Abstract
The title compound, C14H15F3N2O4S·C2H5OH, was prepared by reaction of 4-hydroxybenzaldehyde, ethyl 4,4,4-trifluoro-3-oxobutanoate and thiourea. The hexahydropyrimidine ring adopts a half-chair conformation, the mean plane formed by the ring atoms excluding the C atom bonded to the ethoxycarbonyl group has an r.m.s. deviation of 0.0333 Å, and the dihedral angle between this plane and the benzene ring is 56.76 (5)°. The molecular conformation is stabilized by an intramolecular O—H⋯O hydrogen bond, generating an S(6) ring. The crystal structure is stabilized by intermolecular O—H⋯O, O—H⋯S, N—H⋯O and N—H⋯S hydrogen bonds. The ethyl group of the ester unit is disordered over two positions, with an occupancy ratio of 0.757 (10):0.243 (10).
Related literature
For the bioactivity of dihydropyrimidines, see: Brier et al. (2004 ▶); Cochran et al. (2005 ▶); Moran et al. (2007 ▶); Zorkun et al. (2006 ▶). For the bioactivity of organofluorine compounds, see: Hermann et al. (2003 ▶); Ulrich (2004 ▶). For a related structure, see: Song et al. (2010 ▶).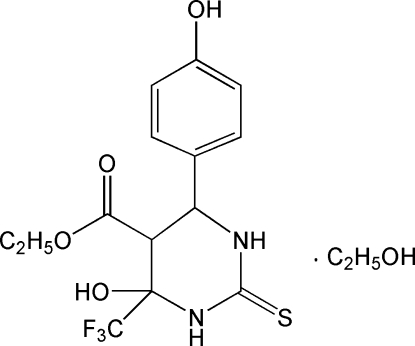
Experimental
Crystal data
C14H15F3N2O4S·C2H6O
M r = 410.41
Monoclinic,

a = 14.7204 (14) Å
b = 9.9772 (12) Å
c = 14.7357 (15) Å
β = 119.716 (11)°
V = 1879.6 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.23 mm−1
T = 113 K
0.20 × 0.16 × 0.10 mm
Data collection
Rigaku Saturn CCD area-detector diffractometer
Absorption correction: multi-scan (CrystalClear; Rigaku, 2009 ▶) T min = 0.955, T max = 0.977
23471 measured reflections
4486 independent reflections
3815 reflections with I > 2σ(I)
R int = 0.047
Refinement
R[F 2 > 2σ(F 2)] = 0.036
wR(F 2) = 0.092
S = 1.03
4486 reflections
280 parameters
24 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.47 e Å−3
Δρmin = −0.27 e Å−3
Data collection: CrystalClear (Rigaku, 2009 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811015376/om2424sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811015376/om2424Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811015376/om2424Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯O2 | 0.853 (19) | 1.987 (19) | 2.7383 (13) | 146.3 (17) |
| O1—H1⋯S1i | 0.853 (19) | 3.029 (19) | 3.4858 (11) | 115.8 (15) |
| O4—H4⋯O5ii | 0.835 (18) | 1.873 (19) | 2.7066 (14) | 175.6 (19) |
| O5—H5⋯S1iii | 0.84 | 2.36 | 3.1970 (11) | 175 |
| N1—H1A⋯S1iii | 0.855 (17) | 2.583 (18) | 3.4307 (12) | 171.5 (14) |
| N2—H2A⋯O1iv | 0.814 (14) | 2.368 (15) | 3.1701 (15) | 168.7 (14) |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  .
.
Acknowledgments
This work was supported by the Natural Science Foundation of Henan Province, China (grant No. 082300420110) and the Natural Science Foundation of Henan Province Education Department, China (grant No. 2007150036).
supplementary crystallographic information
Comment
Dihydropyrimidine (DHPM) derivatives can be used as potential calcium channel blockers (Zorkun et al., 2006), inhibitors of mitotic kinesin Eg5 for treating cancer (Cochran et al., 2005; Brier et al., 2004) and as TRPA1 modulators for treating pain (Moran et al., 2007). In addition, compounds that contain fluorine have special bioactivity, e.g. flumioxazin is a widely used herbicide (Hermann et al., 2003; Ulrich, 2004). This led us to focus our attention on the synthesis and bioactivity of these important fused perfluoroalkylated heterocyclic compounds. During the synthesis of DHPM derivatives, the title compound, an intermediate C14H15F3N2O4S.C2H5OH was isolated and the structure confirmed by X-ray diffraction.
In the structure of the title molecule, the 1,3-diazinane ring adopts a half-chair conformation, the mean plane formed by the ring atoms excluding the C atom bonded to the ethoxy carbonyl group has an r.m.s. deviation of 0.0333 Å, the dihedral angle between the mean plane and benzene ring is 56.76 (5)°. The molecular conformation is stabilized by intramolecular O—H···O hydrogen bond, generating an S(6) ring. The crystal structure is stabilized by intermolecular hydrogen bonds (O—H···O, O—H···S, N—H···O and N—H···S). The ethyl group of the ester unit is disordered over two positions, with a site-occupancy ratio of 0.757 (10):0.243 (10). For a crystal structure related to the title compound, see: Song et al., 2010.
Experimental
The title compound was synthesized by refluxing for 3 h a stirred solution of 4-hydroxybenzaldehyde (2.45 g, 20 mmol), ethyl 4,4,4-trifluoro-3-oxobutanoate (4.42 g, 24 mmol) and thiourea (2.28 g, 30 mmol) in 20 ml of anhydrous ethanol. The reaction was catalyzed by sulfamic acid (0.6 g). The solvent was evaporated in vacuo and the residue was washed with water. The title compound was recrystallized by slow evaporation of a 50% aqueous ethanol solution.
Refinement
H atoms involved in hydrogen-bonding inetractions were located by difference Fourier methods and their positional and isotropic displacement parameters were refined. Other H atoms were placed in calculated positions, with C—H(aromatic) = 0.95 Å and C—H(aliphatic) = 0.98 Å or 0.99 Å, and treated as riding, with Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.
Molecular configuration and atom numbering scheme with displacement ellipsoids drawn at the 30% probability level.
Crystal data
| C14H15F3N2O4S·C2H6O | F(000) = 856 |
| Mr = 410.41 | Dx = 1.450 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 14.7204 (14) Å | Cell parameters from 6768 reflections |
| b = 9.9772 (12) Å | θ = 2.0–27.9° |
| c = 14.7357 (15) Å | µ = 0.23 mm−1 |
| β = 119.716 (11)° | T = 113 K |
| V = 1879.6 (3) Å3 | Prism, colorless |
| Z = 4 | 0.20 × 0.16 × 0.10 mm |
Data collection
| Rigaku Saturn CCD area-detector diffractometer | 4486 independent reflections |
| Radiation source: rotating anode | 3815 reflections with I > 2σ(I) |
| multilayer | Rint = 0.047 |
| Detector resolution: 14.63 pixels mm-1 | θmax = 27.9°, θmin = 2.6° |
| ω and φ scans | h = −18→19 |
| Absorption correction: multi-scan (CrystalClear; Rigaku, 2009) | k = −13→13 |
| Tmin = 0.955, Tmax = 0.977 | l = −19→19 |
| 23471 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.036 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.092 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0506P)2] where P = (Fo2 + 2Fc2)/3 |
| 4486 reflections | (Δ/σ)max = 0.004 |
| 280 parameters | Δρmax = 0.47 e Å−3 |
| 24 restraints | Δρmin = −0.27 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| S1 | 0.42068 (2) | 0.59475 (3) | 0.35546 (2) | 0.01745 (10) | |
| F1 | 0.79908 (6) | 0.50693 (8) | 0.56642 (6) | 0.0289 (2) | |
| F2 | 0.74838 (6) | 0.31639 (8) | 0.59449 (6) | 0.0272 (2) | |
| F3 | 0.85429 (6) | 0.32318 (9) | 0.53355 (6) | 0.0293 (2) | |
| O1 | 0.65426 (7) | 0.24640 (9) | 0.38459 (7) | 0.0190 (2) | |
| H1 | 0.6926 (15) | 0.2229 (18) | 0.3589 (15) | 0.056 (6)* | |
| O2 | 0.79221 (7) | 0.28215 (9) | 0.31283 (7) | 0.0234 (2) | |
| O3 | 0.85300 (8) | 0.49263 (10) | 0.32835 (9) | 0.0356 (3) | |
| O4 | 0.68310 (8) | 0.73637 (10) | −0.05667 (8) | 0.0264 (2) | |
| H4 | 0.7053 (15) | 0.6801 (19) | −0.0829 (14) | 0.048 (6)* | |
| N1 | 0.58962 (8) | 0.44198 (11) | 0.41833 (8) | 0.0161 (2) | |
| N2 | 0.52429 (9) | 0.53998 (11) | 0.25624 (8) | 0.0171 (2) | |
| C1 | 0.51772 (9) | 0.52182 (12) | 0.34218 (9) | 0.0154 (3) | |
| C2 | 0.67753 (10) | 0.38150 (12) | 0.41637 (10) | 0.0156 (3) | |
| C3 | 0.77017 (10) | 0.38279 (13) | 0.52882 (10) | 0.0199 (3) | |
| C4 | 0.70305 (9) | 0.46428 (12) | 0.34352 (9) | 0.0153 (3) | |
| H4A | 0.7255 | 0.5564 | 0.3730 | 0.018* | |
| C5 | 0.78765 (10) | 0.40048 (13) | 0.32782 (10) | 0.0183 (3) | |
| C6 | 0.9245 (3) | 0.4501 (3) | 0.2899 (4) | 0.0349 (8) | 0.757 (10) |
| H6A | 0.8925 | 0.3759 | 0.2391 | 0.042* | 0.757 (10) |
| H6B | 0.9914 | 0.4179 | 0.3490 | 0.042* | 0.757 (10) |
| C7 | 0.9440 (2) | 0.5681 (4) | 0.2384 (2) | 0.0371 (9) | 0.757 (10) |
| H7A | 0.9920 | 0.5418 | 0.2134 | 0.056* | 0.757 (10) |
| H7B | 0.9751 | 0.6413 | 0.2890 | 0.056* | 0.757 (10) |
| H7C | 0.8777 | 0.5981 | 0.1791 | 0.056* | 0.757 (10) |
| C6' | 0.9536 (7) | 0.4377 (10) | 0.3420 (11) | 0.034 (2) | 0.243 (10) |
| H6'A | 0.9568 | 0.3387 | 0.3462 | 0.041* | 0.243 (10) |
| H6'B | 1.0158 | 0.4770 | 0.4028 | 0.041* | 0.243 (10) |
| C7' | 0.9382 (8) | 0.4894 (18) | 0.2394 (9) | 0.056 (3) | 0.243 (10) |
| H7'A | 0.9987 | 0.4651 | 0.2320 | 0.083* | 0.243 (10) |
| H7'B | 0.9309 | 0.5872 | 0.2373 | 0.083* | 0.243 (10) |
| H7'C | 0.8748 | 0.4497 | 0.1820 | 0.083* | 0.243 (10) |
| C8 | 0.60150 (10) | 0.47359 (12) | 0.23647 (9) | 0.0157 (3) | |
| H8A | 0.5765 | 0.3807 | 0.2107 | 0.019* | |
| C9 | 0.61729 (10) | 0.54638 (12) | 0.15579 (9) | 0.0158 (3) | |
| C10 | 0.63212 (10) | 0.47113 (13) | 0.08457 (9) | 0.0171 (3) | |
| H10A | 0.6260 | 0.3763 | 0.0842 | 0.021* | |
| C11 | 0.65560 (10) | 0.53228 (13) | 0.01436 (10) | 0.0179 (3) | |
| H11A | 0.6675 | 0.4794 | −0.0323 | 0.021* | |
| C12 | 0.66173 (10) | 0.67134 (13) | 0.01224 (10) | 0.0179 (3) | |
| C13 | 0.64511 (11) | 0.74801 (13) | 0.08177 (10) | 0.0215 (3) | |
| H13A | 0.6479 | 0.8430 | 0.0799 | 0.026* | |
| C14 | 0.62449 (11) | 0.68552 (13) | 0.15354 (10) | 0.0194 (3) | |
| H14A | 0.6151 | 0.7382 | 0.2020 | 0.023* | |
| O5 | 0.74901 (7) | 0.56068 (11) | 0.84823 (8) | 0.0276 (2) | |
| H5 | 0.7076 | 0.5158 | 0.7956 | 0.041* | |
| C15 | 0.84625 (12) | 0.57515 (17) | 0.84999 (13) | 0.0359 (4) | |
| H15A | 0.8351 | 0.6201 | 0.7854 | 0.043* | |
| H15B | 0.8774 | 0.4859 | 0.8539 | 0.043* | |
| C16 | 0.91849 (13) | 0.65741 (19) | 0.94378 (15) | 0.0484 (5) | |
| H16A | 0.9857 | 0.6682 | 0.9460 | 0.073* | |
| H16B | 0.9295 | 0.6120 | 1.0074 | 0.073* | |
| H16C | 0.8873 | 0.7457 | 0.9391 | 0.073* | |
| H1A | 0.5812 (12) | 0.4270 (16) | 0.4708 (13) | 0.037 (5)* | |
| H2A | 0.4826 (11) | 0.5934 (14) | 0.2148 (11) | 0.018 (4)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.01884 (18) | 0.01825 (17) | 0.01816 (17) | 0.00402 (12) | 0.01138 (14) | 0.00283 (12) |
| F1 | 0.0287 (5) | 0.0258 (4) | 0.0222 (4) | −0.0028 (4) | 0.0051 (4) | −0.0062 (3) |
| F2 | 0.0265 (5) | 0.0362 (5) | 0.0191 (4) | 0.0057 (4) | 0.0116 (4) | 0.0094 (3) |
| F3 | 0.0191 (4) | 0.0438 (5) | 0.0235 (4) | 0.0123 (4) | 0.0093 (4) | 0.0032 (4) |
| O1 | 0.0237 (5) | 0.0137 (5) | 0.0235 (5) | −0.0001 (4) | 0.0147 (4) | −0.0010 (4) |
| O2 | 0.0242 (5) | 0.0206 (5) | 0.0291 (5) | 0.0010 (4) | 0.0160 (4) | −0.0031 (4) |
| O3 | 0.0276 (6) | 0.0282 (6) | 0.0643 (8) | −0.0086 (5) | 0.0330 (6) | −0.0118 (5) |
| O4 | 0.0427 (6) | 0.0200 (5) | 0.0283 (5) | 0.0029 (4) | 0.0267 (5) | 0.0046 (4) |
| N1 | 0.0176 (6) | 0.0183 (5) | 0.0151 (5) | 0.0028 (4) | 0.0100 (5) | 0.0027 (4) |
| N2 | 0.0167 (6) | 0.0204 (6) | 0.0145 (5) | 0.0046 (5) | 0.0080 (5) | 0.0033 (4) |
| C1 | 0.0169 (6) | 0.0133 (6) | 0.0158 (6) | −0.0025 (5) | 0.0080 (5) | −0.0010 (5) |
| C2 | 0.0159 (6) | 0.0148 (6) | 0.0165 (6) | 0.0006 (5) | 0.0084 (5) | 0.0000 (5) |
| C3 | 0.0199 (7) | 0.0216 (7) | 0.0195 (6) | 0.0037 (5) | 0.0108 (6) | 0.0014 (5) |
| C4 | 0.0167 (6) | 0.0143 (6) | 0.0161 (6) | −0.0005 (5) | 0.0090 (5) | −0.0008 (5) |
| C5 | 0.0145 (6) | 0.0229 (7) | 0.0159 (6) | −0.0009 (5) | 0.0063 (5) | −0.0011 (5) |
| C6 | 0.0241 (17) | 0.0424 (15) | 0.050 (2) | −0.0031 (12) | 0.0270 (17) | −0.0049 (16) |
| C7 | 0.0250 (12) | 0.062 (2) | 0.0300 (12) | −0.0070 (13) | 0.0177 (10) | 0.0034 (13) |
| C6' | 0.012 (4) | 0.041 (4) | 0.050 (6) | 0.001 (3) | 0.015 (4) | 0.002 (4) |
| C7' | 0.039 (5) | 0.081 (8) | 0.058 (5) | −0.005 (5) | 0.033 (4) | 0.002 (5) |
| C8 | 0.0175 (6) | 0.0158 (6) | 0.0154 (6) | −0.0005 (5) | 0.0095 (5) | −0.0012 (5) |
| C9 | 0.0155 (6) | 0.0167 (6) | 0.0147 (6) | 0.0008 (5) | 0.0072 (5) | 0.0009 (5) |
| C10 | 0.0196 (7) | 0.0142 (6) | 0.0179 (6) | −0.0006 (5) | 0.0096 (6) | −0.0004 (5) |
| C11 | 0.0209 (7) | 0.0184 (6) | 0.0160 (6) | 0.0000 (5) | 0.0105 (6) | −0.0023 (5) |
| C12 | 0.0193 (7) | 0.0190 (7) | 0.0173 (6) | 0.0018 (5) | 0.0106 (6) | 0.0034 (5) |
| C13 | 0.0292 (8) | 0.0137 (6) | 0.0251 (7) | 0.0012 (5) | 0.0161 (6) | 0.0002 (5) |
| C14 | 0.0253 (7) | 0.0172 (6) | 0.0196 (6) | 0.0010 (5) | 0.0140 (6) | −0.0018 (5) |
| O5 | 0.0236 (5) | 0.0372 (6) | 0.0255 (5) | −0.0044 (4) | 0.0149 (5) | −0.0081 (4) |
| C15 | 0.0264 (8) | 0.0492 (10) | 0.0368 (9) | −0.0002 (7) | 0.0193 (8) | 0.0010 (8) |
| C16 | 0.0275 (9) | 0.0468 (11) | 0.0586 (12) | −0.0065 (8) | 0.0120 (9) | −0.0034 (9) |
Geometric parameters (Å, °)
| S1—C1 | 1.6987 (13) | C7—H7C | 0.9800 |
| F1—C3 | 1.3373 (15) | C6'—C7' | 1.504 (14) |
| F2—C3 | 1.3364 (15) | C6'—H6'A | 0.9900 |
| F3—C3 | 1.3440 (15) | C6'—H6'B | 0.9900 |
| O1—C2 | 1.4123 (15) | C7'—H7'A | 0.9800 |
| O1—H1 | 0.853 (19) | C7'—H7'B | 0.9800 |
| O2—C5 | 1.2090 (15) | C7'—H7'C | 0.9800 |
| O3—C5 | 1.3279 (16) | C8—C9 | 1.5063 (17) |
| O3—C6 | 1.483 (3) | C8—H8A | 1.0000 |
| O3—C6' | 1.497 (9) | C9—C10 | 1.3927 (17) |
| O4—C12 | 1.3666 (15) | C9—C14 | 1.3939 (17) |
| O4—H4 | 0.835 (18) | C10—C11 | 1.3844 (17) |
| N1—C1 | 1.3551 (16) | C10—H10A | 0.9500 |
| N1—C2 | 1.4410 (16) | C11—C12 | 1.3917 (18) |
| N1—H1A | 0.855 (17) | C11—H11A | 0.9500 |
| N2—C1 | 1.3294 (16) | C12—C13 | 1.3940 (18) |
| N2—C8 | 1.4640 (16) | C13—C14 | 1.3849 (18) |
| N2—H2A | 0.814 (14) | C13—H13A | 0.9500 |
| C2—C3 | 1.5380 (18) | C14—H14A | 0.9500 |
| C2—C4 | 1.5413 (17) | O5—C15 | 1.4261 (16) |
| C4—C5 | 1.5149 (17) | O5—H5 | 0.8400 |
| C4—C8 | 1.5472 (17) | C15—C16 | 1.503 (2) |
| C4—H4A | 1.0000 | C15—H15A | 0.9900 |
| C6—C7 | 1.504 (4) | C15—H15B | 0.9900 |
| C6—H6A | 0.9900 | C16—H16A | 0.9800 |
| C6—H6B | 0.9900 | C16—H16B | 0.9800 |
| C7—H7A | 0.9800 | C16—H16C | 0.9800 |
| C7—H7B | 0.9800 | ||
| C2—O1—H1 | 107.7 (13) | O3—C6'—H6'A | 112.7 |
| C5—O3—C6 | 116.66 (15) | C7'—C6'—H6'A | 112.7 |
| C5—O3—C6' | 114.4 (4) | O3—C6'—H6'B | 112.7 |
| C6—O3—C6' | 26.4 (4) | C7'—C6'—H6'B | 112.7 |
| C12—O4—H4 | 108.2 (13) | H6'A—C6'—H6'B | 110.2 |
| C1—N1—C2 | 124.76 (11) | C6'—C7'—H7'A | 109.5 |
| C1—N1—H1A | 116.6 (11) | C6'—C7'—H7'B | 109.5 |
| C2—N1—H1A | 118.6 (11) | H7'A—C7'—H7'B | 109.5 |
| C1—N2—C8 | 123.93 (11) | C6'—C7'—H7'C | 109.5 |
| C1—N2—H2A | 114.8 (10) | H7'A—C7'—H7'C | 109.5 |
| C8—N2—H2A | 121.3 (10) | H7'B—C7'—H7'C | 109.5 |
| N2—C1—N1 | 118.24 (11) | N2—C8—C9 | 112.19 (10) |
| N2—C1—S1 | 120.92 (10) | N2—C8—C4 | 106.10 (10) |
| N1—C1—S1 | 120.84 (9) | C9—C8—C4 | 112.61 (10) |
| O1—C2—N1 | 109.49 (10) | N2—C8—H8A | 108.6 |
| O1—C2—C3 | 107.76 (10) | C9—C8—H8A | 108.6 |
| N1—C2—C3 | 107.48 (10) | C4—C8—H8A | 108.6 |
| O1—C2—C4 | 112.54 (10) | C10—C9—C14 | 118.48 (12) |
| N1—C2—C4 | 108.72 (10) | C10—C9—C8 | 118.55 (11) |
| C3—C2—C4 | 110.72 (10) | C14—C9—C8 | 122.83 (11) |
| F2—C3—F1 | 107.49 (10) | C11—C10—C9 | 121.06 (12) |
| F2—C3—F3 | 106.79 (10) | C11—C10—H10A | 119.5 |
| F1—C3—F3 | 107.01 (11) | C9—C10—H10A | 119.5 |
| F2—C3—C2 | 111.90 (11) | C10—C11—C12 | 119.94 (12) |
| F1—C3—C2 | 112.59 (10) | C10—C11—H11A | 120.0 |
| F3—C3—C2 | 110.75 (10) | C12—C11—H11A | 120.0 |
| C5—C4—C2 | 112.57 (10) | O4—C12—C11 | 122.09 (12) |
| C5—C4—C8 | 108.75 (10) | O4—C12—C13 | 118.32 (12) |
| C2—C4—C8 | 107.24 (10) | C11—C12—C13 | 119.58 (12) |
| C5—C4—H4A | 109.4 | C14—C13—C12 | 119.94 (12) |
| C2—C4—H4A | 109.4 | C14—C13—H13A | 120.0 |
| C8—C4—H4A | 109.4 | C12—C13—H13A | 120.0 |
| O2—C5—O3 | 124.85 (12) | C13—C14—C9 | 120.96 (12) |
| O2—C5—C4 | 124.23 (12) | C13—C14—H14A | 119.5 |
| O3—C5—C4 | 110.86 (11) | C9—C14—H14A | 119.5 |
| O3—C6—C7 | 108.6 (2) | C15—O5—H5 | 109.5 |
| O3—C6—H6A | 110.0 | O5—C15—C16 | 108.56 (13) |
| C7—C6—H6A | 110.0 | O5—C15—H15A | 110.0 |
| O3—C6—H6B | 110.0 | C16—C15—H15A | 110.0 |
| C7—C6—H6B | 110.0 | O5—C15—H15B | 110.0 |
| H6A—C6—H6B | 108.4 | C16—C15—H15B | 110.0 |
| C6—C7—H7A | 109.5 | H15A—C15—H15B | 108.4 |
| C6—C7—H7B | 109.5 | C15—C16—H16A | 109.5 |
| H7A—C7—H7B | 109.5 | C15—C16—H16B | 109.5 |
| C6—C7—H7C | 109.5 | H16A—C16—H16B | 109.5 |
| H7A—C7—H7C | 109.5 | C15—C16—H16C | 109.5 |
| H7B—C7—H7C | 109.5 | H16A—C16—H16C | 109.5 |
| O3—C6'—C7' | 95.3 (7) | H16B—C16—H16C | 109.5 |
| C8—N2—C1—N1 | 3.16 (18) | C8—C4—C5—O2 | −75.56 (15) |
| C8—N2—C1—S1 | −175.89 (9) | C2—C4—C5—O3 | −139.51 (11) |
| C2—N1—C1—N2 | 4.26 (18) | C8—C4—C5—O3 | 101.80 (12) |
| C2—N1—C1—S1 | −176.69 (9) | C5—O3—C6—C7 | 147.6 (2) |
| C1—N1—C2—O1 | −100.03 (14) | C6'—O3—C6—C7 | −120.7 (11) |
| C1—N1—C2—C3 | 143.19 (12) | C5—O3—C6'—C7' | 119.4 (7) |
| C1—N1—C2—C4 | 23.29 (16) | C6—O3—C6'—C7' | 18.2 (10) |
| O1—C2—C3—F2 | −59.19 (13) | C1—N2—C8—C9 | −159.43 (11) |
| N1—C2—C3—F2 | 58.73 (13) | C1—N2—C8—C4 | −36.07 (16) |
| C4—C2—C3—F2 | 177.35 (10) | C5—C4—C8—N2 | −178.07 (10) |
| O1—C2—C3—F1 | 179.61 (10) | C2—C4—C8—N2 | 59.94 (12) |
| N1—C2—C3—F1 | −62.47 (13) | C5—C4—C8—C9 | −54.97 (13) |
| C4—C2—C3—F1 | 56.15 (14) | C2—C4—C8—C9 | −176.96 (10) |
| O1—C2—C3—F3 | 59.84 (13) | N2—C8—C9—C10 | −141.94 (12) |
| N1—C2—C3—F3 | 177.76 (10) | C4—C8—C9—C10 | 98.43 (13) |
| C4—C2—C3—F3 | −63.63 (14) | N2—C8—C9—C14 | 42.35 (16) |
| O1—C2—C4—C5 | −52.75 (14) | C4—C8—C9—C14 | −77.28 (15) |
| N1—C2—C4—C5 | −174.22 (10) | C14—C9—C10—C11 | 1.30 (18) |
| C3—C2—C4—C5 | 67.91 (13) | C8—C9—C10—C11 | −174.60 (11) |
| O1—C2—C4—C8 | 66.82 (13) | C9—C10—C11—C12 | −1.85 (19) |
| N1—C2—C4—C8 | −54.65 (13) | C10—C11—C12—O4 | −178.99 (12) |
| C3—C2—C4—C8 | −172.52 (10) | C10—C11—C12—C13 | 0.62 (19) |
| C6—O3—C5—O2 | 11.0 (3) | O4—C12—C13—C14 | −179.24 (12) |
| C6'—O3—C5—O2 | −18.2 (6) | C11—C12—C13—C14 | 1.14 (19) |
| C6—O3—C5—C4 | −166.3 (2) | C12—C13—C14—C9 | −1.7 (2) |
| C6'—O3—C5—C4 | 164.5 (6) | C10—C9—C14—C13 | 0.48 (19) |
| C2—C4—C5—O2 | 43.13 (17) | C8—C9—C14—C13 | 176.20 (12) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O2 | 0.853 (19) | 1.987 (19) | 2.7383 (13) | 146.3 (17) |
| O1—H1···S1i | 0.853 (19) | 3.029 (19) | 3.4858 (11) | 115.8 (15) |
| O4—H4···O5ii | 0.835 (18) | 1.873 (19) | 2.7066 (14) | 175.6 (19) |
| O5—H5···S1iii | 0.84 | 2.36 | 3.1970 (11) | 175 |
| N1—H1A···S1iii | 0.855 (17) | 2.583 (18) | 3.4307 (12) | 171.5 (14) |
| N2—H2A···O1iv | 0.814 (14) | 2.368 (15) | 3.1701 (15) | 168.7 (14) |
Symmetry codes: (i) −x+1, y−1/2, −z+1/2; (ii) x, y, z−1; (iii) −x+1, −y+1, −z+1; (iv) −x+1, y+1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: OM2424).
References
- Brier, S., Lemaire, D., Debonis, S., Forest, E. & Kozielski, F. (2004). Biochemistry, 43, 13072–13082. [DOI] [PubMed]
- Cochran, J. C., Gatial, J. E., Kapoor, T. M. & Gilbert, S. P. (2005). J. Biol. Chem. 280, 12658–12667. [DOI] [PMC free article] [PubMed]
- Hermann, B., Erwin, H. & Hansjorg, K. (2003). US Patent No. 2 003 176 284.
- Moran, M. M., Fanger, C., Chong, J. A., McNamara, C., Zhen, X. G. & Mandel-Brehm, J. (2007). WO Patent No. 2 007 073 505.
- Rigaku (2009). CrystalClear and CrystalStructure Rigaku/MSC, The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Song, X.-P., Li, G.-C., Wu, C.-Z. & Yang, F.-L. (2010). Acta Cryst. E66, o1083. [DOI] [PMC free article] [PubMed]
- Ulrich, H. (2004). US Patent No. 2 004 033 897.
- Zorkun, I. S., Sarac, S., Celebi, S. & Erol, K. (2006). Bioorg. Med. Chem. 14, 8582–8589. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811015376/om2424sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811015376/om2424Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811015376/om2424Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



