Abstract
The structure of the racemic title compound, C10H15NO4, consists of a tricyclic skeleton comprising a six-membered piperidine ring and five-membered isoxazolidine and tetrahydrofuran rings. The piperidine ring adopts a distorted chair conformation, while the isoxazolidine and tetrahydrofuran rings have envelope conformations.
Related literature
For related piperidine geometry, see: Parkin et al. (2004 ▶). For bicyclic polyhydroisoxazolopyridines, see: Banerji et al. (2006 ▶); Carmona et al. (2009 ▶). For literature related to cycloaddition reactions of cyclic nitrones, see: Ali & Wazeer (1988 ▶); Ali et al. (1988 ▶); Merino (2004 ▶); Chandrasekhar (2005 ▶); Moosa & Ali (2009 ▶, 2010 ▶). For the natural product SB-219383 and its inhibitory activity against tyrosyl tRNA sythetase, see: Houge-Frydrych et al. (2000 ▶); Stefanska et al. (2000 ▶).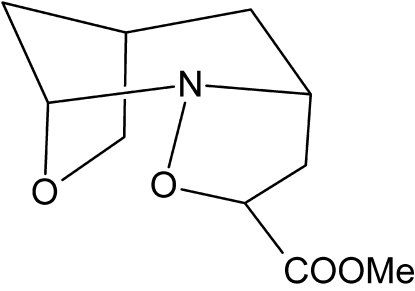
Experimental
Crystal data
C10H15NO4
M r = 213.23
Monoclinic,

a = 11.213 (3) Å
b = 7.1075 (18) Å
c = 12.910 (3) Å
β = 91.546 (5)°
V = 1028.4 (4) Å3
Z = 4
Mo Kα radiation
μ = 0.11 mm−1
T = 294 K
0.20 × 0.10 × 0.05 mm
Data collection
Bruker SMART APEX area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.979, T max = 0.995
13498 measured reflections
2563 independent reflections
1567 reflections with I > 2σ(I)
R int = 0.052
Refinement
R[F 2 > 2σ(F 2)] = 0.057
wR(F 2) = 0.155
S = 1.03
2563 reflections
185 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.24 e Å−3
Δρmin = −0.22 e Å−3
Data collection: SMART (Bruker, 2008 ▶); cell refinement: SAINT (Bruker, 2008 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: SHELXTL (Sheldrick, 2008 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053681101484X/om2422sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053681101484X/om2422Isup2.hkl
Supplementary material file. DOI: 10.1107/S160053681101484X/om2422Isup3.mol
Supplementary material file. DOI: 10.1107/S160053681101484X/om2422Isup4.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors acknowledge King Fahd University of Petroleum and Minerals, Dhahran, Saudi Arabia, for financial support.
supplementary crystallographic information
Comment
1,3-Dipolar cycloaddition reaction of cyclic nitrones with alkenes shows greater stereoselectivity and reactivity compared to their acyclic counterparts for applications in the synthesis of natural products (Merino, 2004; Moosa & Ali, 2009, 2010; Ali et al., 1988; Ali & Wazeer, 1988). Our interest in developing a synthetic methodology to construct the ring skeleton present in a natural product called SB-219383 (Houge-Frydrych et al., 2000), first member of a new class of compounds having inhibitory activity against tyrosyl tRNA sythetase (Stefanska et al., 2000), led to explore the synthesis and cycloaddition of the bicyclic nitrone 1-oxa-5,6-dehydro-6-aza-bicyclo[3,2,1]heptane 6-oxide with methyl acrylate. The structure of a recemic sample of the cycloadduct is reported here. It consists of a tricyclic skeleton corresponding to a cis-invertomer with the nitrogen lone pair in axial position. The two C—N bond lengths of the piperidine ring are N1—C9: 1.453 (2) Å and N1—C5: 1.473 (2) Å. The former bond is shorter presumably as a result of anomeric effect (Chandrasekhar, 2005), inducing a partial double bond character. The bond distances are consistent with those reported for piperidine (Parkin et al., 2004) and bicyclic polyhydroisoxazolopyridines (Banerji et al., 2006; Carmona et al., 2009). The piperidine ring adopts a distorted chair conformation likely due to the strain of the 5-membered rings. Angular constraints imposed by the five-membered tetrahydrofuran ring skeleton led to the squeezing of the C7—C8—C9 angle to 97.8 (2)°; the corresponding bond angle in piperidine, the parent six-membered heterocyclic, is 110.21 (7)°. The angles C5—N1—C9, N1—C5—C6 and C5—C6—C7 on the other end of the six-membered ring were expanded to 114.1 (2)°, 112.3 (2)° and 113.7 (2)°, respectively, while the corresponding angles in piperidine are 111.04 (7)°, 109.84 (7)° and 110.70 (7)° (Parkin et al., 2004). The destabilizing diaxial interactions among the three axially oriented substituents in the piperidine ring is somewhat relieved by moving outward from their ideal axial positions. This leads to a flattening of the chair at N1, C5 and C6. The isoxazolidine ring adopts an envelope conformation with N1, O3, C3 and C4 essentially in the plane while C5 is 0.482 (3) Å out of the plane. The dihedral angle between the previous plane and the plane N1—C5—C4 is 31.9 (2)°. The tetrahydrofuran ring has also an envelope conformation with C8 at 0.733 (3) Å out of the plane C9—O4—C10—C7 which has a dihedral angle of 47.0 (2)° with the plane C7—C8—C9.
Experimental
To a stirred solution of N-hydroxy-4-piperidinemethanol (0.39 g, 3.0 mmol) in chloroform (20 ml) was added mercuric oxide (0.65 g, 12 mmol) and anhydrous magnesium sulfate (150 mg) in 10 min. The reaction mixture was then stirred at room temperature for 6 h. The mixture was filtered over a bed of magnesium sulfate to obtain a solution of the bicyclic nitrone 1-oxa-5,6-dehydro-6-aza-bicyclo[3,2,1]heptane 6-oxide which was used without isolation. The solution of the nitrone and methyl acrylate (2 ml) was stirred at room temperature for 4 h. After removal of the solvent and excess methyl acrylate, the reaction mixture was concentrated and the residual liquid was chromatographed over silica using 9:1 ether/methanol mixture as an eluant to give the cycloadduct as a solid (0.28 g, 44%). Colorless blocks were obtained after crystallization at 0°C from ether/CH2Cl2 (9/1) mixture.
Refinement
Methyl H atoms were included as a rigid group and refined using a riding model with C—H = 0.96 Å and Uiso(H) = 1.5Ueq(C). All other H atoms were located in a difference Fourier map and refined isotropically.
Figures
Fig. 1.
The molecular structure with atom labels and 30% probability displacement ellipsoids for non-H atoms.
Crystal data
| C10H15NO4 | F(000) = 456 |
| Mr = 213.23 | Dx = 1.377 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 13498 reflections |
| a = 11.213 (3) Å | θ = 1.8–28.4° |
| b = 7.1075 (18) Å | µ = 0.11 mm−1 |
| c = 12.910 (3) Å | T = 294 K |
| β = 91.546 (5)° | Block, colourless |
| V = 1028.4 (4) Å3 | 0.20 × 0.10 × 0.05 mm |
| Z = 4 |
Data collection
| Bruker SMART APEX area-detector diffractometer | 2563 independent reflections |
| Radiation source: normal-focus sealed tube | 1567 reflections with I > 2σ(I) |
| graphite | Rint = 0.052 |
| ω scans | θmax = 28.4°, θmin = 1.8° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −14→14 |
| Tmin = 0.979, Tmax = 0.995 | k = −9→9 |
| 13498 measured reflections | l = −17→17 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.057 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.155 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0701P)2 + 0.2128P] where P = (Fo2 + 2Fc2)/3 |
| 2563 reflections | (Δ/σ)max = 0.002 |
| 185 parameters | Δρmax = 0.24 e Å−3 |
| 0 restraints | Δρmin = −0.22 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.14231 (14) | 0.9997 (2) | 0.39231 (12) | 0.0433 (4) | |
| O1 | 0.43183 (15) | 0.7626 (3) | 0.60910 (14) | 0.0821 (6) | |
| O2 | 0.24093 (17) | 0.6880 (3) | 0.59172 (15) | 0.0838 (6) | |
| O3 | 0.23751 (14) | 0.86158 (19) | 0.40074 (11) | 0.0559 (4) | |
| O4 | 0.26276 (13) | 1.22561 (19) | 0.29897 (11) | 0.0516 (4) | |
| C1 | 0.4428 (3) | 0.6375 (5) | 0.6972 (2) | 0.0950 (11) | |
| H1A | 0.3834 | 0.6687 | 0.7465 | 0.142* | |
| H1B | 0.5207 | 0.6507 | 0.7290 | 0.142* | |
| H1C | 0.4316 | 0.5099 | 0.6745 | 0.142* | |
| C2 | 0.3263 (2) | 0.7726 (3) | 0.56322 (17) | 0.0514 (5) | |
| C3 | 0.3243 (2) | 0.9186 (3) | 0.47749 (16) | 0.0484 (5) | |
| C4 | 0.2854 (2) | 1.1087 (3) | 0.51816 (18) | 0.0508 (5) | |
| C5 | 0.15285 (19) | 1.1108 (3) | 0.48841 (15) | 0.0458 (5) | |
| C6 | 0.0935 (2) | 1.3043 (3) | 0.47755 (17) | 0.0515 (5) | |
| C7 | 0.1024 (2) | 1.3900 (3) | 0.36965 (17) | 0.0535 (6) | |
| C8 | 0.0548 (2) | 1.2468 (3) | 0.29092 (19) | 0.0542 (6) | |
| C9 | 0.15798 (18) | 1.1088 (3) | 0.29848 (15) | 0.0441 (5) | |
| C10 | 0.2287 (2) | 1.4113 (3) | 0.3337 (2) | 0.0565 (6) | |
| H4 | 0.400 (2) | 0.929 (3) | 0.4479 (16) | 0.056 (6)* | |
| H5 | 0.3284 (19) | 1.202 (3) | 0.4850 (17) | 0.050 (6)* | |
| H6 | 0.301 (2) | 1.116 (3) | 0.590 (2) | 0.069 (7)* | |
| H7 | 0.1119 (18) | 1.042 (3) | 0.5399 (16) | 0.049 (6)* | |
| H8 | 0.011 (2) | 1.290 (3) | 0.4920 (18) | 0.053 (6)* | |
| H9 | 0.1310 (19) | 1.383 (3) | 0.5271 (17) | 0.051 (6)* | |
| H10 | 0.061 (2) | 1.508 (4) | 0.3653 (17) | 0.061 (6)* | |
| H11 | −0.020 (2) | 1.189 (3) | 0.3108 (17) | 0.060 (7)* | |
| H12 | 0.050 (2) | 1.304 (4) | 0.220 (2) | 0.073 (7)* | |
| H13 | 0.1645 (16) | 1.018 (3) | 0.2423 (15) | 0.041 (5)* | |
| H14 | 0.235 (2) | 1.501 (4) | 0.2774 (19) | 0.072 (7)* | |
| H15 | 0.284 (2) | 1.451 (3) | 0.3903 (18) | 0.056 (6)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0489 (10) | 0.0403 (9) | 0.0410 (9) | −0.0017 (7) | 0.0035 (7) | −0.0061 (7) |
| O1 | 0.0499 (10) | 0.1099 (15) | 0.0864 (13) | −0.0060 (9) | −0.0012 (9) | 0.0487 (11) |
| O2 | 0.0686 (12) | 0.0947 (14) | 0.0873 (13) | −0.0279 (11) | −0.0100 (10) | 0.0354 (11) |
| O3 | 0.0732 (11) | 0.0427 (8) | 0.0513 (9) | 0.0081 (7) | −0.0086 (7) | −0.0092 (7) |
| O4 | 0.0545 (9) | 0.0480 (8) | 0.0532 (9) | −0.0020 (7) | 0.0173 (7) | −0.0050 (7) |
| C1 | 0.0693 (18) | 0.124 (3) | 0.091 (2) | 0.0066 (17) | −0.0051 (15) | 0.058 (2) |
| C2 | 0.0501 (13) | 0.0510 (12) | 0.0534 (13) | −0.0031 (10) | 0.0046 (10) | 0.0037 (10) |
| C3 | 0.0473 (12) | 0.0519 (12) | 0.0462 (12) | −0.0021 (10) | 0.0027 (9) | 0.0016 (9) |
| C4 | 0.0619 (14) | 0.0484 (12) | 0.0420 (12) | −0.0078 (11) | −0.0028 (10) | −0.0022 (10) |
| C5 | 0.0555 (13) | 0.0459 (11) | 0.0363 (10) | −0.0072 (9) | 0.0093 (9) | −0.0032 (9) |
| C6 | 0.0563 (14) | 0.0519 (12) | 0.0470 (12) | 0.0013 (11) | 0.0122 (10) | −0.0102 (10) |
| C7 | 0.0668 (15) | 0.0436 (11) | 0.0505 (12) | 0.0114 (11) | 0.0099 (10) | −0.0018 (10) |
| C8 | 0.0569 (14) | 0.0598 (14) | 0.0460 (13) | 0.0079 (11) | 0.0021 (11) | −0.0017 (10) |
| C9 | 0.0495 (12) | 0.0448 (11) | 0.0382 (11) | 0.0015 (9) | 0.0059 (9) | −0.0069 (9) |
| C10 | 0.0726 (16) | 0.0437 (12) | 0.0539 (14) | −0.0049 (11) | 0.0158 (12) | 0.0010 (10) |
Geometric parameters (Å, °)
| N1—O3 | 1.452 (2) | C4—H5 | 0.93 (2) |
| N1—C9 | 1.453 (2) | C4—H6 | 0.94 (3) |
| N1—C5 | 1.473 (2) | C5—C6 | 1.532 (3) |
| O1—C2 | 1.311 (3) | C5—H7 | 0.95 (2) |
| O1—C1 | 1.446 (3) | C6—C7 | 1.526 (3) |
| O2—C2 | 1.197 (3) | C6—H8 | 0.95 (2) |
| O3—C3 | 1.429 (3) | C6—H9 | 0.94 (2) |
| O4—C9 | 1.439 (2) | C7—C10 | 1.509 (3) |
| O4—C10 | 1.449 (3) | C7—C8 | 1.525 (3) |
| C1—H1A | 0.9600 | C7—H10 | 0.96 (2) |
| C1—H1B | 0.9600 | C8—C9 | 1.518 (3) |
| C1—H1C | 0.9600 | C8—H11 | 0.98 (2) |
| C2—C3 | 1.517 (3) | C8—H12 | 1.01 (3) |
| C3—C4 | 1.518 (3) | C9—H13 | 0.976 (19) |
| C3—H4 | 0.95 (2) | C10—H14 | 0.97 (3) |
| C4—C5 | 1.525 (3) | C10—H15 | 0.99 (2) |
| O3—N1—C9 | 108.55 (14) | C6—C5—H7 | 107.9 (12) |
| O3—N1—C5 | 104.91 (14) | C7—C6—C5 | 113.74 (17) |
| C9—N1—C5 | 114.04 (16) | C7—C6—H8 | 107.9 (14) |
| C2—O1—C1 | 116.43 (19) | C5—C6—H8 | 108.0 (13) |
| C3—O3—N1 | 110.24 (14) | C7—C6—H9 | 110.1 (13) |
| C9—O4—C10 | 107.76 (15) | C5—C6—H9 | 106.8 (13) |
| O1—C1—H1A | 109.5 | H8—C6—H9 | 110.3 (19) |
| O1—C1—H1B | 109.5 | C10—C7—C8 | 100.20 (18) |
| H1A—C1—H1B | 109.5 | C10—C7—C6 | 113.9 (2) |
| O1—C1—H1C | 109.5 | C8—C7—C6 | 108.17 (19) |
| H1A—C1—H1C | 109.5 | C10—C7—H10 | 110.8 (13) |
| H1B—C1—H1C | 109.5 | C8—C7—H10 | 112.4 (14) |
| O2—C2—O1 | 123.6 (2) | C6—C7—H10 | 110.9 (13) |
| O2—C2—C3 | 124.9 (2) | C9—C8—C7 | 97.78 (18) |
| O1—C2—C3 | 111.22 (19) | C9—C8—H11 | 111.8 (14) |
| O3—C3—C2 | 107.94 (18) | C7—C8—H11 | 113.3 (13) |
| O3—C3—C4 | 107.17 (17) | C9—C8—H12 | 110.2 (14) |
| C2—C3—C4 | 110.81 (18) | C7—C8—H12 | 110.5 (15) |
| O3—C3—H4 | 110.3 (13) | H11—C8—H12 | 112 (2) |
| C2—C3—H4 | 110.7 (13) | O4—C9—N1 | 114.92 (16) |
| C4—C3—H4 | 109.9 (13) | O4—C9—C8 | 104.39 (17) |
| C3—C4—C5 | 102.07 (17) | N1—C9—C8 | 106.81 (17) |
| C3—C4—H5 | 108.5 (13) | O4—C9—H13 | 108.0 (11) |
| C5—C4—H5 | 113.0 (13) | N1—C9—H13 | 106.1 (11) |
| C3—C4—H6 | 110.0 (15) | C8—C9—H13 | 117.0 (11) |
| C5—C4—H6 | 113.8 (15) | O4—C10—C7 | 105.13 (18) |
| H5—C4—H6 | 109 (2) | O4—C10—H14 | 110.0 (15) |
| N1—C5—C4 | 105.22 (16) | C7—C10—H14 | 112.6 (15) |
| N1—C5—C6 | 112.30 (17) | O4—C10—H15 | 108.9 (13) |
| C4—C5—C6 | 116.70 (19) | C7—C10—H15 | 112.1 (13) |
| N1—C5—H7 | 106.4 (12) | H14—C10—H15 | 108 (2) |
| C4—C5—H7 | 107.7 (13) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: OM2422).
References
- Ali, S. A., Khan, J. H. & Wazeer, M. I. M. (1988). Tetrahedron, 44, 5911–5920.
- Ali, S. A. & Wazeer, M. I. M. (1988). J. Chem. Soc. Perkin Trans. 1, pp. 597–605.
- Banerji, A., Bandyopadhyay, D., Sengupta, P., Basak, B., Prange, T. & Neuman, A. (2006). Tetrahedron Lett. 47, 3827–3830.
- Bruker, (2008). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Carmona, D., Lamata, M. P., Viguri, F., Rodriguez, R., Lahoz, F. J., Fabra, M. J. & Oro, L. A. (2009). Tetrahedron Asymmetry, 20, 1197–1205.
- Chandrasekhar, S. (2005). Arkivoc, xiii, 37–66.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Houge-Frydrych, C. S. V., Readshaw, S. A. & Bell, D. J. (2000). J. Antibiot. 53, 351–356. [DOI] [PubMed]
- Merino, P. (2004). Science of Synthesis, Vol. 27, Heteroatom Analogues of Aldehydes and Ketones, edited by A. Padwa, pp. 511–580. Stuttgart: Thieme.
- Moosa, B. A. & Ali, S. A. (2009). Tetrahedron, 65, 8231–8243.
- Moosa, B. A. & Ali, S. A. (2010). Arkivoc, x, 132–148.
- Parkin, A., Oswald, I. D. H. & Parsons, S. (2004). Acta Cryst. B60, 219–227. [DOI] [PubMed]
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Stefanska, A. L., Coates, N. J., Mensah, L. M., Pope, A. J., Ready, S. J. & Warr, S. R. (2000). J. Antibiot. 53, 345–350. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053681101484X/om2422sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053681101484X/om2422Isup2.hkl
Supplementary material file. DOI: 10.1107/S160053681101484X/om2422Isup3.mol
Supplementary material file. DOI: 10.1107/S160053681101484X/om2422Isup4.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



