Abstract
The genetic basis for biosynthesis of the (α1→4)-linked N-acetyl-d-glucosamine 1-phosphate capsule of Neisseria meningitidis serogroup X was defined. The biosynthesis gene cassette was a ∼4.2-kb region located between ctrA of the capsule transport operon and galE, which encodes the UDP-glucose-4-epimerase. This location was identical to the locations of the biosynthesis cassettes in other meningococcal serogroups. Three open reading frames unique to meningococcus serogroup X were identified. Deletion-insertion mutation and colony immunoblotting confirmed that these three genes were essential for serogroup X capsule expression, and the genes were designated xcbA, xcbB, and xcbC (serogroup X capsule biosynthesis). Reverse transcriptase PCR indicated that the xcbABC genes form an operon and are cotranscribed divergently from ctrA. XcbA exhibited 52% amino acid similarity to SacB, the putative capsule polymerase of meningococcus serogroup A, suggesting that it plays a role as the serogroup X capsule polymerase. An IS1016 element was found within the intergenic region separating ctrA and xcbA in multiple strains, and this element did not interfere with capsule expression.
Neisseria meningitidis is the cause of epidemic bacterial meningitis. Capsular polysaccharide is a major virulence determinant of N. meningitidis (23, 35). Among the 13 meningococcal serogroups classified based on capsular polysaccharide structure, serogroups A, B, C, Y, and W135 are associated with the majority of cases of meningococcal disease. In the African meningitis belt most large epidemics have been caused by serogroup A meningococci, whereas sporadic disease and outbreaks in developed countries are usually caused by serogroup B and C meningococci (1). Serogroup Y meningococci emerged as an important cause of sporadic disease and outbreaks in the United States in the late 1990s (30, 33), and in 2000 serogroup W135 meningococci caused worldwide disease in association with the Hajj pilgrimage (6, 7, 42) and large outbreaks in sub-Saharan Africa (43).
Sporadic cases of meningococcal disease caused by serogroup X meningococci have been reported in both industrialized countries (15, 18, 28, 34) and African countries (11, 31). However, recently, large serogroup X meningitis outbreaks in Niger (5, 10) and Ghana (13) have been reported. A genetic diversity study of N. meningitidis serogroup X isolates in which multilocus sequence typing and pulsed-field gel electrophoresis were used showed that most carrier and disease isolates recovered in the last 30 years in the African meningitis belt belonged to the same clonal group (12), while most European and American isolates were highly diverse. In a longitudinal carriage study designed to investigate the dynamics of meningococcal carriage during an interepidemic period in Ghana, the disappearance of the epidemic serogroup A strain was accompanied by a sharp increase in nasopharyngeal carriage of serogroup X meningococci (13). The carriage rate reached 18% of the population sampled, and this coincided with an outbreak of serogroup X disease. Serogroup X meningococci have also been reported to be very efficient in colonizing military recruits in the United Kingdom (21).
The capsular polysaccharides of serogroup B, C, Y, and W135 meningococci are composed of sialic acid derivatives. Serogroup B and C meningococci express (α2→8)- and (α2→9)-linked polysialic acid, respectively (3, 26), while alternating sequences of d-glucose or d-galactose and sialic acid are expressed by serogroup Y and W135 N. meningitidis. In contrast, the capsule of serogroup A meningococci is composed of (α1→6)-linked N-acetylmannosamine 6-phosphate (27), while N. meningitidis serogroup X synthesizes capsular polymers of (α1→4)-linked N-acetylglucosamine 1-phosphate (4). In order to better understand the evolution of the meningococcal capsule and its role in pathogenesis, the genetic basis of meningococcal capsule expression in serogroups A, B, C, Y, and W135 has been defined previously (37-39). Here we describe the first characterization of a capsule biosynthesis locus in N. meningitidis serogroup X.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The meningococcal strains, plasmids, and primers used in this study are listed in Table 1. Most meningococci were grown on gonococcal (GC) base agar (Difco Laboratories, Detroit, Mich.) or in GC broth at 37°C in the presence of 3.5% CO2; the only exception was the meningococcal mutant grown with kanamycin, which was propagated on brain heart infusion base (Becton Dickinson and Co., Cockeysville, Md.) containing 1.25% fetal bovine serum (GIBCO BRL, Gaithersburg, Md.) at 37°C in the presence of 3.5% CO2. Escherichia coli strains were grown in Luria-Bertani media. Antibiotics (Sigma Chemical Co., St. Louis, Mo.) were used for selection at the following concentrations: 50 μg of kanamycin per ml, 100 μg of spectinomycin per ml, and 100 μg of ampicillin per ml for E. coli; and 80 μg of kanamycin per ml and 60 μg of spectinomycin per ml for N. meningitidis. E. coli TOP10F′ (Invitrogen, San Diego, Calif.) and DH5α were used as the host strains for cloned PCR products and recombinant plasmids created during this study.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description or sequence | Reference or sourcea |
|---|---|---|
| N. meningitidis strains | ||
| NMB | B:2b:P1.2,5:L2 (CDC8201085) | 36 |
| M7 | NMB synA::Tn916, unencapsulated | 41 |
| F8229 | Serogroup A strain | 38 |
| M328 | Serogroup X reference strain | CDC |
| M2526 | Serogroup X blood isolate from Florida, 1996 | CDCb |
| M4222 | Serogroup X sputum isolate from Florida, 1997 | CDCb |
| M4370 | Serogroup X blood isolate from Connecticut, 1997 | CDCb |
| M7575 | Serogroup X blood isolate from Maryland, 2000 | CDC |
| M8210 | Serogroup X blood isolate from North Carolina, 2001 | CDC |
| M328::302 | M328 with insertion-deletion mutation in xcbABC | This study |
| M2526::302 | M2526 with insertion-deletion mutation in xcbABC | This study |
| Plasmids | ||
| pCR2.1 | TA cloning vector, Kanr Ampr | Stratagene |
| pUC18 | Cloning vector, Ampr | 49 |
| pHP45 | Ω(Spr) Ampr | 29 |
| pTA7575 | TA cloning of LJ8-ga1E1 PCR product into pCR2.1 | This study |
| pUC7575 | EcoRI fragment of pTA7575 subcloned into EcoRI site of pUC18 | This study |
| pYT302 | 2.447 bp of NcoI-EcoRV fragment in pUC7575 replaced with Ω(Sp) | This study |
| Primers | ||
| CN1 | 5′-GGCGTTATAATGCTGGTAATTGGATTC-3′ | This study |
| CN2 | 5′-CAAGCACATCTGAGACTCTACAAGG-3′ | This study |
| CN4 | 5′-CGGATCATCATCGGAACATTC-3′ | This study |
| CN5 | 5′-GCGAATACAGCCCACATTCTATCTG-3′ | This study |
| CN6 | 5′-TTGAATTTCTGTGCACTAGATGCG-3′ | This study |
| CN7 | 5′-GCGCATCTAGTGCACAGAAATTC-3′ | This study |
| CN8 | 5′-CCACCCAAGAAGCCGACAAAG-3′ | This study |
| CN9 | 5′-GTACCATCCGGAGCGACTGAAG-3′ | This study |
| CN10 | 5′-AGTTTTGCTAATCCGCTGCTTG-3′ | This study |
| CN12 | 5′-CAGATAGAATGTGGGCTGTATTCGC-3′ | This study |
| GalE1 | 5′-CGTGGCAGGATATTGATGCTGG-3′ | This study |
| LJ8 | 5′-CCACCACCAAACAATACTGCC-3′ | This study |
| RN7 | 5′-CCAGCCGAAGCATAACCATCGC-3′ | This study |
CDC, Centers for Disease Control and Prevention.
See reference 12.
Transformation.
Meningococcal strains were transformed by using the procedure described by Janik et al. (19). Plasmids digested with ScaI were used directly for meningococcal transformation. Transformants were screened by colony PCR by using a cassette-specific primer and a chromosome-specific anchoring primer. E. coli strains were transformed by electroporation by using a Gene-Pulser (Bio-Rad, Hercules, Calif.).
Nucleic acid purification.
Chromosomal DNA was isolated from N. meningitidis by the following procedure. Bacteria were scraped from one confluent overnight growth plate and resuspended in 10 ml of DNA extraction buffer (10 mM NaCl, 20 mM Tris-HCl [pH 8.0], 1 mM EDTA). Proteinase K (Fisher Scientific, Pittsburgh, Pa.) was added to a final concentration of 100 μg/ml, and the suspension was incubated for at least 6 h at 50°C. An equal volume of phenol-chloroform (1:1) was then added, and the solution was mixed on a rocker for 10 min at room temperature; this was followed by 20 min of centrifugation at 10,000 × g. The upper aqueous layer was poured into a clean tube, and centrifugation was repeated. Chromosomal DNA was spooled out of the aqueous layer after addition of 0.1 volume of 3 M sodium acetate and 2 volumes of ethanol. The DNA was rinsed with 70% ethanol, air dried briefly, and then suspended in 2 ml of 10 mM Tris [pH 8.0]-1 mM EDTA. Total RNA was prepared from bacteria grown in GC broth to the mid-log phase by using an RNeasy mini kit (Qiagen, Valencia, Calif.) according to the manufacturer's protocol. Purified total RNA was further treated with RNase-free DNase to remove contaminating chromosomal DNA. PCRs performed with the total RNAs obtained before and after DNase treatment as templates confirmed that the RNA preparation was free of DNA contamination.
PCR, RT-PCR, and colony PCR.
PCRs were performed as previously described (39). A reverse transcriptase PCR (RT-PCR) assay was performed by using a GeneAmp RT-PCR kit (Applied Biosystems, Roche) and the protocol recommended by the manufacturer. A single colony from a plated culture was collected with a sterile toothpick and suspended in sterile water, and 2 μl of the suspension was used as a template for colony PCR performed by using the standard conditions (40).
Construction of pYT302.
The PCR product generated with primers LJ8 and galE1 by using chromosomal DNA from meningococcal serogroup X strain M7575 was cloned into the pCR2.1 vector with a TOPO-TA cloning kit (Invitrogen) to obtain pTA7575. This fragment was subsequently released by EcoRI digestion and subcloned into the EcoRI site of pUC18 to obtain pUC7575. After double digestion of pUC7575 with NcoI and EcoRV to remove approximately 2.4 kb, the plasmid was gel purified, blunted with the Klenow fragment, and ligated to the Ω cassette obtained from SmaI digestion of pHP45Ω (29). Transformants were selected with spectinomycin, and insert-containing clones were identified by colony PCR. PCR and direct sequencing analysis of the resulting plasmid, pYT302, confirmed correct deletion and insertion of the Ω cassette.
Southern blotting.
PCR products were used as templates to generate random primed digoxigenin-labeled probes with the Genius nonradioactive DNA labeling and detection system (Boehringer Mannheim, Indianapolis, Ind.). DNA hybridization was performed by following the manufacturer's suggested procedure (Boehringer Mannheim).
Whole-bacterium immunoblotting.
A detailed immunoblot procedure in which whole cells are used has been described previously (22). Briefly, N. meningitidis cells from plate-grown overnight cultures were suspended in GC broth, and the optical density at 550 nm was determined. Sequential dilutions were made to obtain the required numbers of organisms in 50-μl aliquots. The cell suspensions were applied to a prewetted nitrocellulose membrane by using a BioDot apparatus (Bio-Rad). The membrane was subsequently processed by using the previously described procedure (22). Before the membrane was probed, polyclonal antiserum to N. meningitidis serogroup X (Meningitis and Special Pathogens Branch, Centers for Disease Control and Prevention) was preabsorbed with a suspension of strain M328::302 meningococci to remove nonspecific antibodies that recognize other meningococcal surface antigens. The antiserum was used at a 1:250 dilution. Alkaline phosphatase-conjugated anti-rabbit immunoglobulin G/M monoclonal antibody (ICN/CAPPEL, West Chester, Pa.) was used at a 1:2,500 dilution.
Serum bactericidal assay.
A microdilution serum bactericidal assay was performed by using the procedure described by Kahler et al. (23). Pooled normal human serum was used at a 10% (vol/vol) dilution. The percent survival (log10) was calculated by dividing the number of CFU per milliliter obtained after incubation in serum (at 15 min) by the number of CFU per milliliter at time zero. A Student's t test with a two-tailed hypothesis was used to determine the significance (P ≤ 0.05) for two variables.
Nucleotide sequence accession numbers.
The nucleotide and predicted amino acid sequences of the capsule biosynthesis genes derived from strain M7575 have been deposited in the GenBank database under accession number AY289931. The GenBank accession number for the intergenic region sequence from strain M0328 is AY289932.
RESULTS
Characterization of the nucleotide sequence between ctrA and galE in N. meningitidis serogroup X.
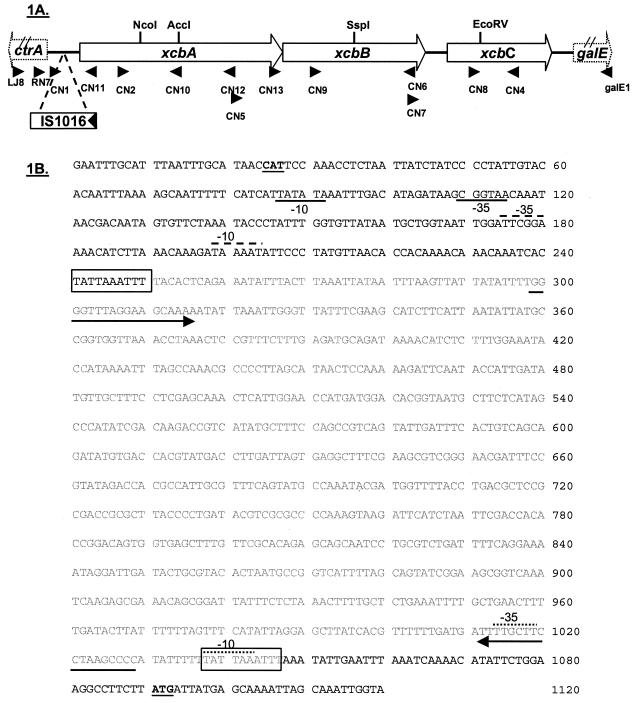
In all meningococcal serogroups characterized thus far, the capsule biosynthesis genetic cassette is flanked by ctrA, the first gene in the capsule transport operon, and galE, a gene involved in lipooligosaccharide biosynthesis (2, 44). To confirm that the biosynthesis genes of serogroup X are located in this region, primers that annealed to ctrA or galE were used to PCR amplify the region from chromosomal DNA prepared from six serogroup X isolates. A ∼4.2-kb DNA fragment was obtained from strain M7575, while the other five strains yielded ∼5-kb PCR products. Two independent PCR products from M7575 (RN7-galE1 and CN1-galE1) were cloned into the pCR2.1 vector, and the resulting plasmids were used as templates to obtain a nucleotide sequence by primer walking (20). The entire sequence was confirmed by 2× coverage sequencing of the two independent PCR clones. In addition, overlapping PCR amplification was performed to determine the location of the additional ∼800 bp present in the five serogroup X strains that produced larger PCR products than M7575 produced. A larger PCR product was obtained when primers CN1 and CN11 were used for amplification (Fig. 1A). Nucleotide sequencing of the larger CN1-CN11 PCR product revealed the presence of an intact IS1016 element (8). The IS1016 open reading frame (ORF) was predicted to encode a 217-amino-acid protein homologous to the IS1016C2 transposase and was oriented in the same direction as ctrA (Fig. 1A). There was no difference in the level of capsule expression between strains with and without IS1016 when they were examined by colony immunoblotting (data not shown).
FIG. 1.
(A) Schematic diagram of the capsule biosynthesis gene cassette in N. meningitidis serogroup X. Three ORFs, xcbA, xcbB, and xcbC, were identified and were transcribed divergently from the first gene of the capsule transport operon, ctrA. An IS1016 element (indicated by an open box with the arrow showing the transcriptional direction of the transposase) was found in some strains. The locations of nucleotide primers used in this study are indicated by arrowheads. (B) Nucleotide sequence of the intergenic region between ctrA and xcbA of strain M0328 (accession number AY289932). The translational start codons of ctrA and xcbA are indicated by boldface type and underlining. The sequence of the IS1016 element is indicated by lightface type. A 10-bp duplication associated with the IS1016 insertion is enclosed in a box. The arrows indicate an imperfect 18-bp inverted repeat flanking the IS1016 element. Probable promoters (−10 and −35) for xcbA in the strains containing IS1016 in this region are indicted by dotted lines above the sequence, while the putative xcbA promoter in the strain without IS1016 is indicated by the dashed line above the sequence. The putative ctrA promoter sequences are indicated by underlining.
Nucleotide sequence analysis of the putative capsule biosynthesis nucleotide sequence of strain M7575 indicated that there were three putative ORFs, which were designated xcbA, xcbB, and xcbC. The ORFs were transcribed divergently from ctrA, a common feature in all meningococcal serogroups (Fig. 1A). The IS1016 element was located in the region between the ctrA and xcbA genes. xcbA was 1,458 bp long and was predicted to encode a protein containing 486 residues. xcbA in strain M7575 was separated from ctrA by a 266-bp intergenic region, which exhibited no sequence similarity to the serogroup A and serogroup B intergenic regions (39). xcbB, which overlapped xcbA by 35 bp, was 1,050 bp long and was predicted to encode a 350-amino-acid protein. xcbC, which was separated from xcbB by 140 bp, was 768 nucleotides long and was predicted to encode a putative protein containing 256 amino acids. xcbABC had a G+C content of 35 to 39%.
Both nucleotide and predicted protein sequences were used to search the GenBank database. XcbA exhibited significant homology to the following three meningococcal proteins closely associated with meningococcal capsule loci: a hypothetical protein (P value, 2e-68; 40% identity and 58% similarity) encoded by a gene located between rfbD and lipA in the capsule locus of serogroup B strain B1940 (16), LcbA (P value, 2e-66; 38% identity and 53% similarity), and SacB (P value, 2e-46; 31% identity and 52% similarity) (38). LcbA, a 366-residue protein, is encoded by the first gene of a gene cluster similarly flanked by ctrA and galE in serogroup L (GenBank accession number AF112478). It has been proposed that LcbA is involved in capsule biosynthesis; however, its function and role in capsule expression have not been confirmed. SacB (545 residues) is the putative capsular polymerase encoded by the serogroup A capsule biosynthesis gene cluster (38), and a sacB mutant is nonencapsulated (38). An alignment of the XcbA, LcbA, and SacB sequences is shown in Fig. 2. The homology is spread throughout the protein sequences; no known domain or motif was identified. In addition, four conserved hypothetical proteins in Streptomyces coelicolor A3, a putative capsular polysaccharide synthesis protein in Aeromonas hydrophila, and a capsular polysaccharide synthesis protein (Cps1A) in Actinobacillus pleuropneumoniae also exhibited significant protein sequence similarity to XcbA (P value range, 2e-59 to 2e-41). However, the functions of these proteins have not been demonstrated.
FIG. 2.
Alignment of the amino acid sequences of XcbA, LcbA, and SacB generated by the Clustal W method (45).
ctrABCD and lipAB are conserved in N. meningitidis serogroup X.
The capsule transport genes, ctrABCD and lipAB, are highly conserved in the major disease-causing serogroups, serogroups A, B, C, Y, and W135 (2, 44). Nucleotide probes for ctrABCD and lipAB were amplified by PCR from the chromosomal DNA of a serogroup B strain, NMB, by using primers designed from the MC58 serogroup B sequence (44). Southern blotting performed under high-stringency conditions with the six serogroup X isolates confirmed that each isolate contained ctrABCD and lipAB. PCR amplification of the ctrABCD and lipAB coding sequences, as well as the linkage between these genes, gave product sizes identical to the sizes of the products amplified from serogroup B strain NMB (data not shown). Furthermore, PCR amplification with gltS-specific primers, located at the 3′ end of lipB (Fig. 3), and lipB-specific primers gave products for serogroup X identical to those of serogroup B strains. Thus, ctrABCD and lipAB are conserved in N. meningitidis serogroup X and have a genetic organization similar to that in other well-characterized serogroups (Fig. 3).
FIG. 3.
Schematic diagram of the capsule genetic loci of N. meningitidis serogroup X and serogroup B, showing the similarity of the two serogroups.
xcbA, xcbB, and xcbC are transcribed as an operon.
RT-PCR was performed to determine whether xcbA, xcbB, and xcbC are linked as an operon. The CN4 primer located in xcbC (Fig. 1A) was utilized as the reverse transcription primer to generate cDNA from total RNA isolated from strain M7575. The RT reaction mixture was subsequently used as the template for PCR amplification of an internal fragment of each gene. PCR products were obtained not only for xcbC but also for xcbB and xcbA, indicating that these three genes are transcribed as a single transcript (Fig. 4).
FIG. 4.
RT-PCR demonstrating that xcbA, xcbB, and xcbC constitute an operon. An xcbC internal primer, CN4, was used to generate cDNA from total RNA isolated from strain M7575. Equivalent RT reactions without RT added (−RT) were also performed to check for possible chromosomal DNA contamination. The reverse transcription reaction mixtures were subsequently used as templates in PCR amplifications with primers internal to xcbA (primers CN2 and CN10), xcbB (primers CN9 and CN6), and xcbC (primers CN4 and CN8). PCR products generated with chromosomal DNA were used to assess the expected product size.
xcbABC gene products are required for capsule expression.
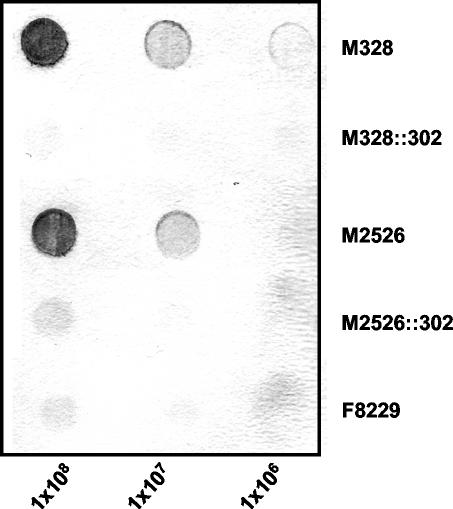
To determine whether the xcbABC gene cassette is required for expression of the serogroup X capsule, a deletion-insertion mutation was introduced into the xcbABC gene cassette. A fragment between the NcoI and EcoRV (Fig. 1A) restriction sites was removed from plasmid pUC7575 containing the 4.2-kb ctrA-galE region of strain M7575 and was replaced with an Ω(Sp) cassette. This mutation resulted in 3′ truncation of xcbA, 5′ deletion of xcbC, and complete deletion of xcbB. The resulting plasmid construct, pYT302, was used to transform the six serogroup X meningococcal isolates. Transformants selected with spectinomycin were successfully generated in strains M328 and M2526, and deletion and insertion of the Ω cassette were confirmed by colony PCR performed with primers CN2 and CN4 and by Southern blot analyses (data not shown). Because pYT302 contained the 4.2-kb xcbABC DNA region without IS1016 from strain M7575, two classes of transformants were obtained in strains carrying IS1016; in one of these classes IS1016 was lost during recombination. Transformants M328::302 and M2526::302, carrying IS1016 in the ctrA-xcbA intergenic region, were tested for capsule expression by whole-bacterium immunoblotting by using serogroup X capsule-specific polyclonal antiserum (Fig. 5). Neither M328::302 nor M2526::302 expressed capsular polysaccharides, further indicating that the xcbABC gene cluster is essential for capsule expression.
FIG. 5.
Whole-bacterium immunoblot for N. meningitidis serogroup X. Aliquots containing 1 × 108, 1 × 107, and 1 × 106 cells of serogroup X meningococcal strains M328 and M2526 and the capsule biosynthesis insertion-deletion mutants of these strains, M328::302 and M2526::302, were dried on a nitrocellulose membrane and probed with serogroup X capsule-specific polyclonal antiserum. Serogroup A strain F8229 was included as a negative control.
Mutations in xcbABC confer sensitivity to normal human serum.
The importance of capsule in conferring resistance to killing by human sera in meningococci has been well documented (23). The capsule-deficient phenotype of xcbABC mutants shown by colony immunoblotting suggested that the mutants should have a serum-sensitive phenotype. Serum bactericidal assays were performed to confirm this prediction. As shown in Fig. 6, the xcbABC mutants were rapidly killed in the presence of 10% normal human serum, whereas the encapsulated parent strain survived. These data further demonstrated that no capsule is present in the xcbABC mutants and that capsule is important for the resistance of N. meningitidis serogroup X to killing by normal human serum. Meningococcal lipooligosaccharide profiles have also been shown to influence serum sensitivity (9, 16, 23, 47). However, no differences in lipooligosaccharide were noted between the parent and capsule-deficient strains when proteinase K digests of whole-cell lysates were analyzed by Tricine polyacrylamide gel electrophoresis (22) (data not shown).
FIG. 6.
Human serum bactericidal assays. N. meningitidis serogroup X strains M328 and M2526 and the capsule biosynthesis insertion-deletion mutants of these strains were exposed to 10% pooled normal human serum (solid bars) or heat-inactivated (56°C, 30 min) human serum (gray bars). The percentage of survival (y axis) is indicated by using a log scale (n ≥ 2 for each variable). The bars indicate means, and the error bars indicate standard deviations.
DISCUSSION
The capsular polysaccharides of N. meningitidis and other bacterial capsules (Haemophilus influenzae, E. coli K1), designated group II capsules, have similar genetic organization and chemical properties (32, 48). Group II capsule loci are usually composed of a unique biosynthesis gene cassette flanked by conserved genes involved in translocation of the capsular polysaccharides (32, 48). Recently, N. meningitidis serogroup X has emerged as a serogroup that has caused large outbreaks of disease in sub-Saharan Africa (10, 13). The capsule of serogroup X, (α1→4)-linked N-acetylglucosamine 1-phosphate, is biochemically similar to the serogroup A capsule, (α1→6)-linked N-acetylmannosamine 1-phosphate. The overall organization of the capsule transport and biosynthesis genes of serogroup X meningococci, as defined in this study, showed similarity to the organization in other meningococcal serogroups characterized to date (2, 44). The capsule locus is located near gltS, as it is in other meningococci. In addition, xcbABC, sacABCD of N. meningitidis serogroup A, and synABCD of N. meningitidis serogroup B have much lower G+C contents (35 to 39, 24 to 35, and 28 to 41%, respectively) than the whole meningococcal genome (52%) (44). The difference in G+C contents suggests that horizontal gene transfer occurred.
The intergenic regions separating the divergently transcribed transport and biosynthesis genes in serogroups A, B, C, Y, and W135 have been characterized in detail and have similar organizations of transcriptional control (37, 38, 46). The sialic acid-containing serogroups (serogroups B, C, Y, and W135) utilize identical 134-bp intergenic regions to initiate transcription (37, 46), while a completely different 218-bp sequence separates the transport and biosynthesis gene clusters in N. meningitidis serogroup A (38). The intergenic regions of these serogroups contain overlapping promoters for controlling the divergent operons. The nucleotide sequence of the 266-bp intergenic region separating the divergently transcribed ctrA and xcbA genes differs from that found in serogroup A or serogroup B. Interestingly, expression of the serogroup X capsule was not affected by the presence of an IS1016 element in the intergenic region (Fig. 1). IS1016 may have inserted into the intergenic region after acquisition of the capsule locus in strain M328. Alternatively, IS1016 may mediate acquisition of the capsule genes and may have been lost in some strains. IS1016 was originally described as flanking the capsule locus of H. influenzae, and it was proposed that this element mobilizes the ∼17-kb capsule gene cluster as a compound transposon in the H. influenzae chromosome (24, 25). Many virulent H. influenzae serotype b strains carry a duplicated capsule locus, and flanking IS1016 elements may facilitate reversible gene amplification through unequal homologous recombination events (25). It has been noted that the presence of outwardly directed promoters in an insertion element or the formation of hybrid promoters between an insertion element and host DNA (14) may provide and/or enhance expression of certain genes, thus conferring a certain survival advantage. In the case of N. meningitidis serogroup X, predicted promoters resembling a σ70 consensus sequence can be identified in the intergenic region both within and outside the IS1016 that could initiate transcription of xcbA (Fig. 1B), without interference with the putative ctrA promoter. There was no difference in the level of serogroup X capsule expression between strains with IS 1016 and strains without IS1016, suggesting that IS1016 does not affect capsule expression. Another insertion element, IS1301, has been shown to mediate on-off switching of capsule expression through reversible insertion and excision within the coding sequence of the first biosynthesis gene, synA, in a serogroup B strain (17).
The serogroup X capsule is a polymer of (α1→4)-linked N-acetylglucosamine 1-phosphate. N-Acetylglucosamine is a common precursor of important bacterial components, such as peptidoglycan [(α1→4)-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid]. The coupling of a C-4 hydroxyl group in N-acetylmuramic acid and the C-1 carbon in UDP-GlcNAc, along with the release of UDP, generates the disaccharide precursor of the peptidoglycan. Analogously, coupling of two UDP-GlcNAc molecules between the C-4 hydroxyl of one UDP-GlcNAc and the C-1 phosphate of the other UDP-GlcNAc through the energy provided by the hydrolysis of UMP is predicted to produce the (1→4) phosphodiester linkage of the serogroup X capsule. A similar sequence of reactions has also been proposed for capsule expression in serogroup A meningococci, whose capsular structure is (α1→6)-linked N-acetylmannosamine (ManNAc) 1-phosphate (38). SacB is believed to be responsible for the polymerization of UDP-ManNAc (38), creating the phosphodiester bond between positions 1 and 6 of individual UDP-ManNAc molecules through the release of UMP. XcbA is probably the capsular polymerase for serogroup X meningococci, considering its homology to SacB. XcbA also showed sequence similarity to the putative biosynthesis protein, LcbA, of N. meningitidis serogroup L, which expresses a capsule composed of (α1-P→3)-linked trisaccharide of GlcNAc.
In summary, the genetic basis of capsule expression in serogroup X meningococci was defined. Like the capsules of other meningococcal serogroups (23), the serogroup X capsule is critical for resistance to normal human serum. With the identification of the unique serogroup X biosynthesis sequence, molecular tools for diagnosis and monitoring the epidemiology and emergence of serogroup X disease can be developed. In addition, this study provided additional information on the evolution of the capsule biosynthesis region of group II encapsulated bacterial pathogens.
Acknowledgments
We thank Lane Pucko for administrative assistance and Larry Martin for technical assistance. We thank Gloria Ajello, Leonard Mayer, and Nancy Rosenstein for their help.
This work was supported by National Institute of Allergy and Infectious Diseases grant AI33517 to D.S.S. and by the Meningitis and Special Pathogens Branch of the Centers for Disease Control and Prevention.
Editor: J. N. Weiser
REFERENCES
- 1.Achtman, M. 1995. Global epidemiology of meningococcal disease, p. 159-175. In K. Cartwright (ed.), Meningococcal disease. John Wiley and Sons, Chichester, United Kingdom.
- 2.Achtman, M., K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M.-A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, B. G. Barrell, and J. Parkhill. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharjee, A. K., H. J. Jennings, C. P. Kenny, A. Martin, and I. C. Smith. 1976. Structural determination of the polysaccharide antigens of Neisseria meningitidis serogroups Y, W-135, and BO1. Can. J. Biochem. 54:1-8. [DOI] [PubMed] [Google Scholar]
- 4.Bundle, D. R., H. J. Jennings, and C. P. Kenny. 1974. Studies on the group-specific polysaccharide of Neisseria meningitidis serogroup X and an improved procedure for its isolation. J. Biol. Chem. 249:4797-4801. [PubMed] [Google Scholar]
- 5.Campagne, G., A. Schuchat, S. Djibo, A. Ousseini, L. Cisse, and J. P. Chippaux. 1999. Epidemiology of bacterial meningitis in Niamey, Niger, 1981-96. Bull. W. H. O. 77:499-508. [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2001. Risk for meningococcal disease associated with the Hajj 2001. Morb. Mortal. Wkly. Rep. 50:97-98. [PubMed] [Google Scholar]
- 7. Centers for Disease Control and Prevention. 2000. Serogroup W-135 meningococcal disease among travelers returning from Saudi Arabia—United States, 2000. Morb. Mortal. Wkly. Rep. 49:345-346. [PubMed] [Google Scholar]
- 8.Dobson, S. R., J. S. Kroll, and E. R. Moxon. 1992. Insertion sequence IS1016 and absence of Haemophilus capsulation genes in the Brazilian purpuric fever clone of Haemophilus influenzae biogroup aegyptius. Infect. Immun. 60:618-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estabrook, M. M., J. M. Griffiss, and G. A. Jarvis. 1997. Sialylation of Neisseria meningitidis lipooligosaccharide inhibits serum bactericidal activity by masking lacto-N-neotetraose. Infect. Immun. 65:4436-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Etienne, J., G. Sperber, A. Adamou, and J. J. Picq. 1990. Epidemiological notes: meningococcal meningitis of serogroup X in Niamey (Niger). Med. Trop. 50:227-229. [PubMed] [Google Scholar]
- 11.Gagneux, S., A. Hodgson, I. Ehrhard, G. Morelli, B. Genton, T. Smith, M. Tanner, F. Binka, M. Achtman, and G. Pluschke. 2000. Microheterogeneity of serogroup A (subgroup III) Neisseria meningitidis during an outbreak in northern Ghana. Trop. Med. Int. Health 5:280-287. [PubMed] [Google Scholar]
- 12.Gagneux, S., T. Wirth, A. Hodgson, I. Ehrhard, G. Morelli, P. Kriz, B. Genton, T. Smith, F. Binka, G. Pluschke, and M. Achtman. 2002. Clonal groupings in serogroup X Neisseria meningitidis. Emerg. Infect. Dis. 8:462-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagneux, S. P., A. Hodgson, T. A. Smith, T. Wirth, I. Ehrhard, G. Morelli, B. Genton, F. N. Binka, M. Achtman, and G. Pluschke. 2002. Prospective study of a serogroup X Neisseria meningitidis outbreak in northern Ghana. J. Infect. Dis. 185:618-626. [DOI] [PubMed] [Google Scholar]
- 14.Galas, D. J., and M. Chandler. 1989. Bacterial insertion sequences, p. 109-162. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 15.Grahlow, W. D., H. W. Ocklitz, and H. Mochmann. 1986. Meningococcal infections in the German Democratic Republic 1971-1984. Infection 14:286-288. [DOI] [PubMed] [Google Scholar]
- 16.Hammerschmidt, S., C. Birkholz, U. Zahringer, B. D. Robertson, J. van Putten, O. Ebeling, and M. Frosch. 1994. Contribution of genes from the capsule gene complex (cps) to lipooligosaccharide biosynthesis and serum resistance in Neisseria meningitidis. Mol. Microbiol. 11:885-896. [DOI] [PubMed] [Google Scholar]
- 17.Hammerschmidt, S., R. Hilse, J. P. van Putten, R. Gerardy-Schahn, A. Unkmeir, and M. Frosch. 1996. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 15:192-198. [PMC free article] [PubMed] [Google Scholar]
- 18.Hansman, D. 1983. Meningococcal disease in South Australia: incidence and serogroup distribution 1971-1980. J. Hyg. 90:49-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janik, A., E. Juni, and G. A. Heym. 1976. Genetic transformation as a tool for detection of Neisseria gonorrhoeae. J. Clin. Microbiol. 4:71-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, D. H., and S. C. Winistorfer. 1993. Genome walking with 2- to 4-kb steps using panhandle PCR. PCR Methods Applic. 2:197-203. [DOI] [PubMed] [Google Scholar]
- 21.Jones, G. R., M. Christodoulides, J. L. Brooks, A. R. Miller, K. A. Cartwright, and J. E. Heckels. 1998. Dynamics of carriage of Neisseria meningitidis in a group of military recruits: subtype stability and specificity of the immune response following colonization. J. Infect. Dis. 178:451-459. [DOI] [PubMed] [Google Scholar]
- 22.Kahler, C. M., R. W. Carlson, M. M. Rahman, L. E. Martin, and D. S. Stephens. 1996. Inner core biosynthesis of lipooligosaccharide (LOS) in Neisseria meningitidis serogroup B: identification and role in LOS assembly of the alpha1,2 N-acetylglucosamine transferase (RfaK). J. Bacteriol. 178:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahler, C. M., L. E. Martin, G. C. Shih, M. M. Rahman, R. W. Carlson, and D. S. Stephens. 1998. The (α2→8)-linked polysialic acid capsule and lipooligosaccharide structure both contribute to the ability of serogroup B Neisseria meningitidis to resist the bactericidal activity of normal human serum. Infect. Immun. 66:5939-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kroll, J. S. 1992. The genetics of encapsulation in Haemophilus influenzae. J. Infect. Dis. 165(Suppl. 1):S93-S96. [DOI] [PubMed] [Google Scholar]
- 25.Kroll, J. S., B. M. Loynds, and E. R. Moxon. 1991. The Haemophilus influenzae capsulation gene cluster: a compound transposon. Mol. Microbiol. 5:1549-1560. [DOI] [PubMed] [Google Scholar]
- 26.Liu, T. Y., E. C. Gotschlich, F. T. Dunne, and E. K. Jonssen. 1971. Studies on the meningococcal polysaccharides. II. Composition and chemical properties of the group B and group C polysaccharide. J. Biol. Chem. 246:4703-4712. [PubMed] [Google Scholar]
- 27.Liu, T. Y., E. C. Gotschlich, E. K. Jonssen, and J. R. Wysocki. 1971. Studies on the meningococcal polysaccharides. I. Composition and chemical properties of the group A polysaccharide. J. Biol. Chem. 246:2849-2858. [PubMed] [Google Scholar]
- 28.Pastor, J. M., A. Fe, M. Gomis, and D. Gil. 1985. Meningococcal meningitis caused by Neisseria meningitidis of the X serogroup. Med. Clin. (Barcelona) 85:208-209. [PubMed] [Google Scholar]
- 29.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 30.Racoosin, J. A., C. G. Whitney, C. S. Conover, and P. S. Diaz. 1998. Serogroup Y meningococcal disease in Chicago, 1991-1997. JAMA 280:2094-2098. [DOI] [PubMed] [Google Scholar]
- 31.Riou, J. Y., S. Djibo, L. Sangare, J. P. Lombart, P. Fagot, J. P. Chippaux, and M. Guibourdenche. 1996. A predictable comeback: the second pandemic of infections caused by Neisseria meningitidis serogroup A subgroup III in Africa, 1995. Bull. W. H. O. 74:181-187. [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts, I. S. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50:285-315. [DOI] [PubMed] [Google Scholar]
- 33.Rosenstein, N. E., B. A. Perkins, D. S. Stephens, L. Lefkowitz, M. L. Cartter, R. Danila, P. Cieslak, K. A. Shutt, T. Popovic, A. Schuchat, L. H. Harrison, and A. L. Reingold. 1999. The changing epidemiology of meningococcal disease in the United States, 1992-1996. J. Infect. Dis. 180:1894-1901. [DOI] [PubMed] [Google Scholar]
- 34.Ryan, N. J., and G. R. Hogan. 1980. Severe meningococcal disease caused by serogroups X and Z. Am. J. Dis. Child. 134:1173. [DOI] [PubMed] [Google Scholar]
- 35.Stephens, D. S., P. A. Spellman, and J. S. Swartley. 1993. Effect of the (α 2→8)-linked polysialic acid capsule on adherence of Neisseria meningitidis to human mucosal cells. J. Infect. Dis. 167:475-479. [DOI] [PubMed] [Google Scholar]
- 36.Stephens, D. S., J. S. Swartley, S. Kathariou, and S. A. Morse. 1991. Insertion of Tn916 in Neisseria meningitidis resulting in loss of group B capsular polysaccharide. Infect. Immun. 59:4097-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swartley, J. S., J. H. Ahn, L. J. Liu, C. M. Kahler, and D. S. Stephens. 1996. Expression of sialic acid and polysialic acid in serogroup B Neisseria meningitidis: divergent transcription of biosynthesis and transport operons through a common promoter region. J. Bacteriol. 178:4052-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swartley, J. S., L. J. Liu, Y. K. Miller, L. E. Martin, S. Edupuganti, and D. S. Stephens. 1998. Characterization of the gene cassette required for biosynthesis of the (α1→6)-linked N-acetyl-d-mannosamine-1-phosphate capsule of serogroup A Neisseria meningitidis. J. Bacteriol. 180:1533-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swartley, J. S., A. A. Marfin, S. Edupuganti, L. J. Liu, P. Cieslak, B. Perkins, J. D. Wenger, and D. S. Stephens. 1997. Capsule switching of Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 94:271-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swartley, J. S., C. F. McAllister, R. A. Hajjeh, D. W. Heinrich, and D. S. Stephens. 1993. Deletions of Tn916-like transposons are implicated in tetM-mediated resistance in pathogenic Neisseria. Mol. Microbiol. 10:299-310. [DOI] [PubMed] [Google Scholar]
- 41.Swartley, J. S., and D. S. Stephens. 1994. Identification of a genetic locus involved in the biosynthesis of N-acetyl-d-mannosamine, a precursor of the (α 2→8)-linked polysialic acid capsule of serogroup B Neisseria meningitidis. J. Bacteriol. 176:1530-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taha, M. K., M. Achtman, J. M. Alonso, B. Greenwood, M. Ramsay, A. Fox, S. Gray, and E. Kaczmarski. 2000. Serogroup W135 meningococcal disease in Hajj pilgrims. Lancet 356:2159. [DOI] [PubMed] [Google Scholar]
- 43.Taha, M. K., I. Parent Du Chatelet, M. Schlumberger, I. Sanou, S. Djibo, F. de Chabalier, and J. M. Alonso. 2002. Neisseria meningitidis serogroups W135 and A were equally prevalent among meningitis cases occurring at the end of the 2001 epidemics in Burkina Faso and Niger. J. Clin. Microbiol. 40:1083-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tzeng, Y. L., J. S. Swartley, Y. K. Miller, R. E. Nisbet, L. J. Liu, J. H. Ahn, and D. S. Stephens. 2001. Transcriptional regulation of divergent capsule biosynthesis and transport operon promoters in serogroup B Neisseria meningitidis. Infect. Immun. 69:2502-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogel, U., H. Claus, G. Heinze, and M. Frosch. 1997. Functional characterization of an isogenic meningococcal α-2,3-sialyltransferase mutant: the role of lipooligosaccharide sialylation for serum resistance in serogroup B meningococci. Med. Microbiol. Immunol. 186:159-166. [DOI] [PubMed] [Google Scholar]
- 48.Whitfield, C., and I. S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307-1319. [DOI] [PubMed] [Google Scholar]
- 49.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]