Abstract
The structure of the title thiourea derivative, C14H16N4O5S, features an almost planar central C2N2OS fragment (r.m.s. deviation = 0.005 Å), an arrangement stabilized by an intramolecular N—H⋯O hydrogen bond. The terminal rings are twisted out of this plane, the dihedral angle formed with the benzene ring being 33.22 (10)°. The cyclohexyl ring is disordered, with two orientations (50:50) being resolved. The mean plane passing through the atoms of each disordered component forms dihedral angles of 65.7 (2) and 82.4 (3)° with the central plane. Centrosymmetric dimers mediated by an eight-membered {⋯HNC=S}2 synthon occur in the crystal.
Related literature
For the biological activity of thiourea derivatives, see: Venkatachalam et al. (2004 ▶); Saeed et al. (2011 ▶). For related thiourea structures, see: Gunasekaran et al. (2010 ▶); Saeed et al. (2010 ▶); Dzulkifli et al. (2011 ▶).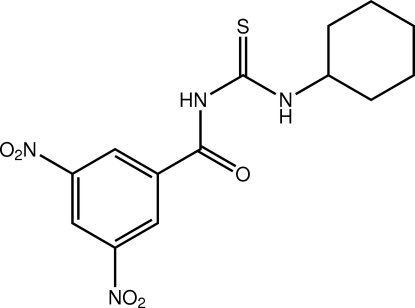
Experimental
Crystal data
C14H16N4O5S
M r = 352.37
Monoclinic,

a = 12.3404 (7) Å
b = 9.0506 (5) Å
c = 14.6534 (6) Å
β = 90.385 (5)°
V = 1636.57 (15) Å3
Z = 4
Mo Kα radiation
μ = 0.23 mm−1
T = 295 K
0.20 × 0.15 × 0.10 mm
Data collection
Agilent Technologies SuperNova Dual diffractometer with an Atlas detector
Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2010 ▶) T min = 0.955, T max = 0.977
7954 measured reflections
3649 independent reflections
1948 reflections with I > 2σ(I)
R int = 0.024
Refinement
R[F 2 > 2σ(F 2)] = 0.061
wR(F 2) = 0.211
S = 1.01
3649 reflections
271 parameters
25 restraints
H-atom parameters constrained
Δρmax = 0.22 e Å−3
Δρmin = −0.28 e Å−3
Data collection: CrysAlis PRO (Agilent, 2010 ▶); cell refinement: CrysAlis PRO; data reduction: CrysAlis PRO; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶) and DIAMOND (Brandenburg, 2006 ▶); software used to prepare material for publication: publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811013377/hb5839sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811013377/hb5839Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O1 | 0.88 | 1.89 | 2.639 (4) | 142 |
| N1—H1′⋯O1 | 0.88 | 1.99 | 2.639 (4) | 130 |
| N2—H2⋯S1i | 0.88 | 2.65 | 3.449 (3) | 152 |
Symmetry code: (i)  .
.
Acknowledgments
The authors are grateful to Allama Iqbal Open University, Islamabad, Pakistan, for the allocation of research and analytical laboratory facilities. The authors also thank the University of Malaya for supporting this study.
supplementary crystallographic information
Comment
Continuing structural studies (Gunasekaran et al. 2010; Saeed et al. 2010; Dzulkifli et al., 2011) of thiourea derivatives are motivated by their biological potential (Venkatachalam et al., 2004; Saeed et al., 2011) and led to the investigation of the title compound, (I).
The molecular structure of (I), Fig. 1, is highly twisted with dihedral angles formed between the central chromophore (r.m.s. = 0.0054 Å for C7,C8,N1,N2,O1 & S1) and the benzene ring being 33.22 (10) °. Two orientations of equal weight were found for the cyclohexyl ring, each with a chair conformation, and these make angles of 65.74 (24) and 82.42 (30) °, respectively, with the central plane. The N—H atoms are anti as are the S and O atoms. As a consequence, the N1—H atom forms an intramolecular hydrogen bond with the carbonyl-O1 atom to close a pseudo six-membered ring, Table 1; there are two values cited owing to the disorder in the molecule. The nitro groups are effectively co-planar with the benzene ring to which they are bonded as seen in the values of the O2—N3—C11—C10 and O4—N4—C13—C12 torsion angles of 1.2 (5) and -7.0 (5) °, respectively.
The most prominent feature of the crystal packing is the formation of centrosymmetric eight-membered {···HNC═S}2 synthon leading to dimeric aggregates, Fig. 2 and Table 1.
Experimental
A solution of 3,5-dinitrobenzoyl chloride (0.01 mol) in anhydrous acetone (75 ml) and 3% tetrabutylammonium bromide (TBAB), as a phase-transfer catalyst (PTC), in anhydrous acetone was added drop-wise to a suspension of dry potassium thiocyanate (0.01 mol) in acetone (50 ml). The reaction mixture was refluxed for 50 min. After cooling to room temperature, a solution of cyclohexylamine (0.01 mol) in anhydrous acetone (25 ml) was added drop-wise and the resulting mixture refluxed for 3 h. Hydrochloric acid (0.1 N, 300 ml) was added and the solution was filtered. The solid product was washed with water and purified by re-crystallization from ethanol; Yield: 1.50 g (88%) and M.pt. 409 K. IR (KBr, cm-1): 3215 ν(NH), 1673 (C=O), 1527 (benzene ring), 1138 ν(C═S). Anal. Calcd. for C14H16N4O5S: C, 47.72; H, 4.58; N, 15.90; S, 9.10%. Found: C, 47.51; H, 4.75; N, 15.88; S, 9.11%.
Refinement
Carbon-bound H-atoms were placed in calculated positions [C—H 0.93 to 0.97 Å, Uiso(H) 1.2Ueq(C)] and were included in the refinement in the riding model approximation. The two amino H-atoms were similarly placed [N–H 0.88 Å, Uiso(H) 1.2Ueq(N)]. The cyclohexyl ring is disordered over two positions; the disorder could not be refined, and was assumed to be a 1:1 type of disorder. The 1,2-related C–C distances were restrained to 1.54±0.01 Å and the 1,3-related ones to 2.51±0.01 Å. The pair of N–Ccyclohexyl and N–C'cyclohexyl distances were restrained to within 0.01 Å of each other.
Figures
Fig. 1.
The molecular structure of (I) showing displacement ellipsoids at the 35% probability level. Only one orientation of the disordered cyclohexyl ring is shown.
Fig. 2.
Supramolecular dimer in (I) mediated by N—H···S hydrogen bonding shown as orange dashed lines. Only one orientation of the disordered cyclohexyl ring is shown.
Crystal data
| C14H16N4O5S | F(000) = 736 |
| Mr = 352.37 | Dx = 1.430 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 2522 reflections |
| a = 12.3404 (7) Å | θ = 2.6–29.2° |
| b = 9.0506 (5) Å | µ = 0.23 mm−1 |
| c = 14.6534 (6) Å | T = 295 K |
| β = 90.385 (5)° | Prism, colorless |
| V = 1636.57 (15) Å3 | 0.20 × 0.15 × 0.10 mm |
| Z = 4 |
Data collection
| Agilent Technologies SuperNova Dual diffractometer with an Atlas detector | 3649 independent reflections |
| Radiation source: SuperNova (Mo) X-ray Source | 1948 reflections with I > 2σ(I) |
| Mirror | Rint = 0.024 |
| Detector resolution: 10.4041 pixels mm-1 | θmax = 27.5°, θmin = 2.7° |
| ω scans | h = −11→16 |
| Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2010) | k = −11→9 |
| Tmin = 0.955, Tmax = 0.977 | l = −19→18 |
| 7954 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.061 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.211 | H-atom parameters constrained |
| S = 1.01 | w = 1/[σ2(Fo2) + (0.0901P)2 + 0.5204P] where P = (Fo2 + 2Fc2)/3 |
| 3649 reflections | (Δ/σ)max = 0.001 |
| 271 parameters | Δρmax = 0.22 e Å−3 |
| 25 restraints | Δρmin = −0.28 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| S1 | 0.41365 (9) | 0.64167 (12) | 0.59283 (6) | 0.0914 (4) | |
| O1 | 0.2058 (2) | 0.5972 (4) | 0.33847 (18) | 0.1069 (9) | |
| O2 | 0.1940 (3) | 0.2554 (4) | 0.0798 (2) | 0.1410 (13) | |
| O3 | 0.3315 (3) | 0.2306 (4) | −0.0052 (3) | 0.1351 (13) | |
| O4 | 0.6723 (3) | 0.4548 (4) | 0.0872 (2) | 0.1237 (11) | |
| O5 | 0.6733 (2) | 0.6067 (4) | 0.2004 (2) | 0.1140 (10) | |
| N1 | 0.2226 (2) | 0.6699 (3) | 0.5124 (2) | 0.0864 (9) | |
| H1 | 0.1881 | 0.6629 | 0.4598 | 0.104* | 0.50 |
| H1' | 0.1777 | 0.6614 | 0.4657 | 0.104* | 0.50 |
| N2 | 0.3606 (2) | 0.5812 (3) | 0.42290 (16) | 0.0689 (7) | |
| H2 | 0.4294 | 0.5564 | 0.4197 | 0.083* | |
| N3 | 0.2883 (4) | 0.2769 (4) | 0.0621 (3) | 0.0964 (10) | |
| N4 | 0.6288 (3) | 0.5175 (4) | 0.1510 (2) | 0.0907 (9) | |
| C1 | 0.1543 (7) | 0.7215 (9) | 0.5861 (6) | 0.074 (3) | 0.50 |
| H1A | 0.1990 | 0.7735 | 0.6313 | 0.088* | 0.50 |
| C2 | 0.0991 (8) | 0.5900 (9) | 0.6317 (6) | 0.084 (3) | 0.50 |
| H2A | 0.1529 | 0.5203 | 0.6537 | 0.101* | 0.50 |
| H2B | 0.0520 | 0.5398 | 0.5885 | 0.101* | 0.50 |
| C3 | 0.0323 (6) | 0.6513 (8) | 0.7122 (4) | 0.094 (2) | 0.50 |
| H3A | −0.0055 | 0.5709 | 0.7420 | 0.113* | 0.50 |
| H3B | 0.0807 | 0.6964 | 0.7567 | 0.113* | 0.50 |
| C4 | −0.0488 (6) | 0.7647 (9) | 0.6791 (5) | 0.106 (3) | 0.50 |
| H4A | −0.1009 | 0.7171 | 0.6389 | 0.127* | 0.50 |
| H4B | −0.0879 | 0.8040 | 0.7309 | 0.127* | 0.50 |
| C5 | 0.0049 (6) | 0.8899 (8) | 0.6289 (5) | 0.098 (3) | 0.50 |
| H5A | 0.0532 | 0.9427 | 0.6700 | 0.117* | 0.50 |
| H5B | −0.0496 | 0.9586 | 0.6069 | 0.117* | 0.50 |
| C6 | 0.0701 (8) | 0.8278 (11) | 0.5471 (5) | 0.091 (3) | 0.50 |
| H6A | 0.0222 | 0.7764 | 0.5051 | 0.109* | 0.50 |
| H6B | 0.1054 | 0.9075 | 0.5146 | 0.109* | 0.50 |
| C1' | 0.1883 (7) | 0.7287 (12) | 0.6020 (6) | 0.121 (6) | 0.50 |
| H1B | 0.2530 | 0.7692 | 0.6321 | 0.145* | 0.50 |
| C2' | 0.1393 (7) | 0.6133 (12) | 0.6672 (7) | 0.101 (4) | 0.50 |
| H2C | 0.1891 | 0.5312 | 0.6747 | 0.121* | 0.50 |
| H2D | 0.1277 | 0.6576 | 0.7267 | 0.121* | 0.50 |
| C3' | 0.0326 (8) | 0.5580 (10) | 0.6290 (9) | 0.149 (6) | 0.50 |
| H3C | 0.0025 | 0.4846 | 0.6699 | 0.179* | 0.50 |
| H3D | 0.0448 | 0.5112 | 0.5704 | 0.179* | 0.50 |
| C4' | −0.0484 (6) | 0.6857 (12) | 0.6172 (9) | 0.146 (5) | 0.50 |
| H4C | −0.0635 | 0.7300 | 0.6760 | 0.175* | 0.50 |
| H4D | −0.1159 | 0.6487 | 0.5917 | 0.175* | 0.50 |
| C5' | 0.0006 (8) | 0.8018 (12) | 0.5529 (10) | 0.150 (5) | 0.50 |
| H5C | 0.0107 | 0.7586 | 0.4930 | 0.180* | 0.50 |
| H5D | −0.0492 | 0.8842 | 0.5467 | 0.180* | 0.50 |
| C6' | 0.1077 (7) | 0.8568 (10) | 0.5890 (8) | 0.106 (3) | 0.50 |
| H6C | 0.0968 | 0.9063 | 0.6469 | 0.127* | 0.50 |
| H6D | 0.1376 | 0.9281 | 0.5466 | 0.127* | 0.50 |
| C7 | 0.3244 (3) | 0.6318 (3) | 0.5079 (2) | 0.0695 (8) | |
| C8 | 0.3013 (3) | 0.5664 (4) | 0.3450 (2) | 0.0755 (9) | |
| C9 | 0.3604 (3) | 0.5071 (4) | 0.2637 (2) | 0.0701 (8) | |
| C10 | 0.3015 (3) | 0.4227 (4) | 0.2026 (2) | 0.0757 (9) | |
| H10 | 0.2288 | 0.4025 | 0.2131 | 0.091* | |
| C11 | 0.3514 (3) | 0.3689 (3) | 0.1262 (2) | 0.0753 (9) | |
| C12 | 0.4585 (3) | 0.3965 (3) | 0.1073 (2) | 0.0751 (9) | |
| H12 | 0.4916 | 0.3582 | 0.0556 | 0.090* | |
| C13 | 0.5142 (3) | 0.4833 (3) | 0.1684 (2) | 0.0700 (8) | |
| C14 | 0.4681 (3) | 0.5400 (3) | 0.2462 (2) | 0.0691 (8) | |
| H14 | 0.5082 | 0.5988 | 0.2860 | 0.083* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0966 (7) | 0.1080 (8) | 0.0695 (5) | 0.0228 (6) | 0.0037 (5) | −0.0177 (5) |
| O1 | 0.0737 (17) | 0.145 (3) | 0.1016 (18) | 0.0229 (17) | −0.0108 (14) | −0.0007 (17) |
| O2 | 0.127 (3) | 0.161 (3) | 0.135 (3) | −0.052 (3) | −0.019 (2) | −0.027 (2) |
| O3 | 0.144 (3) | 0.126 (3) | 0.136 (3) | −0.003 (2) | −0.014 (2) | −0.066 (2) |
| O4 | 0.117 (2) | 0.121 (2) | 0.134 (2) | −0.0209 (19) | 0.045 (2) | −0.043 (2) |
| O5 | 0.108 (2) | 0.131 (2) | 0.1037 (19) | −0.0431 (19) | 0.0195 (17) | −0.0334 (18) |
| N1 | 0.0751 (18) | 0.095 (2) | 0.0897 (19) | 0.0212 (16) | 0.0215 (15) | 0.0104 (16) |
| N2 | 0.0680 (15) | 0.0767 (16) | 0.0620 (14) | 0.0098 (13) | 0.0057 (12) | 0.0030 (12) |
| N3 | 0.111 (3) | 0.080 (2) | 0.098 (2) | −0.012 (2) | −0.019 (2) | −0.0087 (18) |
| N4 | 0.102 (2) | 0.086 (2) | 0.0843 (19) | −0.0157 (19) | 0.0192 (18) | −0.0099 (17) |
| C1 | 0.058 (5) | 0.071 (6) | 0.093 (5) | 0.015 (4) | 0.026 (4) | 0.001 (4) |
| C2 | 0.097 (8) | 0.077 (5) | 0.078 (6) | 0.007 (6) | 0.011 (6) | −0.005 (5) |
| C3 | 0.115 (6) | 0.100 (5) | 0.069 (4) | −0.024 (5) | 0.029 (4) | −0.017 (4) |
| C4 | 0.085 (5) | 0.131 (8) | 0.103 (6) | −0.007 (5) | 0.020 (5) | −0.071 (6) |
| C5 | 0.085 (5) | 0.095 (6) | 0.114 (6) | 0.018 (4) | 0.022 (5) | −0.037 (5) |
| C6 | 0.070 (6) | 0.099 (7) | 0.103 (7) | 0.023 (6) | −0.007 (5) | −0.006 (5) |
| C1' | 0.088 (8) | 0.119 (11) | 0.157 (11) | 0.041 (7) | 0.051 (7) | 0.032 (8) |
| C2' | 0.087 (7) | 0.128 (8) | 0.087 (7) | −0.004 (6) | 0.009 (5) | 0.007 (6) |
| C3' | 0.106 (8) | 0.160 (12) | 0.180 (12) | −0.038 (8) | −0.043 (8) | 0.069 (10) |
| C4' | 0.071 (5) | 0.162 (10) | 0.203 (13) | −0.001 (7) | −0.014 (7) | 0.077 (10) |
| C5' | 0.099 (8) | 0.124 (9) | 0.227 (15) | −0.010 (8) | −0.057 (9) | 0.049 (10) |
| C6' | 0.090 (7) | 0.092 (7) | 0.136 (9) | 0.007 (5) | −0.004 (6) | 0.000 (6) |
| C7 | 0.076 (2) | 0.0639 (18) | 0.0689 (18) | 0.0095 (16) | 0.0127 (16) | 0.0061 (14) |
| C8 | 0.077 (2) | 0.076 (2) | 0.073 (2) | 0.0066 (18) | −0.0027 (17) | 0.0090 (16) |
| C9 | 0.083 (2) | 0.0665 (18) | 0.0607 (16) | 0.0028 (17) | −0.0076 (15) | 0.0113 (15) |
| C10 | 0.078 (2) | 0.0711 (19) | 0.077 (2) | −0.0019 (17) | −0.0112 (17) | 0.0110 (17) |
| C11 | 0.096 (3) | 0.0574 (18) | 0.0726 (19) | −0.0027 (18) | −0.0183 (18) | 0.0045 (15) |
| C12 | 0.102 (3) | 0.0593 (18) | 0.0641 (18) | 0.0011 (18) | −0.0014 (18) | 0.0019 (15) |
| C13 | 0.083 (2) | 0.0607 (17) | 0.0665 (18) | −0.0054 (16) | −0.0008 (16) | 0.0053 (15) |
| C14 | 0.084 (2) | 0.0631 (18) | 0.0598 (16) | −0.0052 (16) | −0.0035 (16) | 0.0051 (14) |
Geometric parameters (Å, °)
| S1—C7 | 1.659 (4) | C5—H5B | 0.9700 |
| O1—C8 | 1.215 (4) | C6—H6A | 0.9700 |
| O2—N3 | 1.210 (5) | C6—H6B | 0.9700 |
| O3—N3 | 1.200 (4) | C1'—C6' | 1.539 (8) |
| O4—N4 | 1.221 (4) | C1'—C2' | 1.542 (8) |
| O5—N4 | 1.213 (4) | C1'—H1B | 0.9800 |
| N1—C7 | 1.305 (4) | C2'—C3' | 1.512 (8) |
| N1—C1 | 1.452 (6) | C2'—H2C | 0.9700 |
| N1—C1' | 1.481 (8) | C2'—H2D | 0.9700 |
| N1—H1 | 0.8800 | C3'—C4' | 1.537 (9) |
| N1—H1' | 0.8800 | C3'—H3C | 0.9700 |
| N2—C8 | 1.358 (4) | C3'—H3D | 0.9700 |
| N2—C7 | 1.402 (4) | C4'—C5' | 1.537 (8) |
| N2—H2 | 0.8800 | C4'—H4C | 0.9700 |
| N3—C11 | 1.474 (5) | C4'—H4D | 0.9700 |
| N4—C13 | 1.471 (5) | C5'—C6' | 1.505 (8) |
| C1—C6 | 1.524 (8) | C5'—H5C | 0.9700 |
| C1—C2 | 1.527 (8) | C5'—H5D | 0.9700 |
| C1—H1A | 0.9800 | C6'—H6C | 0.9700 |
| C2—C3 | 1.547 (7) | C6'—H6D | 0.9700 |
| C2—H2A | 0.9700 | C8—C9 | 1.501 (5) |
| C2—H2B | 0.9700 | C9—C10 | 1.380 (5) |
| C3—C4 | 1.511 (8) | C9—C14 | 1.388 (4) |
| C3—H3A | 0.9700 | C10—C11 | 1.370 (5) |
| C3—H3B | 0.9700 | C10—H10 | 0.9300 |
| C4—C5 | 1.507 (7) | C11—C12 | 1.375 (5) |
| C4—H4A | 0.9700 | C12—C13 | 1.372 (5) |
| C4—H4B | 0.9700 | C12—H12 | 0.9300 |
| C5—C6 | 1.552 (8) | C13—C14 | 1.376 (4) |
| C5—H5A | 0.9700 | C14—H14 | 0.9300 |
| C7—N1—C1 | 133.3 (5) | C2'—C1'—H1B | 107.3 |
| C7—N1—C1' | 114.9 (5) | C3'—C2'—C1' | 109.8 (7) |
| C7—N1—H1 | 113.3 | C3'—C2'—H2C | 109.7 |
| C1—N1—H1 | 113.3 | C1'—C2'—H2C | 109.7 |
| C7—N1—H1' | 122.6 | C3'—C2'—H2D | 109.7 |
| C1'—N1—H1' | 122.6 | C1'—C2'—H2D | 109.7 |
| C8—N2—C7 | 127.2 (3) | H2C—C2'—H2D | 108.2 |
| C8—N2—H2 | 116.4 | C2'—C3'—C4' | 110.9 (7) |
| C7—N2—H2 | 116.4 | C2'—C3'—H3C | 109.5 |
| O3—N3—O2 | 123.5 (4) | C4'—C3'—H3C | 109.5 |
| O3—N3—C11 | 119.1 (4) | C2'—C3'—H3D | 109.5 |
| O2—N3—C11 | 117.4 (4) | C4'—C3'—H3D | 109.5 |
| O5—N4—O4 | 124.5 (3) | H3C—C3'—H3D | 108.1 |
| O5—N4—C13 | 117.9 (3) | C3'—C4'—C5' | 109.0 (7) |
| O4—N4—C13 | 117.6 (3) | C3'—C4'—H4C | 109.9 |
| N1—C1—C6 | 108.7 (6) | C5'—C4'—H4C | 109.9 |
| N1—C1—C2 | 109.8 (6) | C3'—C4'—H4D | 109.9 |
| C6—C1—C2 | 110.6 (7) | C5'—C4'—H4D | 109.9 |
| N1—C1—H1A | 109.3 | H4C—C4'—H4D | 108.3 |
| C6—C1—H1A | 109.3 | C6'—C5'—C4' | 111.1 (7) |
| C2—C1—H1A | 109.3 | C6'—C5'—H5C | 109.4 |
| C1—C2—C3 | 107.2 (5) | C4'—C5'—H5C | 109.4 |
| C1—C2—H2A | 110.3 | C6'—C5'—H5D | 109.4 |
| C3—C2—H2A | 110.3 | C4'—C5'—H5D | 109.4 |
| C1—C2—H2B | 110.3 | H5C—C5'—H5D | 108.0 |
| C3—C2—H2B | 110.3 | C5'—C6'—C1' | 111.1 (7) |
| H2A—C2—H2B | 108.5 | C5'—C6'—H6C | 109.4 |
| C4—C3—C2 | 110.7 (5) | C1'—C6'—H6C | 109.4 |
| C4—C3—H3A | 109.5 | C5'—C6'—H6D | 109.4 |
| C2—C3—H3A | 109.5 | C1'—C6'—H6D | 109.4 |
| C4—C3—H3B | 109.5 | H6C—C6'—H6D | 108.0 |
| C2—C3—H3B | 109.5 | N1—C7—N2 | 116.4 (3) |
| H3A—C3—H3B | 108.1 | N1—C7—S1 | 125.6 (3) |
| C5—C4—C3 | 112.0 (6) | N2—C7—S1 | 118.0 (2) |
| C5—C4—H4A | 109.2 | O1—C8—N2 | 124.1 (3) |
| C3—C4—H4A | 109.2 | O1—C8—C9 | 119.7 (3) |
| C5—C4—H4B | 109.2 | N2—C8—C9 | 116.2 (3) |
| C3—C4—H4B | 109.2 | C10—C9—C14 | 120.0 (3) |
| H4A—C4—H4B | 107.9 | C10—C9—C8 | 117.2 (3) |
| C4—C5—C6 | 109.7 (6) | C14—C9—C8 | 122.8 (3) |
| C4—C5—H5A | 109.7 | C11—C10—C9 | 119.2 (3) |
| C6—C5—H5A | 109.7 | C11—C10—H10 | 120.4 |
| C4—C5—H5B | 109.7 | C9—C10—H10 | 120.4 |
| C6—C5—H5B | 109.7 | C10—C11—C12 | 122.6 (3) |
| H5A—C5—H5B | 108.2 | C10—C11—N3 | 118.8 (4) |
| C1—C6—C5 | 107.1 (5) | C12—C11—N3 | 118.6 (3) |
| C1—C6—H6A | 110.3 | C13—C12—C11 | 116.7 (3) |
| C5—C6—H6A | 110.3 | C13—C12—H12 | 121.6 |
| C1—C6—H6B | 110.3 | C11—C12—H12 | 121.6 |
| C5—C6—H6B | 110.3 | C12—C13—C14 | 123.1 (3) |
| H6A—C6—H6B | 108.5 | C12—C13—N4 | 119.0 (3) |
| N1—C1'—C6' | 110.4 (8) | C14—C13—N4 | 117.9 (3) |
| N1—C1'—C2' | 115.0 (8) | C13—C14—C9 | 118.3 (3) |
| C6'—C1'—C2' | 109.4 (6) | C13—C14—H14 | 120.8 |
| N1—C1'—H1B | 107.3 | C9—C14—H14 | 120.8 |
| C6'—C1'—H1B | 107.3 | ||
| C7—N1—C1—C6 | 147.6 (6) | C8—N2—C7—N1 | −0.6 (5) |
| C1'—N1—C1—C6 | 134 (2) | C8—N2—C7—S1 | −179.3 (3) |
| C7—N1—C1—C2 | −91.4 (9) | C7—N2—C8—O1 | 0.6 (6) |
| C1'—N1—C1—C2 | −105 (2) | C7—N2—C8—C9 | −179.1 (3) |
| N1—C1—C2—C3 | 177.3 (7) | O1—C8—C9—C10 | −31.7 (5) |
| C6—C1—C2—C3 | −62.8 (10) | N2—C8—C9—C10 | 148.0 (3) |
| C1—C2—C3—C4 | 57.7 (10) | O1—C8—C9—C14 | 145.0 (4) |
| C2—C3—C4—C5 | −56.9 (9) | N2—C8—C9—C14 | −35.3 (4) |
| C3—C4—C5—C6 | 57.5 (9) | C14—C9—C10—C11 | 1.7 (5) |
| N1—C1—C6—C5 | −175.5 (8) | C8—C9—C10—C11 | 178.6 (3) |
| C2—C1—C6—C5 | 63.9 (10) | C9—C10—C11—C12 | −0.5 (5) |
| C4—C5—C6—C1 | −59.6 (10) | C9—C10—C11—N3 | 179.1 (3) |
| C7—N1—C1'—C6' | 141.7 (5) | O3—N3—C11—C10 | 179.4 (4) |
| C1—N1—C1'—C6' | −49.4 (18) | O2—N3—C11—C10 | 1.2 (5) |
| C7—N1—C1'—C2' | −94.0 (7) | O3—N3—C11—C12 | −1.1 (5) |
| C1—N1—C1'—C2' | 74.9 (19) | O2—N3—C11—C12 | −179.3 (4) |
| N1—C1'—C2'—C3' | −66.7 (10) | C10—C11—C12—C13 | −0.8 (5) |
| C6'—C1'—C2'—C3' | 58.1 (11) | N3—C11—C12—C13 | 179.6 (3) |
| C1'—C2'—C3'—C4' | −59.8 (12) | C11—C12—C13—C14 | 0.9 (5) |
| C2'—C3'—C4'—C5' | 58.6 (13) | C11—C12—C13—N4 | −179.4 (3) |
| C3'—C4'—C5'—C6' | −57.3 (14) | O5—N4—C13—C12 | 172.3 (3) |
| C4'—C5'—C6'—C1' | 57.8 (13) | O4—N4—C13—C12 | −7.0 (5) |
| N1—C1'—C6'—C5' | 69.9 (10) | O5—N4—C13—C14 | −8.0 (5) |
| C2'—C1'—C6'—C5' | −57.5 (11) | O4—N4—C13—C14 | 172.6 (3) |
| C1—N1—C7—N2 | 177.5 (5) | C12—C13—C14—C9 | 0.3 (5) |
| C1'—N1—C7—N2 | −177.5 (5) | N4—C13—C14—C9 | −179.4 (3) |
| C1—N1—C7—S1 | −3.8 (7) | C10—C9—C14—C13 | −1.6 (5) |
| C1'—N1—C7—S1 | 1.1 (6) | C8—C9—C14—C13 | −178.3 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1 | 0.88 | 1.89 | 2.639 (4) | 142 |
| N1—H1'···O1 | 0.88 | 1.99 | 2.639 (4) | 130 |
| N2—H2···S1i | 0.88 | 2.65 | 3.449 (3) | 152 |
Symmetry codes: (i) −x+1, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB5839).
References
- Agilent (2010). CrysAlis PRO Agilent Technologies, Yarnton, England.
- Brandenburg, K. (2006). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Dzulkifli, N. N., Farina, Y., Yamin, B. M., Baba, I. & Tiekink, E. R. T. (2011). Acta Cryst. E67, o872. [DOI] [PMC free article] [PubMed]
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Gunasekaran, N., Karvembu, R., Ng, S. W. & Tiekink, E. R. T. (2010). Acta Cryst. E66, o2601. [DOI] [PMC free article] [PubMed]
- Saeed, S., Rashid, N., Jones, P. G. & Tahir, A. (2011). J. Heterocycl. Chem. 48, 74–84.
- Saeed, S., Rashid, N. & Wong, W.-T. (2010). Acta Cryst. E66, o980. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Venkatachalam, T. K., Mao, C. & Uckun, F. M. (2004). Bioorg. Med. Chem. 12, 4275–4284. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811013377/hb5839sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811013377/hb5839Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




