Abstract
The fibronectin-binding repeats of the SfbI protein of Streptococcus pyogenes constitute the minimal domain able to confer protection against lethal infection. We investigated the presence of B- and T-cell epitopes within this region in congenic mice. One linear B-cell epitope was recognized by BALB/b and BALB/k mice, whereas two epitopes were found in BALB/c animals. A unique T-cell epitope was recognized by all three mouse strains. All identified epitopes clustered in a 30-amino-acid fragment. These results suggest that this polypeptide may be suitable for incorporation into a polyepitope-based vaccine formulation against S. pyogenes.
The human pathogen Streptococcus pyogenes can cause localized infections resulting in noninvasive diseases like pharyngitis, impetigo, or scarlet fever. These infections can lead to severe poststreptococcal diseases, such as rheumatic fever and acute poststreptococcal glomerulonephritis. In addition, S. pyogenes can cause highly invasive diseases, such as sepsis, necrotizing fasciitis, and toxic shock-like syndrome (3, 25, 32). The incidence of S. pyogenes infections and their sequelae, as well as the emergence of strains that are resistant to macrolides (e.g., erythromycin), has been steadily increasing (7, 11, 13, 18). In the last decade, several approaches have been pursued to develop a vaccine against S. pyogenes. The M protein, a major virulence factor of S. pyogenes, constitutes the best-characterized vaccine candidate (2, 4, 17). However, M protein mainly triggers serotype-specific immunity and may lead to cross-reaction with host tissues. In the last few years the C5a peptidase, a surface-bound peptidase that cleaves C5a (10), and the extracellular cysteine protease, which cleaves fibronectin and converts interleukin-1β precursor to biologically active interleukin-1β (12), have been proposed as candidate antigens. The use of peptides encompassing conserved B- and T-cell epitopes of the M protein constitutes a new approach to prevent diseases caused by different M serotypes and the potential side effects resulting from immune cross-recognition (1, 4, 5, 6, 20). However, peptides are usually poorly immunogenic and need to be administered with depot-forming adjuvants (19, 30) or bacterial toxins or their derivatives (2, 24). They can also be generated using approaches in which multiple copies of the epitopes are displayed (17, 31).
It has been shown that intranasal vaccination with the fibronectin-binding protein I (SfbI) of S. pyogenes stimulates humoral and cellular immune responses, which are able to protect against lethal challenge with S. pyogenes (9, 22). The SfbI protein plays a central role in the virulence process; is highly conserved, surface located, and expressed by a large number of clinical isolates from different serotypes (approximately 73%); and does not lead to cross-reactivity with host tissues (16, 26-29). Additional studies have identified the domain encompassing the fibronectin-binding repeats (148 amino acids) as the minimal fragment able to confer protection (Fig. 1). In an attempt to further characterize the vaccine-relevant immune determinants of the SfbI protein, we mapped the linear B-cell and T-cell epitopes located within the fibronectin-binding repeats.
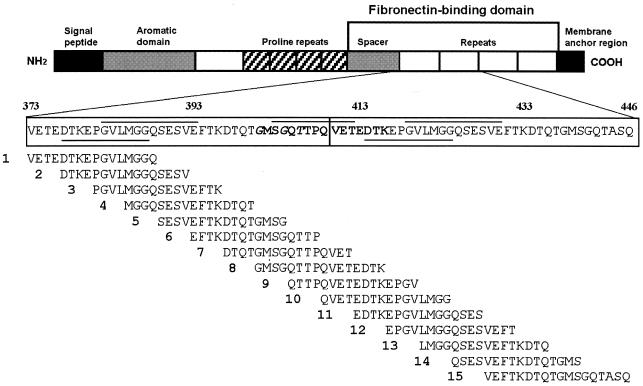
FIG. 1.
Peptides spanning two fibronectin-binding repeats used for epitope-mapping studies. Linear B-cell epitopes recognized in BALB/b and BALB/k mice are underlined, whereas those detected in BALB/c mice are indicated by an overline. The T-cell epitope found in all tested mice is indicated in boldface, with the predicted anchor residues in italics.
Identification of linear B-cell epitopes within the fibronectin-binding repeats of the SfbI protein.
A vaccine should be able to stimulate efficient immune responses in individuals, who are genetically different. Thus, initial studies were performed to investigate whether efficient humoral immune responses against the fibronectin-binding domain were stimulated in congenic BALB/c, BALB/b, and BALB/k mice, after intranasal immunization (11 μg on days 1, 14, and 21) with a polypeptide encompassing the fibronectin-binding domain. The obtained results demonstrated that similar immunoglobulin G (IgG) responses were obtained in all mouse strains, which were characterized by a dominant IgG1 isotype response pattern (data not shown). Thus, subsequent work was carried out to characterize the linear B-cell epitopes recognized by sera from vaccinated animals by using two complementary approaches.
Enzyme-linked immunosorbent assay (ELISA) studies were performed using streptavidin-coated microtiter plates (Pierce), which were incubated with 16-mer biotinylated overlapping peptides (5 μg/well) spanning the first two fibronectin-binding repeats of the SfbI protein with an offset of 4 amino acids (Fig. 1). Plates were then incubated with the sera from immunized animals, and bound antibodies were detected as previously described (23). As shown in Fig. 1 and 2, only one epitope was recognized by sera from BALB/b and BALB/k mice, which corresponds to the NH2-terminal part of each repeat (positions 377 to 387 and 414 to 424, respectively). To further confirm these results, immunoblot studies were performed using cellulose paper membranes (1Chr; 3MM; Whatman) spotted with 15-mer peptides (8), which encompass all four fibronectin-binding repeats (positions 373 to 515 of the SfbI sequence, accession no. X67947) with an offset of 3 amino acids (Table 1. In brief, after overnight incubation in blocking buffer (2 ml of Genosys blocking buffer, 5% saccharose, 8 ml of T-TBS [50 mM Tris base-buffered saline plus 0.05% Tween 20, pH 7.0]), membranes were further incubated with sera from immunized mice diluted 1:100 in blocking buffer for 3.5 h. After three washes with T-TBS, membranes were treated with alkaline phosphatase-conjugated goat anti-mouse antibodies (Dianova, Hamburg, Germany) diluted 1:500 in blocking buffer for 1.5 h. After washing, bound antibodies were detected by incubating membranes in 50 μl of 1 M MgCl2-40 μl of 5-bromo-4-chloro-3-indolylphosphate-60 μl of 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide in 10 ml of CBS (8 g of NaCl, 0.2 g of KCl, 2.1 g of 10 mM citric acid-1-hydrate in 1 liter). Densitometric analysis of the spots was carried out using the Multi-Analyst analysis software (Bio-Rad Laboratories, Inc.), and results were expressed as relative densities (optical density per square millimeter).
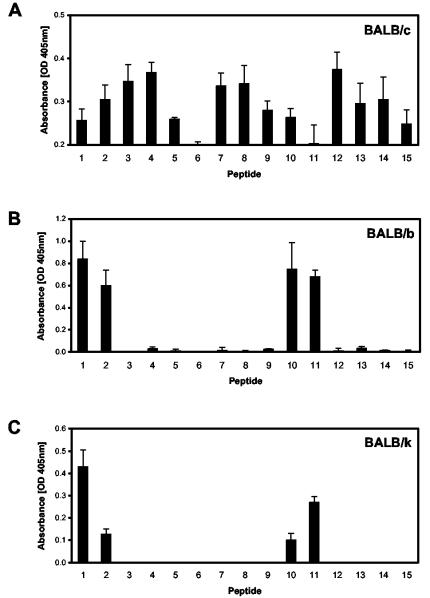
FIG. 2.
Identification of B-cell linear epitopes within the fibronectin-binding repeats of the SfbI protein. Peptide-specific serum IgG responses stimulated after vaccination were determined by ELISA in BALB/c, BALB/b, and BALB/k mice (n = 5). Standard errors of the means are indicated by vertical lines. OD, optical density.
TABLE 1.
Peptides spanning the four fibronectin-binding repeats used to identify linear B-cell epitopes by immunoblot analysis
| No. | Positions | Peptide sequence | No. | Positions | Peptide sequence | |
|---|---|---|---|---|---|---|
| 1 | 361-375 | KLPNETGFSGNMVET | ||||
| 2 | 364-378 | NETGFSGNMVETEDT | ||||
| 3 | 367-381 | GFSGNMVETEDTKEP | ||||
| 4 | 370-384 | GNMVETEDTKEPGVL | ||||
| 5 | 373-387 | VETEDTKEPGVLMGG | ||||
| 6 | 376-390 | EDTKEPGVLMGGQSE | ||||
| 7 | 379-393 | KEPGVLMGGQSESVE | ||||
| 8 | 382-396 | GVLMGGQSESVEFTK | ||||
| 9 | 385-399 | MGGQSESVEFTKDTQ | ||||
| 10 | 388-402 | QSESVEFTKDTQTGM | ||||
| 11 | 391-405 | SVEFTKDTQTGMSGQ | ||||
| 12 | 394-408 | FTKDTQTGMSGQTTP | ||||
| 13 | 397-411 | DTQTGMSGQTTPQVE | ||||
| 14 | 400-414 | TGMSGQTTPQVETED | ||||
| 15 | 403-417 | SGQTTPQVETEDTKE | ||||
| 16 | 406-420 | TTPQVETEDTKEPGV | ||||
| 17 | 409-423 | QVETEDTKEPGVLMG | ||||
| 18 | 412-426 | TEDTKEPGVLMGGQS | ||||
| 19 | 415-429 | TKEPGVLMGGQSESV | ||||
| 20 | 418-432 | PGVLMGGQSESVEFT | ||||
| 21 | 421-435 | LMGGQSESVEFTKDT | ||||
| 22 | 424-438 | GQSESVEFTKDTQTG | ||||
| 23 | 427-441 | ESVEFTKDTQTGMSG | ||||
| 24 | 430-444 | EFTKDTQTGMSGQTA | ||||
| 25 | 433-447 | KDTQTGMSGQTASQV | ||||
| 26 | 436-450 | QTGMSGQTASQVETE | ||||
| 27 | 439-453 | MSGQTASQVETEDTK | ||||
| 28 | 442-456 | QTASQVETEDTKEPG | ||||
| 29 | 445-459 | SQVETEDTKEPGVLM | ||||
| 30 | 448-462 | ETEDTKEPGVLMGGQ | ||||
| 31 | 451-465 | DTKEPGVLMGGQSES | ||||
| 32 | 454-468 | EPGVLMGGQSESVEF | ||||
| 33 | 457-471 | VLMGGQSESVEFTKD | ||||
| 34 | 460-474 | GGQSESVEFTKDTQT | ||||
| 35 | 463-477 | SESVEFTKDTQTGMS | ||||
| 36 | 466-480 | VEFTKDTQTGMSGQT | ||||
| 37 | 469-483 | TKDTQTGMSGQTTPQ | ||||
| 38 | 472-486 | TQTGMSGQTTPQVET | ||||
| 39 | 475-489 | GMSGQTTPQVETEDT | ||||
| 40 | 478-492 | GQTTPQVETEDTKEP | ||||
| 41 | 481-495 | TPQVETEDTKEPGVL | ||||
| 42 | 484-498 | VETEDTKEPGVLMGG | ||||
| 43 | 487-501 | EDTKEPGVLMGGQSE | ||||
| 44 | 490-504 | KEPGVLMGGQSESVE | ||||
| 45 | 493-507 | GVLMGGQSESVEFTK | ||||
| 46 | 496-510 | MGGQSESVEFTKDTQ | ||||
| 47 | 499-513 | QSESVEFTKDTQTGM | ||||
| 48 | 502-516 | SVEFTKDTQTGMSGF | ||||
| 49 | 505-519 | FTKDTQTGMSGFSET | ||||
| 50 | 508-522 | DTQTGMSGFSETVTI | ||||
| 51 | 511-525 | TGMSGFSETVTIVED | ||||
| 52 | 514-528 | SGFSETVTIVEDTRP |
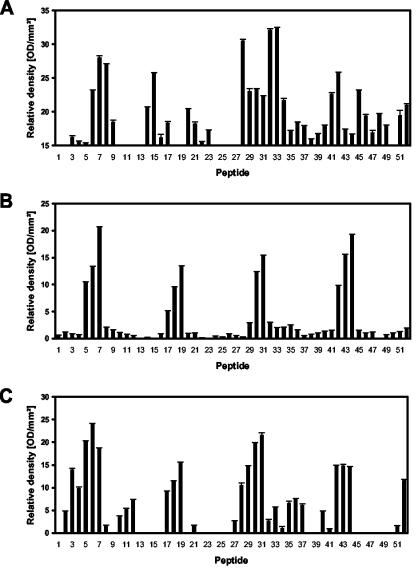
The obtained results demonstrated that the linear B-cell epitope recognized by BALB/b and BALB/k mice corresponds to the sequence DTKEPGVLMGG (Fig. 1 and 3). Peptides encompassing this region showed similar antibody binding rates, whereas binding was drastically reduced when the NH2-terminal aspartic acid or the COOH-terminal glutamic acid was lost (Fig. 1). In contrast, two epitopes were recognized by sera from mice of the BALB/c haplotype. The first one was similar to the one recognized by BALB/b and BALB/k mice and is located between positions 382 and 393 and positions 419 and 430 (GVLMGGQSESVE), respectively. Peptides encompassing this sequence showed the strongest antibody binding rates in both ELISA and Western blot studies (Fig. 2A and 3A), and antibody binding rates were dramatically reduced by loss of the NH2-terminal glycine or the COOH-terminal glutamic acid. The second linear B-cell epitope recognized in BALB/c mice is located in the interrepeat region between positions 403 and 412 (SGQTTPQVET), and also between positions 440 and 449, in which two mismatches are located (SGQTASQVET). Thus, it seems that these two residues are not essential for antibody recognition. By using the antigenic index algorithm (PROTEAN software from DNAStar, Inc., Madison, Wis.) it was shown that all detected epitopes were localized in regions of high probability for antigen determinants. Preliminary studies suggest that similar regions are also recognized by sera from patients convalescing from S. pyogenes infections. When tested by ELISA, antibodies were present in the sera (n = 10), which were able to recognize linear B-cell epitopes encompassed within positions 389 to 416 of SfbI. However, to prove the statistical significance of these initial data, it would be essential to analyze a larger number of samples obtained from patients with a different form of the disease.
FIG. 3.
Identification of B-cell linear epitopes within the fibronectin-binding repeats of the SfbI protein. Peptide-specific serum IgG responses stimulated after vaccination were determined by immunoblot analysis in BALB/c (A), BALB/b (B), and BALB/k (C) mice (n = 5). Standard errors of the means are indicated by vertical lines. OD, optical density.
Identification of T-cell epitopes within the fibronectin-binding repeats of the SfbI protein.
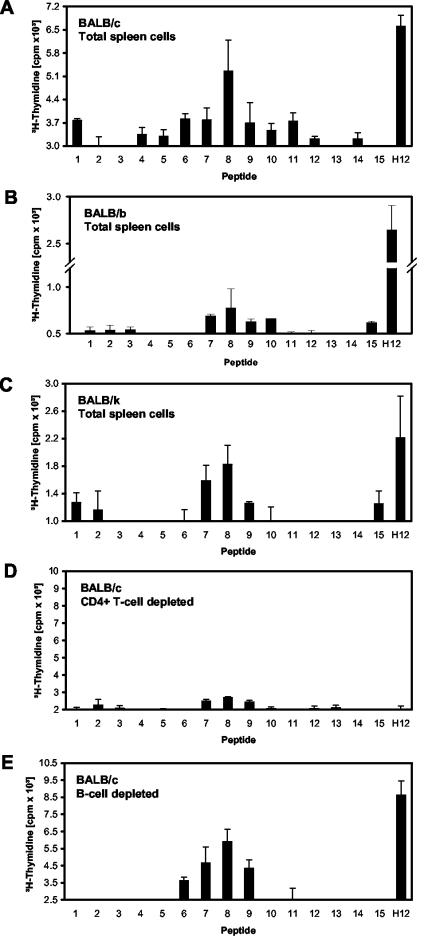
To identify T-cell epitopes located within the fibronectin-binding repeats, spleen cells recovered from immunized mice were restimulated in vitro using the 15 individual peptides described for Fig. 1. Preliminary analysis performed using different concentrations of each peptide in the range of 0 to 50 μg/ml showed that optimal responses were obtained when the peptides were tested at 50 μg/ml. Proliferation was assessed after 4 days of culture by measuring the incorporation of [3H]thymidine (14, 15). The obtained results suggest the presence of one T-cell epitope located in the crossover from one repeat to the next (GMSGQTTPQVETEDTK), since the peptide encompassing positions 401 to 416 stimulated the strongest cellular response (Fig. 4A). Similar results were observed when cells from BALB/b and BALB/k vaccinated mice were tested (Fig. 4B and C). To further verify the nature of the stimulated cells, proliferation studies were performed using spleen cells depleted from either B cells or CD4+ T cells. While no effect could be observed when B cells were depleted, depletion of CD4+ cells significantly decreased the proliferation rates (Fig. 4D and E). This confirmed that CD4+ T cells constitute the stimulated cellular population. Interestingly, a motif within this sequence is also predicted to be recognized in the human system by the SYFPEITHI algorithm (21).
FIG. 4.
Identification of T-cell epitopes within the fibronectin-binding repeats of the SfbI protein. Peptide-specific proliferative responses from immunized mice were evaluated using total spleen cells (A to C) and after depletion of either CD4+ T cells (D) or B cells (E). Results are expressed as mean counts per minute of triplicate samples. Standard errors of the means are indicated by vertical lines.
In conclusion, our results indicate that efficient immune responses are stimulated in congenic mice from different haplotypes after intranasal immunization with a polypeptide encompassing the fibronectin-binding domains of the SfbI protein. This fragment also constitutes the minimal domain able to confer protective immunity. Furthermore, similar linear B-cell epitopes, as well as a T-cell epitope, were recognized in all vaccinated mouse strains. Thus, all experimentally determined linear B- and T-cell epitopes are cross-recognized and encompassed within a 30-amino-acid fragment. The obtained results provide critical information for the exploitation of the fibronectin-binding domain of the SfbI protein in the development of polyepitope-based vaccines against S. pyogenes.
Acknowledgments
This work was in part supported by the DFG grants GU 482/2-1 and GU 482/2-2.
REFERENCES
- 1.Ada, G., and A. Ramsay. 1996. Vaccines, vaccination and the immune response, p. 138-141. Lippincott-Raven, Philadelphia, Pa.
- 2.Bessen, D., and V. A. Fischetti. 1988. Influence of intranasal immunization with synthetic peptides corresponding to conserved epitopes of M protein on mucosal colonization by group A streptococci. Infect. Immun. 56:2666-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisno, A. L. 1991. Group A streptococcal infections and acute rheumatic fever. N. Engl. J. Med. 325:783-793. [DOI] [PubMed] [Google Scholar]
- 4.Brandt, E. R., W. A. Hayman, B. Currie, J. Carapetis, D. C. Jackson, K.-A. Do, and M. F. Good. 1999. Functional analysis of IgA antibodies specific for a conserved epitope within the M protein of group A streptococci from Australian aboriginal endemic communities. Int. Immunol. 11:569-576. [DOI] [PubMed] [Google Scholar]
- 5.Brandt, E. R., T. Teh, W. A. Relf, R. I. Hobb, and M. F. Good. 2000. Protective and nonprotective epitopes from amino termini of M proteins from Australian aboriginal isolates and reference strains of group A streptococci. Infect. Immun. 68:6587-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandt, E. R., K. S. Sriprakash, R. I. Hobb, W. A. Hayman, W. Zeng, M. R. Batzloff, D. C. Jackson, and M. F. Good. 2000. New multi-determinant strategy for a group A streptococcal vaccine designed for the Australian Aboriginal population. Nat. Med. 6:455-459. [DOI] [PubMed] [Google Scholar]
- 7.Cornaglia, G., M. Ligozzi, A. Mazzariol, M. Valentini, G. Orefici, R. Fontana, et al. 1996. Rapid increase of resistance to erythromycin and clindamycin in Streptococcus pyogenes in Italy, 1993-1995. Emerg. Infect. Dis. 2:339-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank, R. 1992. Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 48:9217-9232. [Google Scholar]
- 9.Guzmán, C. A., S. R. Talay, G. Molinari, E. Medina, and G. S. Chhatwal. 1999. Protective immune response against Streptococcus pyogenes in mice after intranasal vaccination with the fibronectin-binding protein SfbI. J. Infect. Dis. 179:901-906. [DOI] [PubMed] [Google Scholar]
- 10.Ji, Y., B. Carlson, A. Kondagunta, and P. P. Cleary. 1997. Intranasal immunization with C5a peptidase prevents nasopharyngeal colonization of mice by the group A Streptococcus. Infect. Immun. 65:2080-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson, D. R., D. L. Stevens, and E. L. Kaplan. 1992. Epidemiologic analysis of group A streptococcal serotypes associated with severe systemic infections, rheumatic fever, or uncomplicated pharyngitis. J. Infect. Dis. 166:374-382. [DOI] [PubMed] [Google Scholar]
- 12.Kapur, V., J. T. Maffei, R. S. Greer, L. L. Li, G. J. Adams, and J. M. Musser. 1994. Vaccination with streptococcal extracellular cysteine protease (interleukin-1β convertase) protects mice against challenge with heterologous group A streptococci. Microb. Pathog. 16:443-450. [DOI] [PubMed] [Google Scholar]
- 13.LaPenta, D., C. Rubens, E. Chi, and P. P. Cleary. 1994. Group A streptococci efficiently invade human respiratory epithelial cells. Proc. Natl. Acad. Sci. USA 91:12115-12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Medina, E., S. R. Talay, G. S. Chhatwal, and C. A. Guzman. 1998. Fibronectin-binding protein I of Streptococcus pyogenes promotes T cell-independent proliferation of murine B lymphocytes and enhances the expression of MHC class II molecules on antigen-presenting cells. Int. Immunol. 10:1657-1664. [DOI] [PubMed] [Google Scholar]
- 15.Medina, E., S. R. Talay, G. S. Chhatwal, and C. A. Guzmán. 1998. Fibronectin-binding protein I of Streptococcus pyogenes is a promising adjuvant for antigens delivered by mucosal route. Eur. J. Immunol. 28:1069-1077. [DOI] [PubMed] [Google Scholar]
- 16.Molinari, G., S. R. Talay, P. Valentin Weigand, M. Rohde, and G. S. Chhatwal. 1997. The fibronectin-binding protein of Streptococcus pyogenes, SfbI, is involved in the internalization of group A streptococci by epithelial cells. Infect. Immun. 65:1357-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nardin, E. H., G. A. Oliveira, J. M. Calvo-Calle, and R. Nussenzweig. 1995. The use of multiple antigen peptides in the analysis and induction of protective immune responses against infectious diseases. Adv. Immunol. 50:105-150. [DOI] [PubMed] [Google Scholar]
- 18.Neu, H. C. 1992. The crisis in antibiotic resistance. Science 257:1064-1073. [DOI] [PubMed] [Google Scholar]
- 19.Olsen, A. W., P. R. Hansen, A. Holm, and P. Andersen. 2000. Efficient protection against Mycobacterium tuberculosis by vaccination with a single subdominant epitope from the ESAT-6 antigen. Eur. J. Immunol. 30:1724-1732. [DOI] [PubMed] [Google Scholar]
- 20.Pruksakorn, S., A. Galbraith, R. A. Houghten, and M. F. Good. 1992. Conserved T and B cell epitopes on the M protein of group A streptococci. Induction of bactericidal antibodies. J. Immunol. 149:2729-2735. [PubMed] [Google Scholar]
- 21.Rammensee, H., J. Bachmann, N. P. Emmerich, O. A. Bachor, and S. Stevanovic. 1999. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50:213-219. [DOI] [PubMed] [Google Scholar]
- 22.Schulze, K., E. Medina, S. R. Talay, R. J. Towers, G. S. Chhatwal, and C. A. Guzmán. 2001. Characterization of the domain of fibronectin-binding protein I of Streptococcus pyogenes responsible for elicitation of a protective immune response. Infect. Immun. 69:622-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulze, K., and C. A. Guzmán. 2003. Identification of the domains of the fibronectin-binding protein I of Streptococcus pyogenes responsible for adjuvanticity. FEMS Immunol. Med. Microbiol. 37:173-177. [DOI] [PubMed] [Google Scholar]
- 24.Simmons, C. P., T. Hussell, T. Sparer, G. Walzl, P. Openshaw, and G. Dougan. 2001. Mucosal delivery of a respiratory syncytial virus CTL peptide with enterotoxin-based adjuvants elicits protective, immunopathogenic, and immunoregulatory antiviral CD8+ T cell responses. J. Immunol. 166:1106-1113. [DOI] [PubMed] [Google Scholar]
- 25.Stevens, D. L. 1995. Streptococcal toxic-shock syndrome: spectrum of disease, pathogenesis, and new concepts in treatment. Emerg. Infect. Dis. 1:69-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talay, S. R., P. Valentin Weigand, P. G. Jerlstrom, K. N. Timmis, and G. S. Chhatwal. 1992. Fibronectin-binding protein of Streptococcus pyogenes: sequence of the binding domain involved in adherence of streptococci to epithelial cells. Infect. Immun. 60:3837-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talay, S. R., P. Valentin Weigand, K. N. Timmis, and G. S. Chhatwal. 1994. Domain structure and conserved epitopes of Sfb protein, the fibronectin-binding adhesin of Streptococcus pyogenes. Mol. Microbiol. 13:531-539. [DOI] [PubMed] [Google Scholar]
- 28.Talay, S. R., A. Zock, M. Rohde, G. Molinari, M. Oggioni, G. Pozzi, C. A. Guzmán, and G. S. Chhatwal. 2000. Cooperative binding of human fibronectin to SfbI protein triggers streptococcal invasion into respiratory epithelial cells. Cell. Microbiol. 2:521-535. [DOI] [PubMed] [Google Scholar]
- 29.Valentin Weigand, P., S. R. Talay, A. Kaufhold, K. N. Timmis, and G. S. Chhatwal. 1994. The fibronectin binding domain of the Sfb protein adhesin of Streptococcus pyogenes occurs in many group A streptococci and does not cross-react with heart myosin. Microb. Pathog. 17:111-120. [DOI] [PubMed] [Google Scholar]
- 30.Vordermeier, H.-M., D. P. Harris, C. Moreno, M. Singh, and J. Ivanyi. 1995. The nature of the immunogen determines the specificity of antibodies and T cells to selected peptides of the 38 kDa mycobacterial antigen. Int. Immunol. 7:559-566. [DOI] [PubMed] [Google Scholar]
- 31.Wang, C. W., D. J. Looney, M. L. Li, A. M. Walfield, J. Ye, B. Hosein, J. P. Tam, and F. Wong-Staal. 1991. Long-term high-titer neutralizing activity induced by octameric synthetic HIV-1 antigen. Science 254:285-288. [DOI] [PubMed] [Google Scholar]
- 32.World Health Organization. 1980. Community control of rheumatic heart disease in developing countries. 1. A major public health problem. W. H. O. Chron. 34:336-345. [PubMed] [Google Scholar]