Abstract
In the molecular structure of the title compound, C9H11NO3S, the N—H and C=O bonds are anti to each other, while the amide H atom is syn with respect to the ortho-methyl group in the benzene ring. The C—S—N—C torsion angle is −58.2 (2)°, indicating a twist in the molecule. In the crystal, N—H⋯O hydrogen bonds link the molecules into chains along the c axis.
Related literature
For the sulfanilamide moiety in sulfonamide drugs, see: Maren (1976 ▶). For hydrogen bonding modes of sulfonamides, see: Adsmond & Grant (2001 ▶). For our study of the effect of substituents on the structures of N-(aryl)-amides, see: Gowda et al. (2004 ▶). For background to the structures of N-(substituted phenylsulfonyl)-substituted-amides, see: Gowda et al. (2010 ▶); Shakuntala et al. (2011 ▶) and for the oxidative strengths of N-chloro, N-arylsulfonamides, see: Gowda & Kumar (2003 ▶).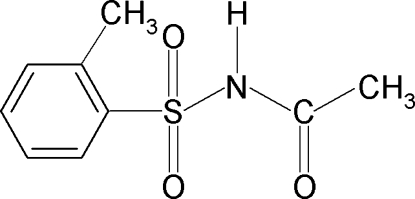
Experimental
Crystal data
C9H11NO3S
M r = 213.25
Tetragonal,

a = 7.9804 (5) Å
c = 16.749 (1) Å
V = 1066.69 (11) Å3
Z = 4
Mo Kα radiation
μ = 0.29 mm−1
T = 293 K
0.40 × 0.18 × 0.12 mm
Data collection
Oxford Diffraction Xcalibur diffractometer with a Sapphire CCD detector
Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▶) T min = 0.895, T max = 0.967
4245 measured reflections
1944 independent reflections
1690 reflections with I > 2σ(I)
R int = 0.016
Refinement
R[F 2 > 2σ(F 2)] = 0.035
wR(F 2) = 0.086
S = 1.08
1944 reflections
132 parameters
2 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.14 e Å−3
Δρmin = −0.17 e Å−3
Absolute structure: Flack (1983 ▶), 824 Friedel pairs
Flack parameter: 0.03 (9)
Data collection: CrysAlis CCD (Oxford Diffraction, 2009 ▶); cell refinement: CrysAlis RED (Oxford Diffraction, 2009 ▶); data reduction: CrysAlis RED; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2009 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811014218/tk2737sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811014218/tk2737Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯O3i | 0.86 (2) | 1.95 (2) | 2.770 (3) | 162 (3) |
Symmetry code: (i)  .
.
Acknowledgments
KS thanks the University Grants Commission, Government of India, New Delhi, for the award of a research fellowship under its faculty improvement program.
supplementary crystallographic information
Comment
The structures of sulfonamide drugs contain the sulfanilamide moiety (Maren, 1976). The hydrogen bonding preferences of sulfonamides has been investigated (Adsmond & Grant, 2001). The nature and position of the substituents play a significant role on their crystal structures and other aspects of N-(aryl)-amides and N-(aryl)-sulfonamides (Gowda et al., 2003, 2004; Shakuntala et al., 2011). As a part of a study of the effects of substituents on the structures of this class of compounds, the structure of N-(2-methylphenylsulfonyl)-acetamide (I) has been determined (Fig. 1). The conformation of the N—C bond in the C—SO2—NH—C(O) segment has gauche torsions with respect to the S═O bonds, the torsion angles being C7—N1—S1—O2 = 57.5 (3) ° and C7—N1—S1—O1 = -174.5 (2) °.
The N—H and C═O bonds are anti to each other, similar to that observed in N-(phenylsulfonyl)-acetamide (II) (Gowda et al., 2010) and N-(2-chlorophenylsulfonyl)-acetamide (III) (Shakuntala et al., 2011). Further, the conformation of the amide-H atom is syn to the ortho-methyl group in the benzene ring, similar to that observed between the amide-H atom and the ortho-chloro group in (III). The molecule of (I) is bent at the S-atom with a C—S—N—C torsion angle of -58.2 (2) °, compared to the values of -58.8 (4) ° in (II), and -71.7 (3) and 61.2 (3) ° in the two independent molecules of (III).
In the crystal structure, intermolecular N—H···O hydrogen bonds (Table 1) link the molecules into chains along the c axis; part of the crystal structure is shown in Fig. 2.
Experimental
The title compound was prepared by refluxing 2-methylbenzenesulfonamide (0.10 mole) with an excess of acetyl chloride (0.20 mole) for one hour on a water bath. The reaction mixture was cooled and poured into ice cold water. The resulting solid was separated, washed thoroughly with water and dissolved in warm dilute sodium hydrogen carbonate solution. The title compound was re-precipitated by acidifying the filtered solution with glacial acetic acid. It was filtered, dried and recrystallized from ethanol. Colourless rods of the title compound were obtained from a slow evaporation of its ethanol solution.
Refinement
The H atom of the NH group was located in a difference map and later restrained to the distance N—H = 0.86±0.02 Å. The other H atoms were positioned with idealized geometry using a riding model with the aromatic C—H distance = 0.93 Å and methyl C—H = 0.96 Å. All H atoms were refined with isotropic displacement parameters set to 1.2 times of the Ueq of the parent atom.
Figures
Fig. 1.
Molecular structure of (I), showing the atom-labelling scheme. Displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.
View of the crystal packing in (I). Hydrogen bonds are shown as dashed lines.
Crystal data
| C9H11NO3S | Dx = 1.328 Mg m−3 |
| Mr = 213.25 | Mo Kα radiation, λ = 0.71073 Å |
| Tetragonal, P43 | Cell parameters from 1894 reflections |
| Hall symbol: P 4cw | θ = 2.5–27.8° |
| a = 7.9804 (5) Å | µ = 0.29 mm−1 |
| c = 16.749 (1) Å | T = 293 K |
| V = 1066.69 (11) Å3 | Prism, colourless |
| Z = 4 | 0.40 × 0.18 × 0.12 mm |
| F(000) = 448 |
Data collection
| Oxford Diffraction Xcalibur diffractometer with a Sapphire CCD detector | 1944 independent reflections |
| Radiation source: fine-focus sealed tube | 1690 reflections with I > 2σ(I) |
| graphite | Rint = 0.016 |
| Rotation method data acquisition using ω and φ scans | θmax = 26.4°, θmin = 2.6° |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | h = −9→5 |
| Tmin = 0.895, Tmax = 0.967 | k = −6→9 |
| 4245 measured reflections | l = −17→20 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.035 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.086 | w = 1/[σ2(Fo2) + (0.0453P)2 + 0.1227P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.08 | (Δ/σ)max = 0.016 |
| 1944 reflections | Δρmax = 0.14 e Å−3 |
| 132 parameters | Δρmin = −0.17 e Å−3 |
| 2 restraints | Absolute structure: Flack (1983), 824 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: 0.03 (9) |
Special details
| Experimental. CrysAlis RED (Oxford Diffraction, 2009) Empirical absorption correction using spherical harmonics, implemented in SCALE3 ABSPACK scaling algorithm. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.0401 (3) | 0.5352 (3) | 0.16636 (17) | 0.0418 (6) | |
| C2 | 0.0148 (3) | 0.3636 (3) | 0.15630 (19) | 0.0534 (7) | |
| C3 | −0.0178 (4) | 0.2719 (4) | 0.2248 (3) | 0.0758 (11) | |
| H3 | −0.0359 | 0.1571 | 0.2205 | 0.091* | |
| C4 | −0.0243 (5) | 0.3456 (5) | 0.2992 (2) | 0.0844 (12) | |
| H4 | −0.0458 | 0.2805 | 0.3440 | 0.101* | |
| C5 | 0.0008 (4) | 0.5134 (5) | 0.3071 (2) | 0.0730 (10) | |
| H5 | −0.0034 | 0.5631 | 0.3573 | 0.088* | |
| C6 | 0.0326 (4) | 0.6095 (4) | 0.24026 (19) | 0.0571 (7) | |
| H6 | 0.0490 | 0.7244 | 0.2452 | 0.069* | |
| C7 | 0.4165 (3) | 0.6071 (3) | 0.10946 (17) | 0.0462 (6) | |
| C8 | 0.5725 (3) | 0.5282 (5) | 0.0766 (2) | 0.0738 (10) | |
| H8A | 0.6102 | 0.5908 | 0.0310 | 0.089* | |
| H8B | 0.5489 | 0.4149 | 0.0608 | 0.089* | |
| H8C | 0.6582 | 0.5283 | 0.1168 | 0.089* | |
| C9 | 0.0213 (5) | 0.2751 (4) | 0.0766 (3) | 0.0830 (11) | |
| H9A | 0.1276 | 0.2961 | 0.0517 | 0.100* | |
| H9B | −0.0669 | 0.3162 | 0.0429 | 0.100* | |
| H9C | 0.0075 | 0.1568 | 0.0845 | 0.100* | |
| N1 | 0.2818 (3) | 0.6051 (3) | 0.05903 (12) | 0.0413 (5) | |
| H1N | 0.278 (3) | 0.556 (3) | 0.0138 (12) | 0.050* | |
| O1 | −0.0096 (3) | 0.6269 (3) | 0.01708 (12) | 0.0644 (6) | |
| O2 | 0.0971 (3) | 0.8346 (2) | 0.11214 (13) | 0.0632 (6) | |
| O3 | 0.4067 (2) | 0.6678 (3) | 0.17544 (11) | 0.0588 (5) | |
| S1 | 0.09057 (8) | 0.66608 (8) | 0.08490 (5) | 0.04393 (18) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0313 (12) | 0.0414 (14) | 0.0526 (15) | 0.0022 (11) | 0.0085 (11) | −0.0017 (12) |
| C2 | 0.0416 (15) | 0.0414 (15) | 0.077 (2) | 0.0004 (12) | 0.0128 (14) | −0.0002 (14) |
| C3 | 0.068 (2) | 0.0495 (18) | 0.110 (3) | 0.0058 (16) | 0.027 (2) | 0.017 (2) |
| C4 | 0.078 (2) | 0.090 (3) | 0.085 (3) | 0.011 (2) | 0.030 (2) | 0.033 (2) |
| C5 | 0.069 (2) | 0.098 (3) | 0.0526 (19) | 0.0076 (18) | 0.0181 (17) | 0.0058 (18) |
| C6 | 0.0480 (16) | 0.0596 (17) | 0.0637 (18) | 0.0013 (13) | 0.0148 (14) | −0.0082 (16) |
| C7 | 0.0392 (14) | 0.0478 (14) | 0.0515 (17) | −0.0071 (11) | −0.0043 (11) | −0.0001 (13) |
| C8 | 0.0401 (15) | 0.102 (3) | 0.080 (2) | 0.0051 (15) | 0.0006 (17) | −0.012 (2) |
| C9 | 0.084 (2) | 0.0521 (18) | 0.112 (3) | −0.0123 (16) | 0.020 (2) | −0.030 (2) |
| N1 | 0.0379 (11) | 0.0493 (12) | 0.0366 (12) | −0.0012 (9) | 0.0021 (9) | −0.0061 (9) |
| O1 | 0.0510 (12) | 0.0821 (16) | 0.0601 (14) | 0.0057 (10) | −0.0138 (10) | 0.0055 (12) |
| O2 | 0.0717 (13) | 0.0375 (10) | 0.0806 (15) | 0.0068 (9) | 0.0170 (11) | 0.0028 (10) |
| O3 | 0.0577 (12) | 0.0760 (14) | 0.0426 (11) | −0.0075 (10) | −0.0091 (9) | −0.0100 (10) |
| S1 | 0.0385 (3) | 0.0424 (3) | 0.0509 (4) | 0.0049 (3) | 0.0006 (3) | 0.0015 (3) |
Geometric parameters (Å, °)
| C1—C6 | 1.374 (4) | C7—N1 | 1.367 (3) |
| C1—C2 | 1.395 (4) | C7—C8 | 1.500 (4) |
| C1—S1 | 1.765 (3) | C8—H8A | 0.9600 |
| C2—C3 | 1.386 (5) | C8—H8B | 0.9600 |
| C2—C9 | 1.511 (5) | C8—H8C | 0.9600 |
| C3—C4 | 1.378 (5) | C9—H9A | 0.9600 |
| C3—H3 | 0.9300 | C9—H9B | 0.9600 |
| C4—C5 | 1.361 (6) | C9—H9C | 0.9600 |
| C4—H4 | 0.9300 | N1—S1 | 1.659 (2) |
| C5—C6 | 1.380 (5) | N1—H1N | 0.855 (17) |
| C5—H5 | 0.9300 | O1—S1 | 1.424 (2) |
| C6—H6 | 0.9300 | O2—S1 | 1.4213 (19) |
| C7—O3 | 1.209 (3) | ||
| C6—C1—C2 | 121.8 (3) | C7—C8—H8A | 109.5 |
| C6—C1—S1 | 116.8 (2) | C7—C8—H8B | 109.5 |
| C2—C1—S1 | 121.4 (2) | H8A—C8—H8B | 109.5 |
| C3—C2—C1 | 116.5 (3) | C7—C8—H8C | 109.5 |
| C3—C2—C9 | 119.4 (3) | H8A—C8—H8C | 109.5 |
| C1—C2—C9 | 124.1 (3) | H8B—C8—H8C | 109.5 |
| C4—C3—C2 | 122.0 (3) | C2—C9—H9A | 109.5 |
| C4—C3—H3 | 119.0 | C2—C9—H9B | 109.5 |
| C2—C3—H3 | 119.0 | H9A—C9—H9B | 109.5 |
| C5—C4—C3 | 120.2 (3) | C2—C9—H9C | 109.5 |
| C5—C4—H4 | 119.9 | H9A—C9—H9C | 109.5 |
| C3—C4—H4 | 119.9 | H9B—C9—H9C | 109.5 |
| C4—C5—C6 | 119.6 (3) | C7—N1—S1 | 123.94 (18) |
| C4—C5—H5 | 120.2 | C7—N1—H1N | 125.3 (19) |
| C6—C5—H5 | 120.2 | S1—N1—H1N | 109.7 (19) |
| C1—C6—C5 | 119.9 (3) | O2—S1—O1 | 119.00 (13) |
| C1—C6—H6 | 120.0 | O2—S1—N1 | 109.12 (11) |
| C5—C6—H6 | 120.0 | O1—S1—N1 | 104.10 (12) |
| O3—C7—N1 | 121.2 (2) | O2—S1—C1 | 108.68 (13) |
| O3—C7—C8 | 123.9 (3) | O1—S1—C1 | 110.99 (13) |
| N1—C7—C8 | 114.9 (3) | N1—S1—C1 | 103.79 (11) |
| C6—C1—C2—C3 | −0.2 (4) | O3—C7—N1—S1 | −6.3 (4) |
| S1—C1—C2—C3 | 177.5 (2) | C8—C7—N1—S1 | 173.3 (2) |
| C6—C1—C2—C9 | 180.0 (3) | C7—N1—S1—O2 | 57.5 (3) |
| S1—C1—C2—C9 | −2.3 (4) | C7—N1—S1—O1 | −174.5 (2) |
| C1—C2—C3—C4 | −0.3 (5) | C7—N1—S1—C1 | −58.2 (2) |
| C9—C2—C3—C4 | 179.6 (3) | C6—C1—S1—O2 | −6.2 (3) |
| C2—C3—C4—C5 | 0.3 (6) | C2—C1—S1—O2 | 176.0 (2) |
| C3—C4—C5—C6 | 0.1 (5) | C6—C1—S1—O1 | −138.9 (2) |
| C2—C1—C6—C5 | 0.6 (4) | C2—C1—S1—O1 | 43.3 (2) |
| S1—C1—C6—C5 | −177.3 (2) | C6—C1—S1—N1 | 109.8 (2) |
| C4—C5—C6—C1 | −0.5 (5) | C2—C1—S1—N1 | −68.0 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O3i | 0.86 (2) | 1.95 (2) | 2.770 (3) | 162 (3) |
Symmetry codes: (i) −y+1, x, z−1/4.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: TK2737).
References
- Adsmond, D. A. & Grant, D. J. W. (2001). J. Pharm. Sci. 90, 2058–2077. [DOI] [PubMed]
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Gowda, B. T., Foro, S., Nirmala, P. G. & Fuess, H. (2010). Acta Cryst. E66, o1284. [DOI] [PMC free article] [PubMed]
- Gowda, B. T. & Kumar, B. H. A. (2003). Oxid. Commun. 26, 403–425.
- Gowda, B. T., Svoboda, I. & Fuess, H. (2004). Z. Naturforsch. Teil A, 55, 845–852.
- Maren, T. H. (1976). Annu. Rev. Pharmacol Toxicol. 16, 309–327. [DOI] [PubMed]
- Oxford Diffraction (2009). CrysAlis CCD and CrysAlis RED Oxford Diffraction Ltd, Yarnton, England.
- Shakuntala, K., Foro, S. & Gowda, B. T. (2011). Acta Cryst. E67, o1097. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811014218/tk2737sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811014218/tk2737Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




