Abstract
The molecular structure of the title compound, C18H23N3S, shows it to be a derivative of an aminothiophenol possessing a tetramethylguanidine group with a localized C=N double bond of 1.304 (2) Å and a protected thiol functional group as an S-benzyl thioether. The two aromatic ring planes make a dihedral angle of 67.69 (6)°.
Related literature
For synthesis, see: Neuba (2009 ▶); Lindoy & Livingstone (1968 ▶); Herres-Pawlis et al. (2005 ▶). For related structures, see: Neuba et al. (2007a
▶,b
▶,c
▶); Herres et al. (2004 ▶); Raab et al. (2003 ▶, 2002 ▶); Peters et al. (2008 ▶). For complexes of metal centres with bis(tetramethylguanidino)propylene and amine guanidine hybrids, see: Harmjanz (1997 ▶); Waden (1999 ▶); Pohl et al. (2000 ▶); Schneider (2000 ▶); Wittmann (1999 ▶); Wittmann et al. (2001 ▶); Herres et al. (2005 ▶); Herres-Pawlis et al. (2009 ▶); Börner et al. (2007 ▶, 2009 ▶). For sulfur guanidine hybrids based on aminothiophenol and cysteamine, see: Neuba (2009 ▶); Neuba et al. (2008a
▶,b
▶, 2010 ▶, 2011 ▶).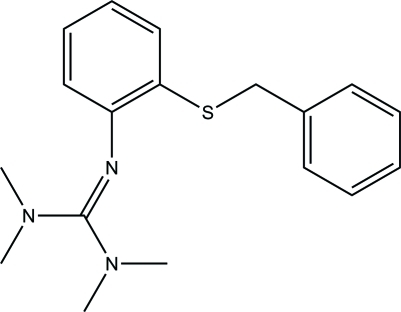
Experimental
Crystal data
C18H23N3S
M r = 313.45
Monoclinic,

a = 7.869 (2) Å
b = 26.850 (7) Å
c = 8.314 (2) Å
β = 106.959 (5)°
V = 1680.2 (8) Å3
Z = 4
Mo Kα radiation
μ = 0.19 mm−1
T = 120 K
0.42 × 0.33 × 0.29 mm
Data collection
Bruker SMART APEX diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2004 ▶) T min = 0.923, T max = 0.946
14278 measured reflections
3988 independent reflections
2816 reflections with I > 2σ(I)
R int = 0.066
Refinement
R[F 2 > 2σ(F 2)] = 0.041
wR(F 2) = 0.091
S = 0.92
3988 reflections
203 parameters
H-atom parameters constrained
Δρmax = 0.24 e Å−3
Δρmin = −0.34 e Å−3
Data collection: SMART (Bruker, 2002 ▶); cell refinement: SAINT (Bruker, 2002 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL and local programs.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811014577/bt5518sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811014577/bt5518Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors thank the German Research Council (DFG) and the Federal Ministry of Education and Research (BMBF) for continuous support of their work. AN thanks the university of Paderborn for granting a doctorate scholarship.
supplementary crystallographic information
Comment
The synthesis and characterization of novel molecules containing nitrogen and sulfur as donor functions and their application in synthesis of sulfur copper complexes is important for biomimetic copper–sulfur chemistry. In search of multifunctional ligands we have extended our studies to guanidyl-type systems with N-donor functions. The first derivative, the ligand bis(tetramethyl-guanidino)propylene as well as amine guanidine hybrids and their complexes with Cu, Fe, Ni, Ag, Mn, Co and Zn have recently been investigated (Harmjanz, 1997; Waden, 1999; Pohl et al., 2000; Schneider, 2000; Wittmann, 1999; Wittmann et al., 2001; Herres-Pawlis et al., 2005, 2009; Herres et al., 2005; Neuba et al., 2008a,b; 2010; Börner et al. 2007, 2009). We have now developed several sulfur guanidine hybrids based on aminothiophenol and cysteamine (Neuba et al., 2007a,b,c; Neuba, 2009). The synthesized sulfur guanidine compounds possess aliphatic and aromatic thioethers or disulfide groups and were used in the synthesis of copper thiolate complexes to mimic active centres like the CuA in cytochrome-c oxidase and N2O-reductase (Neuba et al., 2011). The two aromatic ring planes make a dihedral angle of 67.69 (6)° and the N1—C6—C11—S1—C12—C13 moiety is mostly planar with largest deviation of 0.084 (1) Å for S1 from the best plane. The guanidine plane C1N3 makes an angle of 59.80 (5)° with the attached aromatic ring. The N1═C1 guanidine double bond measures 1.304 (2) Å and is clearly localized. Similar double-bond localization is observed in other guanidine compounds (e.g. Herres et al., 2004; Neuba et al., 2007a,b; Raab et al., 2002, 2003; Peters et al., 2008).
Experimental
The title compound was prepared as follows: a solution of tetramethylchloroformamidinium chlorid (Herres-Pawlis et al., 2005) (5.13 g, 30 mmol) in dry MeCN was added dropwise to an ice-cooled solution of 2-(benzylthio)aniline (Lindoy & Livingstone, 1968) (6.45 g, 30 mmol) and triethylamine (4.18 ml, 3.03 g, 30 mmol) in dry MeCN. After 3 h under reflux, a solution of NaOH (1.2 g, 30 mmol) in water was added. The solvents and NEt3 were then evaporated under vacuum. In order to deprotonate the mono-hydrochloride, 50 wt% KOH (aqueous, 15 ml) was added and the free base was extracted into the MeCN phase (3 × 80 ml). The organic phase was dried with Na2SO4. After filtration, the solvent was evaporated under reduced pressure. The title compound was obtained as white powder (yield 69%, 6.4 g). Colourless crystals suitable for X-ray diffraction were obtained by slow cooling of a hot saturated MeCN solution.
Spectroscopic analysis: 1H NMR (500 MHz, CDCl3, 25°C, δ, p.p.m.): 2.68 (s, 12H, CH3), 4.10 (s, 2H, CH2), 6.59 (d, 1H, CH), 6.80 (t, 1H, CH), 7.03 (t, 1H, CH), 7.15 (d, 1H, CH), 7.21 (t, 1H, CH), 7.28 (t, 2H, CH), 7.36 (d, 2H, CH); 13C NMR (125 MHz, CDCl3, 25°C, δ, p.p.m.): 37.3 (CH2), 39.5 (CH3), 120.5 (CH), 121.7 (CH), 126.1 (CH), 126.9 (CH), 127.2 (CH), 127.6 (CH), 128.4 (CH), 128.7 (Cquat), 129.1 (CH), 136.6 (Cquat), 137.8 (Cquat), 160.0 (Cgua); IR (KBr, ν, cm-1): 3053 (w), 3030 (w), 3003 (w), 2918 (m), 2848 (m), 2790 (w), 1589 (vs (C═N)), 1558 (s (C═N)), 1500 (m (C═N)), 1460 (m), 1425 (m), 1377 (s), 1279 (w), 1232 (w), 1207 (w), 1144 (m), 1066 (m), 1038 (m), 1020 (s), 914 (w), 850 (w), 806 (vw), 777 (m), 715 (s), 696 (m), 682 (w), 621 (w), 571 (w), 545 (w), 498 (vw), 484 (w), 461 (w), 445 (w). EI–MS (m/z (%)): 313.0 (100) [M+], 280.0 (31), 269.1 (8) [M+ - N(CH3)2], 242.0 (5), 237.0 (10) [M+ - Ph], 222.0 (14) [M+ - CH2Ph], 215.0 (20), 190.0 (12) [M+ - SCH2Ph], 179.0 (76), 148.9 (28), 135.9 (20), 124.0 (9) [SCH2Ph+], 91.0 (76), 72.0 (20).
Refinement
H atoms were clearly identified in difference syntheses, idealized and refined riding on the C atoms with C—H = 0.95 (aromatic) or 0.98–0.99 Å, and with isotropic displacement parameters Uiso(H) = 1.2Ueq(C) or 1.5Ueq(–CH3 H atoms). All CH3 H atoms were allowed to rotate but not to tip.
Figures
Fig. 1.
Molecular structure with displacement ellipsoids drawn at the 50% probability level.
Crystal data
| C18H23N3S | F(000) = 672 |
| Mr = 313.45 | Dx = 1.239 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 842 reflections |
| a = 7.869 (2) Å | θ = 2.7–27.7° |
| b = 26.850 (7) Å | µ = 0.19 mm−1 |
| c = 8.314 (2) Å | T = 120 K |
| β = 106.959 (5)° | Block, colourless |
| V = 1680.2 (8) Å3 | 0.42 × 0.33 × 0.29 mm |
| Z = 4 |
Data collection
| Bruker SMART APEX diffractometer | 3988 independent reflections |
| Radiation source: sealed tube | 2816 reflections with I > 2σ(I) |
| graphite | Rint = 0.066 |
| φ and ω scans | θmax = 27.9°, θmin = 1.5° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2004) | h = −10→9 |
| Tmin = 0.923, Tmax = 0.946 | k = −35→35 |
| 14278 measured reflections | l = −10→10 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.041 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.091 | H-atom parameters constrained |
| S = 0.92 | w = 1/[σ2(Fo2) + (0.0443P)2] where P = (Fo2 + 2Fc2)/3 |
| 3988 reflections | (Δ/σ)max = 0.001 |
| 203 parameters | Δρmax = 0.24 e Å−3 |
| 0 restraints | Δρmin = −0.34 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.48274 (6) | 0.392868 (13) | 0.76377 (5) | 0.02350 (11) | |
| N1 | 0.64141 (18) | 0.33140 (4) | 0.56770 (16) | 0.0214 (3) | |
| N2 | 0.93294 (17) | 0.29594 (4) | 0.63993 (16) | 0.0232 (3) | |
| N3 | 0.68294 (18) | 0.24922 (4) | 0.50838 (17) | 0.0239 (3) | |
| C1 | 0.7516 (2) | 0.29479 (5) | 0.57258 (18) | 0.0203 (3) | |
| C2 | 1.0512 (2) | 0.27118 (7) | 0.5595 (2) | 0.0362 (4) | |
| H2A | 0.9806 | 0.2524 | 0.4615 | 0.054* | |
| H2B | 1.1294 | 0.2483 | 0.6395 | 0.054* | |
| H2C | 1.1231 | 0.2961 | 0.5232 | 0.054* | |
| C3 | 1.0191 (2) | 0.33296 (6) | 0.7646 (2) | 0.0291 (4) | |
| H3A | 1.0587 | 0.3609 | 0.7089 | 0.044* | |
| H3B | 1.1220 | 0.3179 | 0.8466 | 0.044* | |
| H3C | 0.9349 | 0.3450 | 0.8225 | 0.044* | |
| C4 | 0.4983 (2) | 0.24676 (6) | 0.4081 (2) | 0.0311 (4) | |
| H4A | 0.4223 | 0.2445 | 0.4825 | 0.047* | |
| H4B | 0.4800 | 0.2173 | 0.3354 | 0.047* | |
| H4C | 0.4678 | 0.2768 | 0.3385 | 0.047* | |
| C5 | 0.7491 (2) | 0.20329 (5) | 0.5999 (2) | 0.0309 (4) | |
| H5A | 0.8732 | 0.2079 | 0.6666 | 0.046* | |
| H5B | 0.7414 | 0.1760 | 0.5196 | 0.046* | |
| H5C | 0.6772 | 0.1952 | 0.6746 | 0.046* | |
| C6 | 0.6947 (2) | 0.38116 (5) | 0.56372 (19) | 0.0192 (3) | |
| C7 | 0.7988 (2) | 0.39792 (5) | 0.4653 (2) | 0.0233 (3) | |
| H7A | 0.8470 | 0.3744 | 0.4052 | 0.028* | |
| C8 | 0.8338 (2) | 0.44823 (6) | 0.4527 (2) | 0.0252 (4) | |
| H8A | 0.9042 | 0.4589 | 0.3838 | 0.030* | |
| C9 | 0.7656 (2) | 0.48271 (5) | 0.5412 (2) | 0.0236 (3) | |
| H9A | 0.7905 | 0.5171 | 0.5343 | 0.028* | |
| C10 | 0.6610 (2) | 0.46705 (5) | 0.63978 (19) | 0.0220 (3) | |
| H10A | 0.6153 | 0.4908 | 0.7009 | 0.026* | |
| C11 | 0.6223 (2) | 0.41684 (5) | 0.65010 (18) | 0.0192 (3) | |
| C12 | 0.4411 (2) | 0.44748 (5) | 0.8758 (2) | 0.0243 (4) | |
| H12A | 0.5549 | 0.4615 | 0.9463 | 0.029* | |
| H12B | 0.3797 | 0.4733 | 0.7947 | 0.029* | |
| C13 | 0.3272 (2) | 0.43232 (5) | 0.98479 (19) | 0.0210 (3) | |
| C14 | 0.4001 (2) | 0.40508 (6) | 1.1318 (2) | 0.0274 (4) | |
| H14A | 0.5226 | 0.3966 | 1.1635 | 0.033* | |
| C15 | 0.2967 (3) | 0.39029 (6) | 1.2317 (2) | 0.0317 (4) | |
| H15A | 0.3480 | 0.3719 | 1.3316 | 0.038* | |
| C16 | 0.1184 (3) | 0.40237 (6) | 1.1860 (2) | 0.0332 (4) | |
| H16A | 0.0467 | 0.3919 | 1.2539 | 0.040* | |
| C17 | 0.0442 (2) | 0.42969 (6) | 1.0417 (2) | 0.0329 (4) | |
| H17A | −0.0781 | 0.4384 | 1.0113 | 0.039* | |
| C18 | 0.1484 (2) | 0.44444 (6) | 0.9414 (2) | 0.0266 (4) | |
| H18A | 0.0967 | 0.4630 | 0.8420 | 0.032* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0297 (2) | 0.01664 (17) | 0.0296 (2) | −0.00079 (16) | 0.01715 (18) | −0.00202 (15) |
| N1 | 0.0224 (7) | 0.0167 (6) | 0.0277 (7) | −0.0022 (5) | 0.0116 (6) | −0.0044 (5) |
| N2 | 0.0206 (7) | 0.0215 (6) | 0.0263 (7) | −0.0002 (5) | 0.0047 (6) | −0.0055 (5) |
| N3 | 0.0234 (7) | 0.0164 (6) | 0.0296 (7) | −0.0001 (5) | 0.0043 (6) | −0.0045 (5) |
| C1 | 0.0237 (9) | 0.0187 (7) | 0.0199 (8) | −0.0011 (6) | 0.0089 (7) | −0.0020 (6) |
| C2 | 0.0267 (10) | 0.0419 (10) | 0.0414 (11) | 0.0044 (8) | 0.0121 (9) | −0.0069 (8) |
| C3 | 0.0288 (10) | 0.0255 (8) | 0.0281 (9) | −0.0054 (7) | 0.0008 (8) | −0.0025 (7) |
| C4 | 0.0253 (10) | 0.0251 (8) | 0.0400 (10) | −0.0031 (7) | 0.0049 (8) | −0.0076 (7) |
| C5 | 0.0350 (10) | 0.0193 (7) | 0.0367 (10) | 0.0001 (7) | 0.0076 (9) | −0.0016 (7) |
| C6 | 0.0175 (8) | 0.0181 (7) | 0.0219 (8) | −0.0009 (6) | 0.0058 (7) | −0.0018 (6) |
| C7 | 0.0221 (9) | 0.0230 (8) | 0.0277 (8) | 0.0002 (6) | 0.0119 (7) | −0.0037 (6) |
| C8 | 0.0228 (9) | 0.0273 (8) | 0.0279 (9) | −0.0026 (7) | 0.0111 (7) | 0.0035 (7) |
| C9 | 0.0242 (9) | 0.0168 (7) | 0.0289 (9) | −0.0020 (6) | 0.0061 (7) | 0.0020 (6) |
| C10 | 0.0228 (9) | 0.0173 (7) | 0.0252 (8) | 0.0013 (6) | 0.0059 (7) | −0.0012 (6) |
| C11 | 0.0182 (8) | 0.0195 (7) | 0.0209 (8) | −0.0004 (6) | 0.0071 (7) | −0.0002 (6) |
| C12 | 0.0305 (10) | 0.0174 (7) | 0.0280 (9) | 0.0022 (6) | 0.0131 (8) | −0.0036 (6) |
| C13 | 0.0258 (9) | 0.0169 (7) | 0.0218 (8) | 0.0007 (6) | 0.0092 (7) | −0.0061 (6) |
| C14 | 0.0259 (9) | 0.0296 (8) | 0.0267 (9) | 0.0056 (7) | 0.0076 (8) | −0.0013 (7) |
| C15 | 0.0425 (11) | 0.0296 (8) | 0.0243 (9) | 0.0015 (8) | 0.0117 (8) | 0.0015 (7) |
| C16 | 0.0423 (12) | 0.0292 (9) | 0.0372 (10) | −0.0078 (8) | 0.0260 (9) | −0.0093 (7) |
| C17 | 0.0224 (10) | 0.0332 (9) | 0.0451 (11) | 0.0010 (7) | 0.0129 (9) | −0.0113 (8) |
| C18 | 0.0282 (10) | 0.0235 (8) | 0.0281 (9) | 0.0028 (7) | 0.0082 (8) | −0.0025 (6) |
Geometric parameters (Å, °)
| S1—C11 | 1.7661 (15) | C6—C11 | 1.413 (2) |
| S1—C12 | 1.8175 (15) | C7—C8 | 1.389 (2) |
| N1—C1 | 1.3035 (19) | C7—H7A | 0.9500 |
| N1—C6 | 1.4033 (18) | C8—C9 | 1.384 (2) |
| N2—C1 | 1.373 (2) | C8—H8A | 0.9500 |
| N2—C3 | 1.4536 (19) | C9—C10 | 1.386 (2) |
| N2—C2 | 1.455 (2) | C9—H9A | 0.9500 |
| N3—C1 | 1.3804 (18) | C10—C11 | 1.390 (2) |
| N3—C4 | 1.451 (2) | C10—H10A | 0.9500 |
| N3—C5 | 1.4628 (19) | C12—C13 | 1.505 (2) |
| C2—H2A | 0.9800 | C12—H12A | 0.9900 |
| C2—H2B | 0.9800 | C12—H12B | 0.9900 |
| C2—H2C | 0.9800 | C13—C18 | 1.385 (2) |
| C3—H3A | 0.9800 | C13—C14 | 1.396 (2) |
| C3—H3B | 0.9800 | C14—C15 | 1.380 (2) |
| C3—H3C | 0.9800 | C14—H14A | 0.9500 |
| C4—H4A | 0.9800 | C15—C16 | 1.381 (3) |
| C4—H4B | 0.9800 | C15—H15A | 0.9500 |
| C4—H4C | 0.9800 | C16—C17 | 1.382 (2) |
| C5—H5A | 0.9800 | C16—H16A | 0.9500 |
| C5—H5B | 0.9800 | C17—C18 | 1.387 (2) |
| C5—H5C | 0.9800 | C17—H17A | 0.9500 |
| C6—C7 | 1.391 (2) | C18—H18A | 0.9500 |
| C11—S1—C12 | 102.33 (7) | C8—C7—H7A | 119.2 |
| C1—N1—C6 | 121.20 (13) | C6—C7—H7A | 119.2 |
| C1—N2—C3 | 121.30 (13) | C9—C8—C7 | 119.62 (14) |
| C1—N2—C2 | 122.01 (13) | C9—C8—H8A | 120.2 |
| C3—N2—C2 | 114.35 (14) | C7—C8—H8A | 120.2 |
| C1—N3—C4 | 118.43 (13) | C8—C9—C10 | 120.05 (14) |
| C1—N3—C5 | 120.43 (14) | C8—C9—H9A | 120.0 |
| C4—N3—C5 | 113.92 (13) | C10—C9—H9A | 120.0 |
| N1—C1—N2 | 126.77 (14) | C9—C10—C11 | 120.61 (14) |
| N1—C1—N3 | 118.33 (15) | C9—C10—H10A | 119.7 |
| N2—C1—N3 | 114.87 (13) | C11—C10—H10A | 119.7 |
| N2—C2—H2A | 109.5 | C10—C11—C6 | 119.88 (14) |
| N2—C2—H2B | 109.5 | C10—C11—S1 | 124.60 (12) |
| H2A—C2—H2B | 109.5 | C6—C11—S1 | 115.51 (11) |
| N2—C2—H2C | 109.5 | C13—C12—S1 | 108.58 (10) |
| H2A—C2—H2C | 109.5 | C13—C12—H12A | 110.0 |
| H2B—C2—H2C | 109.5 | S1—C12—H12A | 110.0 |
| N2—C3—H3A | 109.5 | C13—C12—H12B | 110.0 |
| N2—C3—H3B | 109.5 | S1—C12—H12B | 110.0 |
| H3A—C3—H3B | 109.5 | H12A—C12—H12B | 108.4 |
| N2—C3—H3C | 109.5 | C18—C13—C14 | 118.55 (15) |
| H3A—C3—H3C | 109.5 | C18—C13—C12 | 121.19 (14) |
| H3B—C3—H3C | 109.5 | C14—C13—C12 | 120.25 (15) |
| N3—C4—H4A | 109.5 | C15—C14—C13 | 120.91 (16) |
| N3—C4—H4B | 109.5 | C15—C14—H14A | 119.5 |
| H4A—C4—H4B | 109.5 | C13—C14—H14A | 119.5 |
| N3—C4—H4C | 109.5 | C14—C15—C16 | 119.81 (16) |
| H4A—C4—H4C | 109.5 | C14—C15—H15A | 120.1 |
| H4B—C4—H4C | 109.5 | C16—C15—H15A | 120.1 |
| N3—C5—H5A | 109.5 | C15—C16—C17 | 120.10 (16) |
| N3—C5—H5B | 109.5 | C15—C16—H16A | 120.0 |
| H5A—C5—H5B | 109.5 | C17—C16—H16A | 120.0 |
| N3—C5—H5C | 109.5 | C16—C17—C18 | 119.97 (17) |
| H5A—C5—H5C | 109.5 | C16—C17—H17A | 120.0 |
| H5B—C5—H5C | 109.5 | C18—C17—H17A | 120.0 |
| C7—C6—N1 | 123.59 (13) | C13—C18—C17 | 120.66 (16) |
| C7—C6—C11 | 118.26 (13) | C13—C18—H18A | 119.7 |
| N1—C6—C11 | 117.77 (13) | C17—C18—H18A | 119.7 |
| C8—C7—C6 | 121.54 (14) | ||
| C6—N1—C1—N2 | 28.2 (2) | C9—C10—C11—S1 | −177.01 (12) |
| C6—N1—C1—N3 | −153.64 (14) | C7—C6—C11—C10 | −2.2 (2) |
| C3—N2—C1—N1 | 21.5 (2) | N1—C6—C11—C10 | −175.37 (14) |
| C2—N2—C1—N1 | −140.14 (17) | C7—C6—C11—S1 | 176.95 (12) |
| C3—N2—C1—N3 | −156.70 (13) | N1—C6—C11—S1 | 3.76 (18) |
| C2—N2—C1—N3 | 41.6 (2) | C12—S1—C11—C10 | −7.87 (16) |
| C4—N3—C1—N1 | 12.7 (2) | C12—S1—C11—C6 | 173.05 (12) |
| C5—N3—C1—N1 | −135.83 (16) | C11—S1—C12—C13 | −178.03 (11) |
| C4—N3—C1—N2 | −168.95 (14) | S1—C12—C13—C18 | −105.18 (15) |
| C5—N3—C1—N2 | 42.6 (2) | S1—C12—C13—C14 | 74.12 (16) |
| C1—N1—C6—C7 | 42.6 (2) | C18—C13—C14—C15 | 0.3 (2) |
| C1—N1—C6—C11 | −144.63 (15) | C12—C13—C14—C15 | −179.05 (14) |
| N1—C6—C7—C8 | 173.60 (15) | C13—C14—C15—C16 | 0.2 (2) |
| C11—C6—C7—C8 | 0.8 (2) | C14—C15—C16—C17 | −0.8 (2) |
| C6—C7—C8—C9 | 0.7 (2) | C15—C16—C17—C18 | 0.9 (2) |
| C7—C8—C9—C10 | −0.9 (2) | C14—C13—C18—C17 | −0.2 (2) |
| C8—C9—C10—C11 | −0.5 (2) | C12—C13—C18—C17 | 179.14 (14) |
| C9—C10—C11—C6 | 2.0 (2) | C16—C17—C18—C13 | −0.4 (2) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT5518).
References
- Börner, J., Flörke, U., Huber, K., Döring, A., Kuckling, D. & Herres-Pawlis, S. (2009). Chem. Eur. J. 15, 2362–2376. [DOI] [PubMed]
- Börner, J., Herres-Pawlis, S., Flörke, U. & Huber, K. (2007). Eur. J. Inorg. Chem. pp. 5645–5651.
- Bruker (2002). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Harmjanz, F. (1997). PhD thesis, University of Oldenburg, Germany.
- Herres, S., Flörke, U. & Henkel, G. (2004). Acta Cryst. C60, o358–o360. [DOI] [PubMed]
- Herres, S., Heuwing, A. J., Flörke, U., Schneider, J. & Henkel, G. (2005). Inorg. Chim. Acta, 358, 1089–1095.
- Herres-Pawlis, S., Neuba, A., Seewald, O., Seshadri, T., Egold, H., Flörke, U. & Henkel, G. (2005). Eur. J. Org. Chem. pp. 4879–4890.
- Herres-Pawlis, S., Verma, P., Haase, R., Kang, P., Lyons, C. T., Wasinger, E. C., Flörke, U., Henkel, G. & Stack, T. D. P. (2009). J. Am. Chem. Soc. 131, 1154–1169. [DOI] [PMC free article] [PubMed]
- Lindoy, L. F. & Livingstone, S. E. (1968). Inorg. Chem. 7, 1149–1154.
- Neuba, A. (2009). PhD thesis, University of Paderborn, Germany.
- Neuba, A., Flörke, U. & Henkel, G. (2007a). Acta Cryst. E63, o3476–o3477.
- Neuba, A., Flörke, U. & Henkel, G. (2007b). Acta Cryst. E63, o4661.
- Neuba, A., Flörke, U. & Henkel, G. (2007c). Acta Cryst. E63, o4683.
- Neuba, A., Flörke, U., Meyer-Klaucke, W., Salomone-Stagni, M., Bill, E., Bothe, E., Höfer, P. & Henkel, G. (2011). Angew. Chem. In the press. [DOI] [PubMed]
- Neuba, A., Haase, R., Bernard, M., Flörke, U. & Herres-Pawlis, S. (2008b). Z. Anorg. Allg. Chem. 634, 2511–2517.
- Neuba, A., Herres-Pawlis, S., Flörke, U. & Henkel, G. (2008a). Z. Anorg. Allg. Chem. 634, 771–777.
- Neuba, A., Herres-Pawlis, S., Seewald, O., Börner, J., Heuwing, J., Flörke, U. & Henkel, G. (2010). Z. Anorg. Allg. Chem. 636, 2641–2649.
- Peters, A., Wild, U., Hubner, O., Kaifer, E. & Himmel, H.-J. (2008). Chem. Eur. J. 14, 7813–7821. [DOI] [PubMed]
- Pohl, S., Harmjanz, M., Schneider, J., Saak, W. & Henkel, G. (2000). J. Chem. Soc. Dalton Trans. pp. 3473–3479.
- Raab, V., Harms, K., Sundermeyer, J., Kovacevic, B. & Maksic, Z. B. (2003). J. Org. Chem. 68, 8790–8797. [DOI] [PubMed]
- Raab, V., Kipke, J., Gschwind, R. M. & Sundermeyer, J. (2002). Chem. Eur. J. 8, 1682–1693. [DOI] [PubMed]
- Schneider, J. (2000). PhD thesis, University of Duisburg, Germany.
- Sheldrick, G. M. (2004). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Waden, H. (1999). PhD thesis, University of Oldenburg, Germany.
- Wittmann, H. (1999). PhD thesis, University of Marburg, Germany.
- Wittmann, H., Raab, V., Schorm, A., Plackmeyer, J. & Sundermeyer, J. (2001). Eur. J. Inorg. Chem. pp. 1937–1948.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811014577/bt5518sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811014577/bt5518Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



