groove (Fig 4a, red arrows) contributing to the intrafilament contacts. Importantly, being bound in this mode leaves the groove available for LexA cleavage13 and for the invasion of dsDNA into the groove.
An extensive interaction between the adjacent DinI molecules in the groove mode may increase the affinity of DinI for the RecA filament. Because of the preferential binding of DinI to RecAΔC17 in the groove mode, one would expect the RecAΔC17-DinI complex to be more stable that the RecA-DinI one. Strikingly, the RecAΔC17-DinI complex was shown to be 100 times more stable than the one formed with unmodified RecA28.
When DNA in the atomic model of the RecAΔC17 filament is positioned as in the crystal structure18, it is proximal to the L2 loop region of RecA (Fig 5b, red spheres) which has been implicated in DNA binding 37,38 and homologous pairing39. The solution structure of DinI shows that six Asp/Glu residues in the C-terminal helix form an extended negatively charged ridge36 (Fig 5c–f, cyan spheres), and this ridge was proposed to compete with ssDNA for binding to the RecA L2 region27. In the CTD mode the negatively charged region of DinI is located at the outer surface of the RecA filament (Fig 5d, cyan spheres), and is far from the L2 loop of RecA (Fig 5d, red spheres). In the groove mode the negatively charged C-terminal helix forms a continuous helical path (Fig 5f, cyan spheres) flanking the L2 region of RecA (Fig 5f, red spheres). Our data show that when DinI is bound to RecA filament in the groove mode, it can compete with the DNA for binding to the L2 loop. This interaction may facilitate the dissociation of RecA subunits from the nucleoprotein filament.
Fig. 5.
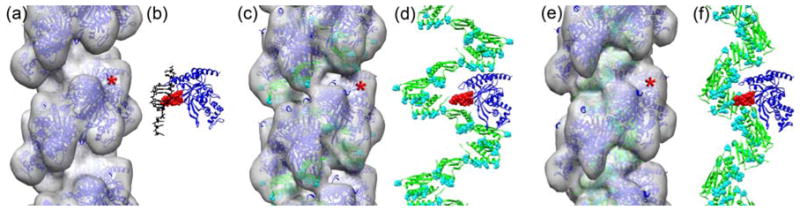
In the atomic model of the pure RecA filament (a, blue ribbons) the dsDNA molecule (b, black ribbons) is proximal to the L2-loop of the RecA (b, red spheres). When DinI binds to RecA in the CTD mode (c,d, green ribbons) the six Asp/Glu residues in the C-terminal helix that form an extended negatively charged ridge36 (c,d, cyan spheres) are distal from the L2-loop of RecA (d, red spheres). In the atomic model of the groove mode the negatively charged C-terminal helix forms a continuous helical pattern (e,f,, cyan spheres) flanking the L2 region of RecA (f, red spheres). The position of the single RecA protomer (blue ribbons in b,d,f) is marked with red asterisks in the filament models (a,c,e).
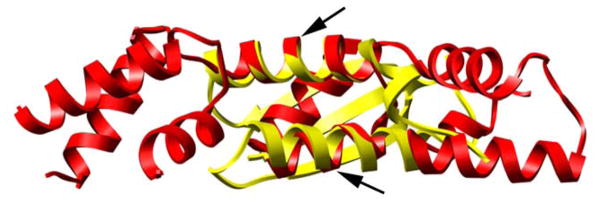
RecX protein is a suppressor of the SOS response and inhibits LexA cleavage as well as the strand exchange reaction by means of depolymerization of RecA filaments21,23. When the RecA filament is stabilized with AMP-PNP, RecX produces a continuous density that forms a block across the deep helical groove of the RecA filament13. Such a mode of binding intuitively explains how RecX can inhibit the LexA cleavage by simply blocking the penetration of LexA into the groove of the filament. It has been suggested recently that the positively charged concave surface of RecX can make an extensive contact with the DNA phosphate backbone40. DinI binds to RecAΔC17 filaments presumably in the groove mode, and its position on the RecA filament in that mode (Fig 4b) is very similar to that found for RecX protein13. Consistent with this picture, DinI inhibits the ability of RecAΔC17 to catalyze the strand exchange reaction28. RecX contains three small domains, each containing three helices. The protein appears to have been the result of a gene triplication, as the three domains show strong structural homology. The two helices in DinI align extremely well with two of the three helices in each of the RecX domains, and the fit of DinI to the central domain of RecX is shown (Fig 6). Whether this represents a convergent evolution or distant homology cannot be determined at this time.
Fig. 6.
The RecX protein (red) consists of three domains, where each domain contains three α-helices40. The structural similarity of each of the three domains has suggested a gene triplication. The DinI protein (yellow) is a single domain, containing two α-helices and a single β-sheet36. Two of the three RecX helices superimpose quite well on the two helices in DinI, raising the possibility of homology for this RecA-binding domain. At the bottom DinI helix containing residues 17-31 is superimposed on the RecX helix containing residues 81-90, while at the top the DinI helix containing residues 57-73 is superimposed on the RecX helix containing residues 63-76.
Our data show that the C-terminus of RecA is involved in modulation of the RecA-DinI interaction. In the groove mode DinI resides deeply in the groove of the RecA filament with its negatively charged C-terminal helix proximal to the L2 loop of RecA. This supports the original suggestion that the negatively charged C-terminal helix of DinI mimics DNA and interferes with the activation of RecA27. We suggest that the negatively charged C-terminus of RecA prevents DinI from entering the groove and protects the RecA filament from depolymerization, since the deletion of the 17 C-terminal residues of RecA favors the groove mode of binding. How can such a regulatory mechanism work in the cell lacking truncated RecA protein? The deletion of the 17 C-terminal residues in RecA in vitro may mimic conformational changes in RecA filament that otherwise are triggered by regulatory proteins such as RecO and RecR. The enhanced binding of DinI to RecA at late stages of the SOS response27 strongly argues for the existence of one or more modulators of the DinI-RecA interaction, since DinI synthesis starts at early stages of the SOS response.
Materials and Methods
Cloning
To clone an E. coli dinI gene that encodes the R43A mutant protein, the wild-type dinI gene in pEAW334 was used as the template in a PCR with the upstream primer consisting of the dinI gene starting at the BsaAI restriction enzyme site at base 115 and including at least 10 bases after the changed bases. For R43A, the CGT starting at base 126 of the dinI gene was changed to GCT. The downstream primer consisted of an EcoRI restriction enzyme site and the last 19 bases of the dinI gene. The PCR product was digested with BsaAI and EcoRI and ligated to pEAW340 cut with the same restriction enzymes. The plasmid designated pEAW340 is the wild-type dinI gene inserted into pUC19 sites. The presence of the dinI mutation was confirmed by direct sequencing of the resulting plasmids. The mutant dinI gene was then subcloned into pET21A cut at the NdeI and EcoRI sites, and the dinI genes of the resulting plasmids were again directly sequenced. The plasmid with dinI R43A was designated pEAW533.
To create His-tagged versions of the mutant and wild type DinI proteins, primers (5′-CCTAAGCCATATGCGAATTGAAGTCACC-3′ and 5′-GGATATCCTCGAGTTCGCTGACAACCAGTC-3′) were used to amplify mutant dinI genes from their respective plasmids (pEAWs 533 and 334) and append flanking NdeI and XhoI restriction sites. PCR products and pET-21b(+) (Novagen) were cut with NdeI and XhoI and were ligated. The resulting plasmids were sequenced to ensure the integrity of the dinI coding region. The above cloning procedure resulted in mutant dinI genes with six His codons at the 3′ end of the dinI coding sequence followed by a stop codon.
DNA substrates
The poly-dT DNA was purchased from Amersham Biosciences. Its average length was 319 nucleotides. The concentration of poly-dT was reported in terms of μM nucleotides (nt).
Proteins
Untagged wild type DinI pr otein was over-expressed in E. coli STL2669/pT7pol26 cells. Ten liters of culture were grown in LB to an OD600 of 0.5, and DinI protein expression was induced with 0.4 mM isopropyl-1-thio-β-D-galactopyranoside. Approximately 15 to 20 g of cells were harvested by centrifugation following 3 hours of incubation at 37 °C. Cells were flash-frozen in liquid N2, and stored at −80°C. Cells were thawed over-night on ice in buffer containing 250 mM Tris-Cl (80% cation, pH 7.8) and 25 % w/v sucrose. The cells were resuspended to 20 % w/v and lysed via sonication with a Fisher Scientific Sonic Dismembrator Model 500 at 60% output, 5 times, for 1 min segments with alternating 0.5 s on/off pulses. Lysed cells were centrifuged for 60 min at 16,000 rpm in a Beckman JLA-16.25 rotor at 4°C. The lysis supernatant was dialyzed against P buffer (10 mM KH2PO4, 10 mM K2HPO4, 0.1 mM EDTA, 10% w/v glycerol, 1 mM DTT) and run over a ceramic hydroxyapatite column. Wild type DinI protein was not retained by the hydroxyapatite and the wash fractions containing DinI were dialyzed against R buffer (20 mM Tris-Cl 80% cation, pH 7.8, 0.1 mM EDTA, 10% w/v glycerol, 1 mM dithiothreitol (DTT)). The DinI was then applied to a Q-sepharose anion exchange column and eluted with a linear gradient from R-buffer to R-buffer + 1 M KCl. Gradient fractions containing DinI protein were dialyzed back into R-buffer and run over a heparin sepharose column. Heparin sepharose flowthrough fractions containing DinI protein were concentrated on a 1 ml Q-sepharose column with a step gradient from R-buffer to R + 1 M KCl. The concentrated DinI was run over a Hi-Prep Sephacryl 16/60 S-100 size exclusion column in R + 200 mM KCl. Fractions containing pure wild type DinI protein were pooled and dialyzed into R-buffer for storage. The wild type DinI protein was free of detectable ssDNA nuclease activity.
To over-express the His-tagged DinI proteins, plasmids bearing the dinI genes were transformed into the E. coli STL327 (DE3) strain which was grown, harvested and frozen as described above for untagged wild type DinI. Five ml of lysis buffer (50 mM potassium phosphate, 300 mM NaCl, 10 mM imidazole, 10% w/v glycerol, pH 8.0) were added per gram of cell pellet to resuspend the pellet. Cells were thawed overnight, on ice, with gentle stirring. Protease inhibitors E-64 and Leupeptin at 0.5 μg/ml, and Pepstatin at 1 μM, were added to fully resuspended cells. The cells were lysed at 4°C for 2 hours with lysozyme at a final concentration of 4 mg/ml. Cells were sonicated and centrifuged as described above. The supernatant was incubated in batch with ~5 ml Ni-NTA slurry pre-equilibrated with lysis buffer for 1 h at 4°C. The slurry was poured into a solid support, then washed with 30 column volumes of wash buffer (50 mM potassium phosphate, 0.3 M NaCl 20 mM imidazole, 10% w/v glycerol, pH 8.0). Histidine-tagged DinI protein was eluted with 5 column volumes of elution buffer (same as wash buffer except with 500 mM imidazole). Wild type histidine-tagged DinI (WT His-DinI) was dialyzed against R buffer and eluted from a 1 ml Q-sepharose column with a linear gradient from R buffer to R-buffer + 1 M KCl. Fractions containing WT His-DinI were run over a Hi-Prep Sephacryl 16/60 S-100 size exclusion column and a ceramic hydroxyapatite column, and concentrated on a 1 ml Q-sepharose column with a step gradient as described above. Eluted WT His-DinI protein was dialyzed against R buffer for storage. His-R43A DinI protein was purified as for the wild type His-tagged protein, except that it was not concentrated on the 1 ml Q-sepharose after the ceramic hydroxyapatite column. All purified proteins were flash frozen with liquid N2 and stored at −80°C. Each was determined to be pure by SDS-PAGE and had no detectable ssDNA nuclease activity at 20 μM DinI after 2 hours of incubation at 37 °C. The theoretical extinction coefficient of 1.44 × 104 M−1 cm−1 at 280 nm for wild type DinI calculated using the ProtParam tool (from the ExPASy web site) was used to determine the concentration of all the DinI proteins.
Each DinI protein was diluted in R-buffer to a concentration between 0.1 and 0.2 mg/ml and was submitted to the Biophysics Instrumentation Facility at UW-Madison to check for proper folding by circular dichroism (CD). All data were collected in 0.1 cm cuvettes at 37 °C, with a 1 nm bandpass at 1 nm intervals with a 5 second averaging time at each point. An R-buffer blank was recorded and subtracted from each spectrum. Conversion to molar ellipiticity was carried out using DinI concentrations measured as described above and a mean residue weight of 110 Da. The CD spectra for the DinI proteins were normalized by arbitrarily setting the molar ellipticity at 218 nm to −1 deg cm2 decimol−1.
ATP hydrolysis assays to monitor the effect of the DinI mutant proteins on RecA
A spectrophotometric, coupled-enzyme assay was used to measure RecA-catalyzed ATP hydrolysis41,42. The ADP produced by hydrolysis was converted back to ATP by the enzyme pyruvate kinase and the substrate phosphoenolpyruvate. The resultant pyruvate was reduced to lactate by lactate dehydrogenase using NADH as a reducing agent. The conversion of NADH to NAD+ was monitored as a decrease in absorbance at 380 nm on a Varian Cary 300 instrument equipped with a temperature controller and 12 position cell changer. The cell pathlength was 0.5 cm. The concentration of oxidized NADH, and thus the concentration of hydrolyzed ATP, was calculated from the NADH extinction coefficient ε380 = 1.21 mM−1 cm−1.
Each ATP hydrolysis assay contained a reaction mixture of 25 mM Tris.OAc (88% cation, pH 7.5), 10 mM magnesium acetate, 4 mM potassium glutamate, 5 % (w/v) glycerol, 1 mM DTT, 3.5 mM phosphoenolpyruvate, 10 U/ml pyruvate kinase, 10 U/ml lactate dehydrogenase, 1.5 mM NADH, 3 μM poly-dT, and 3 mM ATP. DinI proteins, or compensating storage buffer, were incubated with the above reaction mixture at 37 °C for 10 min. The DinI concentration was varied from 0 to 20 μM. Wild type RecA protein was then added to a final concentration of 1 μM. RecA-catalyzed ATP hydrolysis was monitored at 37 °C until the NADH was exhausted. The kcat for RecA-catalyzed ATP hydrolysis at various DinI concentrations was calculated from the rate of ATP hydrolysis divided by the concentration of RecA binding sites on the poly-dT.
Electron microscopy and image analysis
The RecA-dsDNA-AMPPNP, and RecA C17-dsDNA-AMPPNP complexes were incubated in 25 mM triethanolamine-HCl (Fisher) buffer (pH 7.2) at 37° C for 10 min, with [RecA] or [RecA C17]=2 μm, RecA to calf thymus double-stranded DNA (Sigma) ratio of 40:1 (w/w), AMP-PNP (Sigma) 1.5 mM, magnesium acetate (Sigma) 2.5 mM. The DinI complexes were formed in the same manner except that 20 uM DinI was present. Samples were applied to carbon-coated grids and stained with 2% (w/v) uranyl acetate (w/v). Images were recorded on film using a Tecnai 12 electron microscope operating at 80 keV with a nominal magnification of ×30,000. Negatives were scanned with a Nikon Coolscan 8000 densitometer, at a raster of 4.2 Å per pixel. SPIDER43 was used for image processing. Segments of filaments (80 × 80 pixels) of the RecA (n=5,300) or RecAΔ17 (n=5,301) formed on double-stranded DNA (dsDNA) in the presence of AMP-PNP were extracted from the micrographs and reconstructed using the IHRSR method44. Both sets converged to the same helical parameters and yielded a twist of 58.5° and axial rise of 15 Å. Segments (80 × 80 pixels) of RecA-dsDNA-AMP-PNP (n=5,891) or RecAΔ17-dsDNA-AMP-PNP (n=5,773) decorated with DinI protein were extracted from micrographs, and overall reconstructions were calculated (Fig. 3b and c, respectively). Both sets converged to the same helical symmetry of 59°/15 Å with the IHRSR method. The central protomer from the RecA-dsDNA crystal structure (PDB 3CMU) was used to generate a model RecA filament. The DinI solution structure (PDB 1GHH) was used to build up two models – with the DinI attached to the CTD (Fig. 3d, red arrowhead), or located deeply in the groove of the RecA filament (Fig. 3D, cyan arrowhead). The model of the RecA filament along with the two decorated models were used as references in cross-correlation sorting. The three reference volumes were scaled to 4.2 Å per pixel and projected into 80 × 80-pixel images with an azimuthal rotational increment of 4°, generating 270 reference projections (3 × 90). RecA-dsDNA-AMP-PNP-DinI and RecAΔ17-dsDNA-AMP-PNP-DinI segments were cross-correlated with the 270 reference projections. Reconstructions were independently generated for each class (Fig. 3E) starting from a featureless solid cylinder. The two naked classes (n=1,375 and 3,683, respectively) both converged to the symmetry of 59°/15 Å. The two CTD classes (n=3,094 and 766, respectively) yielded symmetries of 58.7°/14.7 Å and 58.4°/14.4 Å, while the groove classes (n=1,422 and 1,324, respectively) converged to 58.8°/14.6 Å and 58.5°/14.8 Å helical parameters. The resolution of the reconstructions was judged to be ~23 Å using the FSC = 0.5 criterion.
UCSF Chimera software45 was used to fit the high resolution structures of RecA and DinI into the experimental density maps. Atomic coordinates from crystal structures were converted to density maps, and these were filtered to the resolution of the experimental map and docked manually.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Kowalczykowski SC, Eggleston AK. Homologous pairing and DNA strand-exchange proteins. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- 2.Little JW, Mount DW. The SOS regulatory system of Escherichia coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- 3.Smith KC, Wang TC. recA-dependent DNA repair processes. BioEssays. 1989;10:12–16. doi: 10.1002/bies.950100104. [DOI] [PubMed] [Google Scholar]
- 4.Story RM, Steitz TA. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 5.Datta S, Prabu MM, Vaze MB, Ganesh N, Chandra NR, Muniyappa K, Vijayan M. Crystal structures of Mycobacterium tuberculosis RecA and its complex with ADP-AlF(4): implications for decreased ATPase activity and molecular aggregation. Nucleic Acids Res. 2000;28:4964–4973. doi: 10.1093/nar/28.24.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xing X, Bell CE. Crystal structures of Escherichia coli RecA in complex with MgADP and MnAMP-PNP. Biochemistry. 2004;43:16142–16152. doi: 10.1021/bi048165y. [DOI] [PubMed] [Google Scholar]
- 7.Xing X, Bell CE. Crystal structures of Escherichia coli RecA in a compressed helical filament. J Mol Biol. 2004;342:1471–1485. doi: 10.1016/j.jmb.2004.07.091. [DOI] [PubMed] [Google Scholar]
- 8.Datta S, Krishna R, Ganesh N, Chandra NR, Muniyappa K, Vijayan M. Crystal structures of Mycobacterium smegmatis RecA and its nucleotide complexes. J Bacteriol. 2003;185:4280–4284. doi: 10.1128/JB.185.14.4280-4284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishna R, Manjunath GP, Kumar P, Surolia A, Chandra NR, Muniyappa K, Vijayan M. Crystallographic identification of an ordered C-terminal domain and a second nucleotide-binding site in RecA: new insights into allostery. Nucleic Acids Res. 2006;34:2186–2195. doi: 10.1093/nar/gkl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roca AI, Cox MM. RecA protein: structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez de Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 12.Little JW. Autodigestion of lexA and phage repressors. Proc Nat Acad Sci, USA. 1984;81:1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.VanLoock MS, Yu X, Yang S, Galkin VE, Huang H, Rajan SS, Anderson WF, Stohl EA, Seifert HS, Egelman EH. Complexes of RecA with LexA and RecX Differentiate Between Active and Inactive RecA Nucleoprotein Filaments. J Mol Biol. 2003;333:345–354. doi: 10.1016/j.jmb.2003.08.053. [DOI] [PubMed] [Google Scholar]
- 14.Kowalczykowski SC, Clow J, Krupp RA. Properties of the duplex DNA-dependent ATPase activity of Escherichia coli RecA protein and its role in branch migration. Proc Nat Acad Sci, USA. 1987;84:3127–3131. doi: 10.1073/pnas.84.10.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lusetti SL, Shaw JJ, Cox MM. Magnesium ion-dependent activation of the RecA protein involves the C terminus. J Biol Chem. 2003;278:16381–16388. doi: 10.1074/jbc.M212916200. [DOI] [PubMed] [Google Scholar]
- 16.Tateishi S, Horii T, Ogawa T, Ogawa H. C-terminal truncated Escherichia coli RecA protein RecA5327 has enhanced binding affinities to single- and double-stranded DNAs. J Mol Biol. 1992;223:115–129. doi: 10.1016/0022-2836(92)90720-5. [DOI] [PubMed] [Google Scholar]
- 17.Eggler AL, Lusetti SL, Cox MM. The C Terminus of the Escherichia coli RecA Protein Modulates the DNA Binding Competition with Single-stranded DNA-binding Protein. J Biol Chem. 2003;278:16389–16396. doi: 10.1074/jbc.M212920200. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–4. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 19.Sano Y. Role of the recA-related gene adjacent to the recA gene in Pseudomonas aeruginosa. J Bacteriol. 1993;175:2451–2454. doi: 10.1128/jb.175.8.2451-2454.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vierling S, Weber T, Wohlleben W, Muth G. Transcriptional and mutational analyses of the Streptomyces lividans recX gene and its interference with RecA activity. J Bacteriol. 2000;182:4005–4011. doi: 10.1128/jb.182.14.4005-4011.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stohl EA, Brockman JP, Burkle KL, Morimatsu K, Kowalczykowski SC, Seifert HS. Escherichia coli RecX inhibits RecA recombinase and coprotease activities in vitro and in vivo. J Biol Chem. 2003;278:2278–2285. doi: 10.1074/jbc.M210496200. [DOI] [PubMed] [Google Scholar]
- 22.Drees JC, Lusetti SL, Chitteni-Pattu S, Inman RB, Cox MM. A RecA filament capping mechanism for RecX protein. Mol Cell. 2004;15:789–798. doi: 10.1016/j.molcel.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 23.Drees JC, Lusetti SL, Cox MM. Inhibition of RecA protein by the Escherichia coli RecX protein: modulation by the RecA C terminus and filament functional state. J Biol Chem. 2004;279:52991–52997. doi: 10.1074/jbc.M409050200. [DOI] [PubMed] [Google Scholar]
- 24.Lewis LK, Harlow GR, Gregg-Jolly LA, Mount DW. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J Mol Biol. 1994;241:507–523. doi: 10.1006/jmbi.1994.1528. [DOI] [PubMed] [Google Scholar]
- 25.Yasuda T, Nagata T, Ohmori H. Multicopy suppressors of the cold-sensitive phenotype of the pcsA68 (dinD68) mutation in Escherichia coli. J Bacteriol. 1996;178:3854–3859. doi: 10.1128/jb.178.13.3854-3859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yasuda T, Morimatsu K, Kato R, Usukura J, Takahashi M, Ohmori H. Physical interactions between DinI and RecA nucleoprotein filament for the regulation of SOS mutagenesis. EMBO J. 2001;20:1192–1202. doi: 10.1093/emboj/20.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voloshin ON, Ramirez BE, Bax A, Camerini-Otero RD. A model for the abrogation of the SOS response by an SOS protein: a negatively charged helix in DinI mimics DNA in its interaction with RecA. Genes Dev. 2001;15:415–427. doi: 10.1101/gad.862901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lusetti SL, Voloshin ON, Inman RB, Camerini-Otero RD, Cox MM. The DinI protein stabilizes RecA protein filaments. J Biol Chem. 2004 doi: 10.1074/jbc.M403064200. [DOI] [PubMed] [Google Scholar]
- 29.Craig NL, Roberts JW. Function of nucleotide triphosphate and polynucleotide in E. coli RecA protein- directed cleavage of phage lambda repressor. J Biol Chem. 1981;256:8039–8044. [PubMed] [Google Scholar]
- 30.Campbell MJ, Davis RW. On the in vivo function of the RecA ATPase. J Mol Biol. 1999;286:437–445. doi: 10.1006/jmbi.1998.2457. [DOI] [PubMed] [Google Scholar]
- 31.VanLoock MS, Yu X, Yang S, Lai AL, Low C, Campbell MJ, Egelman EH. ATP-mediated conformational changes in the RecA filament. Structure (Camb ) 2003;11:187–196. doi: 10.1016/s0969-2126(03)00003-0. [DOI] [PubMed] [Google Scholar]
- 32.Yu X, Egelman EH. Removal of the RecA C-terminus results in a conformational change in the RecA-DNA filament. J Struct Biol. 1991;106:243–254. doi: 10.1016/1047-8477(91)90074-7. [DOI] [PubMed] [Google Scholar]
- 33.Weinstock GM, McEntee K, Lehman IR. Hydrolysis of nucleoside triphosphates catalyzed by the recA protein of Escherichia coli. Steady state kinetic analysis of ATP hydrolysis. J Biol Chem. 1981;256:8845–8849. [PubMed] [Google Scholar]
- 34.De Zutter JK, Knight KL. The hRad51 and RecA proteins show significant differences in cooperative binding to single-stranded DNA. J Mol Biol. 1999;293:769–780. doi: 10.1006/jmbi.1999.3200. [DOI] [PubMed] [Google Scholar]
- 35.Egelman EH. Problems in fitting high resolution structures into electron microscopic reconstructions. HFSP J. 2008;2:324–331. doi: 10.2976/1.2992221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramirez BE, Voloshin ON, Camerini-Otero RD, Bax A. Solution structure of DinI provides insight into its mode of RecA inactivation. Protein Sci. 2000;9:2161–2169. doi: 10.1110/ps.9.11.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Adzuma K. Differential proximity probing of two DNA binding sites in the Escherichia coli recA protein using photo-cross-linking methods. Biochemistry. 1996;35:3563–3571. doi: 10.1021/bi952438v. [DOI] [PubMed] [Google Scholar]
- 38.Malkov VA, Camerini-Otero RD. Photocross-links between single-stranded DNA and Escherichia coli RecA protein map to loops L1 (amino acid residues 157-164) and L2 (amino acid residues 195-209) J Biol Chem. 1995;270:30230–30233. doi: 10.1074/jbc.270.50.30230. [DOI] [PubMed] [Google Scholar]
- 39.Wang L, Voloshin ON, Stasiak A, Camerini-Otero RD. Homologous DNA pairing domain peptides of RecA protein: intrinsic propensity to form beta-structures and filaments. J Mol Biol. 1998;277:1–11. doi: 10.1006/jmbi.1997.1591. [DOI] [PubMed] [Google Scholar]
- 40.Ragone S, Maman JD, Furnham N, Pellegrini L. Structural basis for inhibition of homologous recombination by the RecX protein. EMBO J. 2008;27:2259–2269. doi: 10.1038/emboj.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindsley JE, Cox MM. Assembly and disassembly of RecA protein filaments occur at opposite filament ends. Relationship to DNA strand exchange. J Biol Chem. 1990;265:9043–9054. [PubMed] [Google Scholar]
- 42.Morrical SW, Lee J, Cox MM. Continuous association of Escherichia coli single-stranded DNA binding protein with stable complexes of recA protein and single- stranded DNA. Biochemistry. 1986;25:1482–1494. doi: 10.1021/bi00355a003. [DOI] [PubMed] [Google Scholar]
- 43.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 44.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 45.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.