Abstract
Clearance of the intracellular bacterial pathogen Listeria monocytogenes requires antigen-specific CD8+ T cells. Recently it was shown that activation of class Ib major histocompatibility complex (MHC)-restricted CD8+ T cells alone is sufficient for immune protection against listeriae. A major component of the class Ib MHC-restricted T-cell response is T cells that recognize formylated peptide antigens presented by M3 molecules. Although three N-formylated peptides derived from L. monocytogenes are known to bind to M3 molecules, fMIGWII is the immunodominant epitope presented by M3 during infection of mice. The source of fMIGWII peptide is the L. monocytogenes lemA gene, which encodes a 30-kDa protein of unknown function. In this report, we describe the generation of two L. monocytogenes lemA deletion mutants. We show that lemA is not required for growth of listeriae in tissue culture cells or for virulence during infection of mice. Surprisingly, we found that fMIGWII-specific T cells were still primed following infection with lemA mutant listeriae, suggesting that L. monocytogenes contains at least one additional antigen that is cross-reactive with the fMIGWII epitope. This cross-reactive antigen appears to be a small protease-resistant molecule that is secreted by L. monocytogenes.
Listeria monocytogenes is a gram-positive bacterium that causes food-borne illness, primarily in pregnant women and neonates. L. monocytogenes has also been used for decades as a model organism to better understand the interactions of host cells with intracellular bacterial pathogens. The mouse model of listeriosis has been used extensively to characterize the immune response to intracellular infection, in particular the role of CD8+ T cells.
Components of innate immunity are primarily responsible for clearing sublethal doses of bacteria from the host during primary L. monocytogenes infection of mice (4, 14, 23). However, antigen-specific memory CD8+ T cells are stimulated during this initial infection that can protect mice from secondary challenges with doses of Listeria that would otherwise be lethal. Listeria-specific protective immune responses are not generated in mice that lack CD8+ T cells (9, 17). The central role of CD8+ T cells has been the impetus for many studies directed at identifying which Listeria antigens are recognized by CD8+ T cells and which class I major histocompatibility complex (MHC) molecules present these antigens during the course of L. monocytogenes infection.
The class Ia MHC-restricted T-cell response to Listeria has been well characterized, and several Kd-restricted epitopes derived from L. monocytogenes have been identified. However, it was recently discovered that mice lacking class Ia MHC molecules also have the capacity to clear L. monocytogenes infection, suggesting that class Ib MHC-restricted CD8+ T cells are sufficient for the generation of a protective immune response against Listeria (5, 19). A large part of the class Ib MHC response may be M3 restricted, as suggested by a recent study that used M3-specific antibodies to block antigen presentation to CD8+ T cells (20).
M3 is a nonclassical MHC protein that displays minimal polymorphism within laboratory strains of mice. It is expressed on a wide variety of cell types at a much lower level than class Ia MHC molecules (21). M3 preferentially binds short, hydrophobic peptides with a formylmethionine (fM) at the N terminus. In eukaryotic cells, the only source of formylated peptides is one of 13 mitochondrial proteins. The paucity of epitopes that are able to bind to M3 results in retention of M3 molecules within the Golgi network (3). In contrast, all prokaryotic protein synthesis is initiated with N-formylmethionine, and therefore it has been postulated that M3 molecules have evolved specifically to present bacterial peptide antigens. In fact, the cell surface level of M3 increases significantly upon infection with L. monocytogenes, presumably due to the greatly increased number of formylated peptides available within the cell (3).
During L. monocytogenes infection, M3-restricted CD8+ T cells that recognize at least three formylated peptides thought to be derived from Listeria are activated: fMIVIL, fMIGWII, and fMIVTLF (13, 16). The magnitude of Listeria-specific M3-restricted T-cell responses seems to vary considerably from mouse to mouse even with genetically identical mice, but fMIGWII is always the immunodominant epitope presented by M3 molecules during Listeria infection (6, 20). Adoptive transfer of an fMIGWII-specific T-cell clone into naïve mice resulted in some degree of protection against subsequent challenge with L. monocytogenes (18).
Lenz et al. screened a library of Escherichia coli expressing L. monocytogenes genome fragments to identify the product of the L. monocytogenes lemA (Listeria epitope with M3) gene as the source of fMIGWII peptide (10). lemA encodes a 30-kDa protein of unknown function with no significant homology to any known protein in currently available databases. The lemA gene may be transcribed as part of an operon containing lemA and the gene located immediately downstream, which codes for a 33-kDa protein with homology to heat shock proteins and has been termed lemB (10). The N terminus of LemA is predicted to orient outside the bacterium, which may explain how the N-formyl group avoids bacterial cytosolic formylases and is retained on the peptide.
We generated lemA mutant strains of L. monocytogenes in order to determine whether an fMIGWII-specific T-cell response is a required component of class Ib MHC-restricted CD8+ T-cell-mediated immunity against Listeria. In this report, we provide evidence that lemA is not essential for the metabolism or virulence of L. monocytogenes and that, more surprisingly, it is also not required for the priming of fMIGWII-specific CD8+ T cells in mice. These findings led to the characterization of a small, protease-resistant antigen in L. monocytogenes that appears to be cross-reactive with the fMIGWII epitope.
MATERIALS AND METHODS
Bacteria.
L. monocytogenes 10403s was used as the parental strain for all mutations generated in this study. The reference strain DP-L3903, an erythromycin-resistant derivative of 10403s (1), was generously provided by Dan Portnoy (Berkeley). Listeria strains were grown at 30°C without agitation in brain heart infusion (BHI) medium (Difco) supplemented with 2 μg of erythromycin per ml where indicated. For infection of mice, frozen culture stocks were thawed, grown with agitation at 37°C to early log phase in BHI broth, and diluted in phosphate-buffered saline (PBS) prior to injection. L. monocytogenes 10403s has a 50% lethal dose (LD50) of ≈104 CFU in BALB/c mice.
Construction of lemA mutant strains of L. monocytogenes.
Mutagenesis by overlap extension PCR was used to construct L. monocytogenes strain SD9-1, in which codons 2 to 16 of lemA (including the fMIGWII epitope) were deleted and replaced with a SacI restriction site (see Fig. 1B). Briefly, a fragment spanning the region 1,038 bp upstream of lemA to the ATG start codon of lemA was amplified from L. monocytogenes 10403s genomic DNA with primers that added a 5′ BamHI site and a 3′ SacI site. The primer sequences used were 5′-AGAGGATCCTGGTATTGACGCAGTAATCGTTTC-3′ and 5′-GGGGAGCTCCATAATTAATCTCCTCCT-3′. After restriction digestion, the fragment was subcloned into the temperature-sensitive suicide vector pKSV7 (22), resulting in plasmid pONT2.
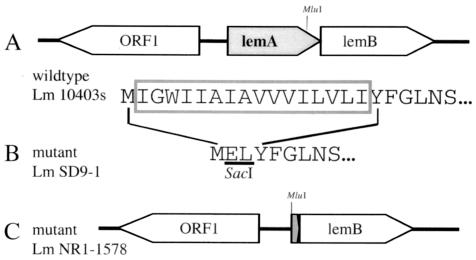
FIG. 1.
Physical and genetic maps of lemA mutant L. monocytogenes strains. (A) Open reading frames found in wild-type L. monocytogenes 10403s; arrows indicate direction of transcription. The N-terminal amino acid sequence of LemA is shown below; the boxed region indicates the amino acids deleted in the ΔMIGWII mutant strain SD9-1. (B) N-terminal amino acid sequence of L. monocytogenes SD9-1 LemA. Underlined amino acids represent the SacI recognition site that replaced the 16 amino acids shown above in the wild-type sequence. (C) Physical map of the chromosomal deletion found in ΔlemA mutant L. monocytogenes NR1-1578.
A second fragment spanning bp 49 of lemA to a region 855 bp downstream of lemA was amplified from L. monocytogenes 10403s genomic DNA with primers that added a 5′ SacI site and a 3′ EcoRI site. The primer sequences used were 5′-GGGGAGCTCTATTTCGGTCTATACAAC-3′ and 5′-AGGGAATTCTAATTGGCGGATGTGAGTC-3′. After restriction digestion, the fragment was subcloned in pONT2. The resultant plasmid, pONT4, contains an in-frame deletion of codons 2 to 16 of lemA and is referred to as the ΔMIGWII mutation. pONT4 was cleaved with SacI and MluI to remove a 487-bp fragment of lemA (see Fig. 1C). The digested plasmid was treated with Pfu polymerase (Stratagene) to create blunt ends and then religated to form recombinant plasmid pNR1. pNR1 contains the wild-type ribosome-binding sequence and ATG start codon fused to remnants of the SacI binding site as well as the last 27 bp of lemA, which are no longer in-frame, and is referred to as the ΔlemA mutation.
Replacement of wild-type chromosomal sequences with either the ΔMIGWII (strain SD9-1) or ΔlemA (strain NR1-1578) mutation was accomplished by allelic exchange, as described previously (2). Chromosomal mutations were confirmed by comparing the DNA sequences of fragments amplified from L. monocytogenes 10403s, L. monocytogenes SD9-1, and L. monocytogenes NR1-1578 genomic DNA.
Mice.
BALB/c/By/J mice were obtained from the Jackson Laboratory and used at 6 to 12 weeks of age. C.B10-H2b/LilMcd/J (BALB/c congenic at the H-2b locus; herein referred to as C.B10 mice) were originally obtained from the Jackson Laboratory and then maintained as a colony in a specific-pathogen-free barrier facility at Harvard Medical School. Class Ia MHC-deficient (Kb−/− Db−/−) C.B10 mice were described previously (5).
Cell lines and cell culture.
EL-4 mouse thymoma cells, J774 mouse macrophage-like cells, L2 mouse fibroblasts, L929 mouse fibroblasts, and Henle 407 human epithelial cells were obtained from the American Type Culture Collection. L2 cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (DMEM-10); all other cells were maintained in a medium (RP-10) consisting of RPMI 1640 supplemented with l-glutamine, HEPES, 50 μM 2-mercaptoethanol, and 10% fetal calf serum. All cells were incubated at 37°C with 7% CO2. Antibiotics were used at the following concentrations: penicillin, 50 units/ml; streptomycin, 50 μg/ml; and gentamicin, 25 and 50 μg/ml.
Bone marrow-derived macrophages were harvested from the femurs of BALB/c mice, cultured in DMEM supplemented with 20% fetal calf serum and 20% L929 cell supernatant, and the medium was replenished every 3 to 4 days. The fMIGWII-specific T-cell line S172 was prepared by stimulating splenocytes from a Listeria-immune Kb−/− Db−/− C.B10 mouse on irradiated (2,000 rads) syngeneic splenocytes coated with 10 nM synthetic N-formylated MIGWII peptide (fMIGWII). The T-cell line was maintained by weekly restimulation, first in RP-10 and after 2 weeks in RP-10 supplemented with supernatant from concanavalin A-stimulated rat splenocytes and 50 mM α-methylmannoside (CTL medium).
Intracellular growth assay.
Cells were seeded on 12-mm round coverslips and incubated overnight in RP-10 without antibiotics to reach confluence. Bacteria were grown to early log phase in BHI broth, washed once with PBS, and used to infect cell monolayers. Cells were infected (on coverslips in triplicate) at a multiplicity of infection of 0.1 (J774 and bone marrow-derived macrophages) or 2.0 (Henle 407), incubated for 1 h, and washed three times with warm PBS, and then RP-10 containing 25 μg of gentamicin sulfate per ml was added. At that time and at each subsequent time point, the number of bacteria associated with each coverslip was determined by placing the coverslip in sterile distilled H2O, vortexing vigorously for 30 s, and plating dilutions on BHI agar.
Plaque assay.
L2 fibroblasts were grown to confluence in six-well dishes and infected with L. monocytogenes at various multiplicities of infection. The cells were incubated for 1 h and then washed three times with warm PBS. Overlays consisting of DMEM-10, 1% agarose, and gentamicin sulfate (10 μg/ml) were added, and the cells were incubated for an additional 3 days. Plaques were visualized by the addition of neutral red overlays (1% agarose and 0.2% neutral red in DMEM-10). Plaque size was measured by analyzing digital images of the overlays.
Competitive index assay.
Growth of the ΔlemA mutant was compared to growth of wild-type bacteria during both primary and secondary Listeria infections in a competitive index assay as previously described (1). Briefly, BALB/c mice were infected intravenously with a 1:1 mixture of DP-L3903 and NR1-1578. For primary infections, mice were given a total of 103 CFU and sacrificed 72 h later; for secondary infections, mice were given a total of 105 CFU and sacrificed 48 h later. Spleens and livers were harvested aseptically, homogenized and diluted in 0.2% NP-40, and plated on BHI agar. At least 100 colonies per organ were replica plated or patched onto BHI agar containing 2 μg of erythromycin per ml. Competitive indices were calculated by dividing the total CFU of test strain NR1-1578 (erythromycin sensitive) by the total CFU of reference strain DP-L3903 (erythromycin resistant).
ELISPOT assay.
The frequency of antigen-specific CD8+ T cells in the spleens of mice was determined by ELISPOT assay. Flat-bottomed 96-well filtration plates (0.45-μm cellulose ester membrane; Millipore) were coated with rat anti-mouse gamma interferon (IFN-γ) antibody (10 μg/ml; clone R4-6A2, Pharmingen) and then blocked with medium containing 5% fetal calf serum. Splenocytes (1 × 105 to 5 × 105/well) were incubated with stimulator cells (105 EL-4 cells/well) pretreated with 1 μM synthetic peptide for 1 h in CTL medium. N-Formylated peptides fMIVIL, fMIGWII, and fMIVTLF were purchased from Biosynthesis Inc. (Lewisville, Tex.). After 22 to 26 h of incubation, the plates were washed with PBS-0.25% Tween 20 (PBS-T), and any remaining cells were lysed with distilled water. After incubation with a biotinylated rat anti-mouse IFN-γ antibody (XMG1.2, Pharmingen), the plates were washed with PBS-T and incubated with streptavidin-labeled peroxidase in PBS-5% fetal calf serum for 1 h at room temperature. Plates were developed by adding 3,3′-diaminobenzidine tetrahydrochloride dihydrate (Bio-Rad, Melville, N.Y.) in Tris buffer plus hydrogen peroxide for 30 min at room temperature, and spots were detected on the membranes with the aid of a dissecting microscope. Each spot represents an area in which a single T cell recognized its cognate antigen and was stimulated to locally secreted IFN-γ. The number of antigen-specific cells was determined by subtracting the number of spots observed for EL-4 cells alone from the number of spots observed for peptide-coated EL-4 cells.
Cytotoxicity assay.
L. monocytogenes strains were grown with aeration in BHI broth at 37°C for 24 h. Supernatants from these cultures were collected, and 50 μl was mixed with 100 μl of RP-10 containing 106 EL-4 target cells. Sodium 51chromate (100 μCi) was added, and the cells were incubated for 1 h. Where indicated, the culture supernatants were treated with 1 mg of proteinase K per ml overnight at 55°C or filtered through a Centricon-10 filter (Amicon; 10,000 molecular weight cutoff passivated with 1% milk buffer). Heat-killed listeriae were prepared by incubating stationary-phase Listeria cultures at 55°C for 4 h; loss of viable bacteria was confirmed by plating on BHI agar. Heat-killed L. monocytogenes organisms were incubated with target cells for a total of 2 h; 51chromate was added during the final hour of incubation. Target cells were washed three times with RPMI 1640, resuspended in RP-10, and added (104 cells/well) to 96-well plates. Serial dilutions of S172 T cells were then added in a final volume of 200 μl/well, and the plates were incubated for 4 h. Spontaneous release was determined in wells containing target cells with no T cells. Maximum release was determined by the addition of 1% Triton X-100. The cytotoxic activity of the T cells was evaluated by measuring 51Cr in the supernatant with a Wallac (Gaithersburg, Md.) 1470 Wizard gamma counter. Percent specific lysis was calculated with the formula [(release by T cells − spontaneous release) ÷ (maximum release − spontaneous release)] × 100.
Intracellular cytokine staining.
Intracellular cytokine staining was performed with the Cytofix/Cytoperm Plus (with GolgiPlug) kit according to the manufacturer's instructions (Pharmingen). EL-4 cells were pretreated with culture supernatants for 30 min at 37°C. S172 T cells were added at an effector-to-target cell ratio of 5. Cells were stained with fluorescently conjugated monoclonal antibodies (Pharmingen) specific for CD8α (clone 53-6.7) and IFN-γ (XMG1.2) and analyzed with CellQuest software on a FACScan flow cytometer (Becton Dickinson). Phycoerythrin-conjugated rat IgG1 (R3-34) was used as an isotype control antibody. Dead cells and monocytes were excluded by forward and side scatter gating. For each sample, 25,000 events were collected.
RESULTS
Generation of L. monocytogenes ΔlemA and ΔMIGWII mutants.
The L. monocytogenes gene lemA encodes a 30-kDa protein of unknown function that was first identified as the source of the immunodominant M3-binding epitope fMIGWII (10). lemA is located immediately adjacent to a gene that has been designated lemB; it is not yet known whether these two genes constitute an operon (Fig. 1A).
We generated two lemA deletion mutant strains of L. monocytogenes. The first mutant strain, NR1-1578, contains a chromosomal deletion spanning almost the entire lemA gene and is referred to as the ΔlemA mutant (Fig. 1C). It was not known at the outset whether deletion of lemA would affect the virulence of L. monocytogenes, resulting in a strain that was unable to establish systemic infection in mice. Since the goal of this study was to create a mutant Listeria strain that could be used to infect mice so that we could then assess specific T-cell responses, a second mutant in which just the fMIGWII epitope sequence was deleted was also created. L. monocytogenes SD9-1 lemA contains an in-frame deletion of 16 amino acids at the N terminus of the product and is referred to as the ΔMIGWII mutant (Fig. 1B). There was no difference in the ability of either mutant strain to grow in broth culture compared to the parental strain L. monocytogenes 10403s (data not shown).
Intracellular growth of ΔlemA mutant L. monocytogenes is not impaired.
One of the hallmarks of L. monocytogenes pathogenesis is the ability of the bacteria to escape from a phagocytic vacuole into the host cell cytoplasm, where they are able to multiply with a doubling time of approximately 1 h in vitro (15). To determine whether the lemA gene product was required for intracellular survival or growth, we infected macrophages and epithelial cells with either mutant or wild-type L. monocytogenes and determined the total number of intracellular bacteria at various times after infection. As shown in Fig. 2, the numbers of wild-type and mutant bacteria associated with J774 macrophages, primary bone marrow-derived macrophages, and Henle 407 epithelial cells 1 h postinfection were approximately the same, suggesting that the ΔlemA mutant strain did not have a defect in cell entry or survival. Intracellular growth of the ΔlemA mutant occurred in a logarithmic manner over an 8-h time period and reached peak levels that were similar to that observed for wild-type L. monocytogenes (Fig. 2). These data suggest that lemA is not required for intracellular growth in macrophages or epithelial cells.
FIG. 2.
Intracellular growth of wild-type and ΔlemA mutant listeriae. Cells were infected with either wild-type L. monocytogenes 10403s (solid squares) or ΔlemA mutant listeriae (open squares) at a multiplicity of infection of either 0.1 (J774 cells and bone marrow-derived macrophages [BMMΦ]) or 2.0 (Henle 407 cells). One hour later, the cells were washed extensively, and medium containing gentamicin (25 μg/ml) was added. Cells were lysed in sterile water at the time points indicated, and dilutions were plated on BHI agar to determine the total number of cell-associated bacteria per coverslip. Average values ± standard deviation are given. Representative data from one of three separate experiments are shown.
The ΔlemA mutant competes with wild-type Listeria during growth in spleen and liver.
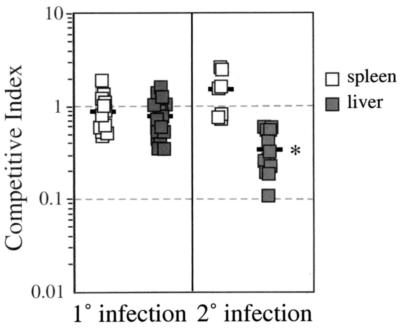
Although the ΔlemA mutant was able to survive and replicate within tissue culture cells, it was possible that the strain would have impaired ability to cause systemic infection after intravenous inoculation of mice. We used a competitive index assay to determine whether the ΔlemA mutant had a subtle growth defect that could be observed only during in vivo infection of mice. As shown in Fig. 3, there was no significant difference in the growth of the ΔlemA mutant compared to wild-type listeriae during primary infection of BALB/c mice (mean competitive indices of 0.9 for spleen and 0.8 for liver).
FIG. 3.
Competitive index assay analysis of primary and secondary infection with wild-type and ΔlemA mutant L. monocytogenes. Groups of four to six naïve and Listeria-immune mice were infected with a 1:1 mixture of DP-L3903 (wild type) and NR1-1578 (ΔlemA). Mice were sacrificed 3 days after primary infections (0.1 LD50 of total bacteria) or 2 days after secondary infections (0.1 LD50 of wild-type listeriae followed by 5 LD50 of total bacteria 3 weeks later). Spleens and livers were harvested and homogenized, and dilutions were plated on BHI agar with or without erythromycin. A competitive index of 1.0 indicates that the two strains were recovered in equal numbers. Data compiled from three separate experiments are shown; bars indicate average values for each experimental group. *, P < 0.01 as determined by a two-sided Mann-Whitney test.
To assess the ability of the mutant strain to replicate in immune animals, we infected mice with 0.1 LD50 of wild-type L. monocytogenes and then challenged those mice 3 weeks later with 5 LD50 of a 1:1 mixture of mutant and wild-type bacteria. On average, there was a 2.8-fold defect in the ability of the ΔlemA mutant to proliferate in the liver during secondary infection (Fig. 3; competitive indices ranging from 0.1 to 0.6). In contrast, wild-type and mutant listeriae were recovered from the spleen in approximately equal numbers during secondary infection. These data suggest that the lemA gene product may play a role in intracellular survival in hepatocytes during secondary infection and that it is not required for growth in splenocytes.
Cell-to-cell spread of the L. monocytogenes ΔlemA mutant is not impaired.
Auberbach et al. previously showed that certain actA mutant strains of L. monocytogenes also display a growth defect in liver but not spleen during infection of immunized animals (1). ActA is essential for actin-based motility in the host cell, a phenomenon that increases the ability of Listeria to spread to neighboring cells. To determine if LemA was required for efficient cell-to-cell spread, we tested the ability of the ΔlemA mutant to form plaques in a fibroblast monolayer. L2 cells were infected with either wild-type or ΔlemA Listeria for 3 days, and the plaques that formed in the cell monolayers were visualized after the addition of neutral red. As shown in Fig. 4, there was no significant difference in the size of the plaques formed by the two strains. The mean plaque size formed by the ΔlemA mutant was 93% ± 4% of the plaque size formed by L. monocytogenes 10403s. Therefore, LemA does not appear to be necessary for efficient cell-to-cell spread during in vitro infection of mouse fibroblasts.
FIG. 4.
Cell-to-cell spread of ΔlemA mutant and wild-type L. monocytogenes. Monolayers of L2 fibroblasts in six-well dishes were infected with approximately 2.0 × 105 ΔlemA (A) or wild-type (B) L. monocytogenes for 3 days. Plaques were visualized by the addition of 0.2% neutral red. Representative data from one of three separate experiments are shown.
fMIGWII-specific T cells are activated during infection with ΔlemA mutant L. monocytogenes.
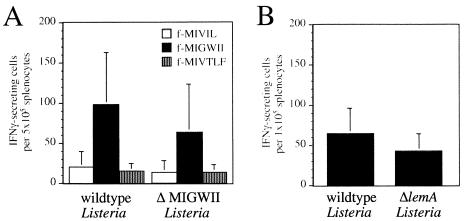
The results described above established that the ΔlemA mutant was fully capable of causing systemic infection in either naïve or immune animals. We therefore proceeded to ask whether protective immunity could be established in mice immunized with ΔlemA mutant Listeria. To verify that the fMIGWII-specific T-cell response was abolished following infection with the lemA mutants, mice were immunized with a sublethal dose of either wild-type or mutant L. monocytogenes. Six days later, the activation of M3-restricted, Listeria-specific T cells in the spleens of these animals was measured by IFN-γ ELISPOT assay. To our surprise, fMIGWII-specific T-cell responses of only slightly reduced magnitude were stimulated in mice immunized with either ΔMIGWII or ΔlemA mutant Listeria compared to mice immunized with wild-type Listeria (Fig. 5). fMIGWII-specific T cells were below the limit of detection in uninfected mice (data not shown). As expected, the fMIVIL- and fMIVTLF-specific T-cell responses were unaffected following immunization with ΔMIGWII listeriae, suggesting that the infection proceeded as normal and that only the LemA antigen was altered in the mutant Listeria strain (Fig. 5A). These results suggested that L. monocytogenes expresses a second antigen that can cross-react with fMIGWII-specific T-cell receptors.
FIG. 5.
Infection of mice with ΔlemA mutant strains of L. monocytogenes results in the activation of fMIGWII-specific T cells. (A) Groups of C.B10 mice (two to four) were given intravenous injections of 103 CFU of either 10403s (wild-type listeriae) or SD9-1 (ΔMIGWII listeriae). Six days later, the mice were sacrificed, and the number of fMIVIL-, fMIGWII-, and fMIVTLF-specific T cells present in the spleen of each mouse was determined by IFN-γ ELISPOT assay. The average number of IFN-γ-secreting cells per 5 × 105 splenocytes ± standard deviation is shown. Data compiled from three separate experiments are shown. (B) Groups of four C.B10 mice were infected with 103 CFU of either 10403s or NR1-1578 (ΔlemA). Six days later, the mice were sacrificed, and the number of fMIGWII-specific T cells present in the spleen each mouse was determined by IFN-γ ELISPOT assay.
fMIGWII-specific T cells recognize an antigen found in the culture supernatant of ΔlemA Listeria.
Previous reports have shown that simple treatment of cells with either heat-killed listeriae or supernatant from Listeria cultures results in M3-restricted recognition by fMIGWII-specific T cells (8, 10, 13). To determine whether the antigen that cross-reacts with fMIGWII was similarly present in Listeria culture supernatants or heat-killed preparations of Listeria, we generated an fMIGWII-specific CD8+ T-cell line that could be used to recognize both fMIGWII and the cross-reactive antigen in chromium release assays.
Splenocytes harvested from a class Ia MHC-deficient mouse infected 2 weeks earlier with L. monocytogenes were stimulated in vitro with irradiated, syngeneic splenocytes coated with 10 nM fMIGWII peptide as a source of antigen. A CD8+ fMIGWII-specific T-cell line (line S172) cultured from this mouse specifically lysed J774 macrophage cells treated with heat-killed wild-type listeriae (Fig. 6A). Specific lysis of cells treated with heat-killed ΔlemA listeriae was also observed, although the level of recognition of these target cells was significantly lower. Similar results were obtained when EL-4 thymoma cells were treated with supernatants from wild-type and ΔlemA Listeria cultures (Fig. 6B), suggesting that processing in macrophages is not required for presentation of the cross-reactive antigen.
FIG. 6.
ΔlemA Listeria culture supernatants and heat-killed bacteria target cells for recognition by fMIGWII-specific T cells. Cells were treated with wild-type (wt) or mutant (ΔlemA) L. monocytogenes (Lm) preparations and then used as targets for lysis by fMIGWII-specific T cells (line S172) in a standard 51Cr release assay. (A) J774 cells were treated with heat-killed (HK) L. monocytogenes for 2 h. (B) EL-4 cells were incubated for 1 h with supernatants from L. monocytogenes cultures. The E:T (effector-to-target cell) ratio indicates the number of T cells added for every target cell in the assay. (C) EL-4 cells were treated for 30 min with supernatants from L. monocytogenes cultures and then added to S172 T cells. The percentage of line S172 cells that were CD8+ and secreting IFN-γ was determined by intracellular cytokine staining. Representative data from one of three separate experiments are shown.
We confirmed this observation by looking at the ability of line S172 T cells to secrete IFN-γ in response to EL-4 cells pretreated with Listeria culture supernatants. As shown in Fig. 6C, fMIGWII-specific T cells recognized EL-4 cells treated with either wild-type or ΔlemA Listeria culture supernatant. Again, the response to the ΔlemA supernatant was significantly lower than that to the wild-type supernatant. Taken together, these results suggest that small amounts of the cross-reactive antigen are secreted or released from L. monocytogenes in a manner similar to that of the fMIGWII epitope in LemA.
The fMIGWII cross-reactive antigen is a small protease-resistant molecule.
To further characterize the nature of the fMIGWII cross-reactive antigen, we treated both wild-type and ΔlemA mutant Listeria culture supernatants with proteinase K and used these preparations to target EL-4 cells for recognition by line S172 T cells. A significant portion of the targeting activity found in the wild-type culture supernatant was lost after proteinase K digestion (Fig. 7). However, the residual antigen found in protease-digested wild-type culture supernatant resulted in approximately the same level of specific lysis as observed for the untreated ΔlemA culture supernatant. In contrast, proteinase K digestion of the ΔlemA mutant supernatant resulted in greater recognition by fMIGWII-specific T cells. The cross-reactive antigen was not lost when undigested ΔlemA culture supernatant was filtered through a 10,000-molecular-weight-cutoff filter, suggesting that it is a small molecule. These results indicate that a small protease-resistant antigen is released from both wild-type and ΔlemA mutant Listeria during growth in BHI broth and that this antigen cross-reacts with fMIGWII-specific T cells.
FIG. 7.
T cells that recognize fMIGWII also recognize a small protease-resistant molecule present in L. monocytogenes culture supernatant. EL-4 cells were treated with 50 μl of 10403s (wild-type L. monocytogenes [wt Lm]) or NR1-1578 (ΔlemA L. monocytogenes [ΔlemA Lm]) culture supernatant preparations either untreated, digested with proteinase K (protK), or filtered through a 10,000-molecular-weight-cutoff filter and then used as targets for recognition by fMIGWII-specific T cells (line S172) in a 51Cr release assay. The effector-to-target cell ratio used was 30:1. Average values ± standard deviation from one of two separate experiments are shown.
DISCUSSION
lemA does not appear to be an essential gene in L. monocytogenes. Strain NR1-1578, which has a large chromosomal deletion encompassing almost all of the lemA gene, did not show a significant growth defect either in vitro or in vivo during primary Listeria infection. The slight defect in the ability of the ΔlemA mutant to grow in the livers of mice during secondary infection may suggest that LemA plays a role in intracellular survival in hepatocytes or in avoiding the enhanced recall immune response that occurs during secondary challenge. However, since this virulence defect was small, any essential function that LemA serves during infection of mice must be at least partially compensated for by another L. monocytogenes gene product.
The most striking finding in this report is that removing the fMIGWII epitope embedded within LemA does not abolish the fMIGWII-specific T-cell response that occurs during Listeria infection. This suggests that there is at least one cross-reactive antigen present in L. monocytogenes that can activate fMIGWII-specific T cells. We provide evidence here that this cross-reactive antigen is a small, protease-resistant molecule that may be actively secreted by L. monocytogenes. Further work will be required to determine whether this antigen is a short hydrophobic N-formylated peptide with sequence similarity to fMIGWII or whether this cross-reactive molecule represents a new class of antigens that can be presented by M3 molecules.
The peptide-binding grooves of MHC molecules are remarkably conserved, with only a few polymorphic residues involved in determining which peptides will bind with high affinity. Analysis of the crystal structure of M3 revealed that an N-formyl group promotes strong binding to the second position of the groove, with room for only six more residues, most of which point towards the backbone of the groove (24). Thus, structural constraints would make it seem likely that the cross-reactive antigen is a closely related hydrophobic peptide similar in sequence to fMIGWII. However, antigen-presenting molecules in the CD1 family have been shown to present lipids to T cells, providing a precedent for class Ib MHC-restricted presentation of nonpeptide antigens. Whether the L. monocytogenes cross-reactive antigen described here is a peptide or nonpeptide moiety, it is also possible that it binds to a distant site on the M3 molecule, outside of the peptide-binding groove.
Nataraj et al. previously described a protease-resistant particulate antigen thought to be a phospholipid that purified with heat-killed Listeria-associated antigen (HAA) (12). It was later thought that HAA was in fact LemA because T-cell clones that recognized HAA also recognized synthetic fMIGWII peptide at concentrations of less than 1 nM (7). Our results are consistent with their data and suggest that fMIGWII-specific T cells can cross-react with both the 6-mer peptide fMIGWII and the protease-resistant molecule described both here and in the previous work.
Nataraj et al. have shown that the hydrophobic amino terminus of lemA is protease resistant in vitro, so it is possible that the hydrophobic nature of the cross-reactive antigen prevents access by proteinase K, either due to association with a bacterial lipid or due to the secondary structure of the protein (12). However, there are two important differences between our studies. First, in the previous studies the authors used T cells stimulated on heat-killed Listeria to identify the HAA. We used an fMIGWII-specific T-cell line that was stimulated in vitro on syngeneic splenocytes coated with synthetic fMIGWII peptide. A T-cell line stimulated on peptide should be greatly enriched for T cells that recognize the specific peptide epitope, unlike T cells stimulated on the heterogenous mixture of antigens found in a heat-killed bacterial preparation. Second, we were readily able to detect the fMIGWII cross-reactive antigen in Listeria culture supernatants, while Nataraj et al. found that HAA was predominately found in bacterial cell wall and membrane preparations.
It is interesting that proteinase K treatment of the ΔlemA culture supernatant resulted in increased recognition by T cells and specific release of chromium when applied to target cells. This finding suggests that the cross-reactive antigen can be taken up more readily by cells when a protease-sensitive portion of it is digested away. In fact, we did see better presentation of the ΔlemA culture supernatant antigen on professional antigen-presenting cells such as primary bone marrow-derived macrophages compared to EL-4 cells (S. E. F. D'Orazio and M. N. Starnbach, unpublished observations). This suggests that the processing requirements for the cross-reactive antigen may be different than for LemA and that uptake by a phagocytic cell facilitates antigen processing. It has been shown that presentation of epitopes by M3 molecules is blocked by inhibitors of endosomal acidification and by brefeldin A, which blocks Golgi transport (3). Presumably antigen is taken up in endosomes or phagosomes and some proteolysis occurs within these vesicles, resulting in peptides available to bind M3. There appear to be both TAP-dependent and -independent pathways for presentation of exogenous antigen on M3 molecules (3, 10, 11), so it is not clear whether the antigens bind in the endoplasmic reticulum or in a post-Golgi compartment.
If the cross-reactive antigen is a short hydrophobic peptide(s) with sequence homology to fMIGWII, the availability of the complete L. monocytogenes genome sequence should aid in the identification of the antigen. In fact, a search of the L. monocytogenes genome database has revealed several candidate genes that may serve as fMIGWII cross-reactive antigens, and we are currently working to examine the relevance of each of these gene products. However, although there are a seemingly large number of potential M3-binding peptides to be found within any bacterial species, it is difficult to predict M3 epitopes by simply analyzing primary amino acid sequence data. Cytosolic formylases are very efficient at removing the N-terminal formyl group from bacterial proteins, so it is essential to understand the topology of a candidate antigen to know whether the formylated methionine is likely to be retained on an amino-terminal peptide. Defining all of the types of L. monocytogenes antigens that can stimulate CD8+ T cells will ultimately lead to a better understanding of how intracellular bacterial pathogens are recognized and eliminated by the immune system.
Acknowledgments
We thank Darren Higgins and Laurel Lenz for strains, plasmids, technical advice, and helpful discussions. We also thank L. Lenz for critical review of the manuscript.
This work was supported by National Institutes of Health grants AI41526 and AI055962 to M.N.S.
Editor: B. B. Finlay
REFERENCES
- 1.Auerbach, V., L. L. Lenz, and D. A. Portnoy. 2001. Devlopment of a competitive index assay to evaluate the virulence of Listeria monocytogenes actA mutants during primary and secondary infection of mice. Infect. Immun. 69:5953-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camilli, A., L. G. Tilney, and D. A. Portnoy. 1993. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 8:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiu, N. M., T. Chun, M. Fay, M. mandal, and C.-R. Wang. 1999. The majority of H2-M3 is retained intracellularly in a peptide-receptive state and traffics to the cell surface in the presence of N-formylated peptides. J. Exp. Med. 190:423-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Czuprynski, C. J., and M. Haak-Frendscho. 1997. Non-specific resistance meachnisms to listeriosis: implications for experimental and naturally occuring infection. Immunol. Rev. 158:47-56. [DOI] [PubMed] [Google Scholar]
- 5.D'Orazio, S. E. F., D. H. Gould, H. L. Ploegh, and M. N. Starnbach. 2003. Class Ia MHC-deficient BALB/c mice generate CD8+ T-cell mediated protective immunity against Listeria monocytogenes infection. J. Immunol. 171:291-298. [DOI] [PubMed] [Google Scholar]
- 6.Kerksiek, K. M., D. H. Busch, and E. G. Pamer. 2001. Variable immunodominance hierarchies for H2-M3-restricted n-formyl peptides following bacterial infection. J. Immunol. 166:1132-1140. [DOI] [PubMed] [Google Scholar]
- 7.Kurlander, R., and C. Nataraj. 1997. Characterization of the murine H2-M3wt-restricted CD8 response against a hydrophobic, protease-resistant, phospholipid associated antigen from Listeria monocytogenes. Immunol. Rev. 158:123-128. [DOI] [PubMed] [Google Scholar]
- 8.Kurlander, R. J., S. M. Shawar, M. L. Brown, and R. R. Rich. 1992. Specialized role for a murine class Ib MHC molecule in prokaryotic defenses. Science 257:678-679. [DOI] [PubMed] [Google Scholar]
- 9.Ladel, C. H., I. E. A. Flesch, J. Arnoldi, and S. H. E. Kaufmann. 1994. Studies with MHC-deficient knock-out mice reveal impact of both MHC I- and MHC II-dependent T-cell responses on Listeria monocytogenes infection J. Immunol. 153:3116-3122. [PubMed] [Google Scholar]
- 10.Lenz, L. L., B. Dere, and M. J. Bevan. 1996. Identification of an H2-M3-restricted Listeria epitope: implications for antigen presentation by M3. Immunity 5:63-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levitt, J., D. Howell, J. Rodgers, and R. Rich. 2001. Exogenous peptides enter the endoplasmic reticulum af TAP-deficient cells and induce the maturation of nascent class I molecules. Eur. J. Immunol. 31:1181-1190. [DOI] [PubMed] [Google Scholar]
- 12.Nataraj, C., M. L. Brown, R. M. Poston, S. M. Shawar, R. R. Rich, K. F. Lindahl, and R. J. Kurlander. 1996. H2-M3wt-restricted, Listeria monocytogenes-specific CD8 T cells recognize a novel, hydrophobic, protease-resistant, periodate-sensitive antigen. Int. Immunol. 8:367-378. [DOI] [PubMed] [Google Scholar]
- 13.Pamer, E. G., C.-R. Wang, L. Flaherty, K. F. Lindahl, and M. J. Bevan. 1992. H2-M3 presents a Listeria monocytogenes peptide to cytotoxic T lymphocytes. Cell 70:215-223. [DOI] [PubMed] [Google Scholar]
- 14.Portnoy, D. A. 1992. Innate immunity to a facultative intracellular bacterial pathogen. Current Opinion in Immunology. 4:20-24. [DOI] [PubMed] [Google Scholar]
- 15.Portnoy, D. A., P. S. Jacks, and D. J. Hinrichs. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J. Exp. Med. 167:1459-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Princiotta, M. F., L. L. Lenz, M. J. Bevan, and U. D. Staerz. 1998. H2-M3 restricted presentation of a Listeria-derived leader peptide J. Exp. Med. 187:1711-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts, A. D., D. J. Ordway, and I. M. Orme. 1993. Listeria monocytogenes infection in β2 microglobulin-deficient mice. Infect. Immun. 61:1113-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rolph, M. S., and S. H. E. Kaufmann. 2000. Partially TAP-independent protection against Listeria monocytogenes by H2-M3-restricted CD8+ T cells. J. Immunol. 165:4575-4580. [DOI] [PubMed] [Google Scholar]
- 19.Seaman, M. S., B. Perarnau, K. F. Lindahl, F. A. Lemmonier, and J. Forman. 1999. Response to Listeria monocytogenes in mice lacking MHC class Ia molecules. J. Immunol. 162:5429-5436. [PubMed] [Google Scholar]
- 20.Seaman, M. S., C.-R. Wang, and J. Forman. 2000. MHC Class Ib-restricted cytotoxic T lymphcytes provide protection against primary and secondary Listeria monocytogenes infection. J. Immunol. 165:5192-5201. [DOI] [PubMed] [Google Scholar]
- 21.Shawar, S. M., J. M. Vyas, J. R. Rodgers, and R. R. Rich. 1994. Antigen presentation by major histocompatibility complex class Ib molecules. Annu. Rev. Immunol. 12:839-880. [DOI] [PubMed] [Google Scholar]
- 22.Smith, K., and P. Youngman. 1992. Use of a new integrational vector to investigate compartment-specific expression of Bacillus subtilis spoIIM gene. Biochimie 74:705-711. [DOI] [PubMed] [Google Scholar]
- 23.Unanue, E. R. 1997. Studies in listeriosis show the strong symbiosis between the innate cellular system and the T-cell response. Immunol. Rev. 158:11-25. [DOI] [PubMed] [Google Scholar]
- 24.Wang, C., A. Castano, P. Peterson, C. Slaughter, and K. Lindahl. 1995. Nonclassical binding of formylated peptide in crystal structure of the MHC class Ib molecule H2-M3. Cell 82:655-664. [DOI] [PubMed] [Google Scholar]