Abstract
In the title compound, C10H13NO2S, the thiophene and isoxazoline rings are almost coplanar, the dihedral angle between their least-squares planes being 2.08 (1)°. The O—H atoms of the methyl hydroxy group and the N atom of the isoxazole ring are orientated in the same direction to allow for the formation of intermolecular O—H⋯N hydrogen bonds that lead to a supramolecular chain along the a axis.
Related literature
For the synthesis, biological activity and mode of action of herbicides, see; Ryu et al. (2005 ▶); Hwang et al. (2005 ▶); Koo et al. (2007 ▶); Koo & Hwang (2008 ▶). For relevant reviews of herbicides, see; Boger et al. (2002 ▶); Bryant & Bite (2010 ▶).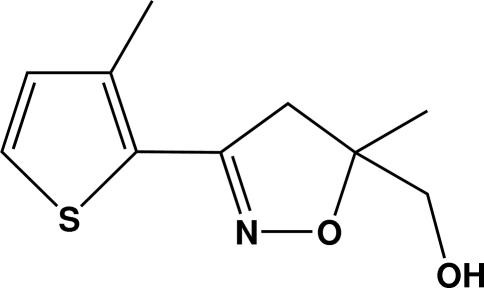
Experimental
Crystal data
C10H13NO2S
M r = 211.27
Orthorhombic,

a = 7.3672 (9) Å
b = 8.8534 (11) Å
c = 16.0632 (19) Å
V = 1047.7 (2) Å3
Z = 4
Mo Kα radiation
μ = 0.28 mm−1
T = 296 K
0.39 × 0.20 × 0.11 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.898, T max = 0.970
11038 measured reflections
2619 independent reflections
2096 reflections with I > 2σ(I)
R int = 0.025
Refinement
R[F 2 > 2σ(F 2)] = 0.047
wR(F 2) = 0.149
S = 1.08
2619 reflections
127 parameters
H-atom parameters constrained
Δρmax = 0.47 e Å−3
Δρmin = −0.36 e Å−3
Absolute structure: Flack (1983 ▶), 1087 Friedel pairs
Flack parameter: 0.02 (14)
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: XP in SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811011639/tk2733sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811011639/tk2733Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O13—H13⋯N7i | 0.82 | 2.17 | 2.905 (3) | 150 |
Symmetry code: (i)  .
.
Acknowledgments
This work was supported by the R&D Program of MKE/KEIT [10035240, Development of new herbicides for resistant weeds with mutated genes].
supplementary crystallographic information
Comment
Weed control is very important for the improvment of agricultural efficiency Boger et al., 2002; Bryant et al., 2010). A number of herbicides have been used for the purpose of weed killing. Recently a new isoxazoline herbicide MRC-01 has been developed (Ryu et al., 2005; Hwang et al., 2005; Koo et al., 2007; Koo & Hwang, 2008; Bryant & Bite, 2010). MRC-01 was synthesized by the reaction of [5-methyl-3-(3-methylthiophen -2-yl)-4,5-dihydroisoxazol-5-yl]methanol and 2,6-difluorobenzylbromide in the presence of base. The key intermediate [5-methyl-3-(3-methylthiophen-2-yl)-4,5-dihydroisoxazol-5-yl]methanol was used as racemic compound but could be separated into enantiomers by employing chiral HPLC column technology. Herein, we report the crystal structure of title compound (Fig. 1). The thiophene ring and the isoxazole ring are almost coplanar with the dihedral angle being 2.08 (1) °. The conformation of the O—H of the methyl hydroxy group and the N atom of the isoxazole ring are in the same direction to allow intermolecular hydrogen bonds to form. In the crystal structure (Fig. 2), the molecules are linked by these O—H···N hydrogen bonds into a one-dimensional chain running along the a axis.
Experimental
The title compound was obtained by a chiral separation of racemic [5-methyl-3-(3-methylthiophen-2-yl)-4,5-dihydroisoxazol-5-yl]methanol employing chiral prep-HPLC under the condition shown below. HPLC conditions: Column: (R,R) WHELK-01 (25 cm x 10.0 mm). Regis.Co.; Eluent: 25% 2-propanol + 75% n-hexane; Flow Rate 4.0 ml/min; Detection: 254 nm; Injection volume: 0.1 ml. The first eluting fraction was concentrated under reduced pressure to provide the title compound [α]D -59.95 (c = 1, dichloromethane). Single crystals suitable for X-ray diffraction were prepared by recrystallization from its ethyl acetate solution at room temperature.
Refinement
All hydrogen atoms were placed in calculated positions using a riding model, with C—H = 0.93–0.97 Å and O—H = 0.82 Å, and with Uiso(H) = 1.2–1.5 Ueq(C, O).
Figures
Fig. 1.
The molecular structure of the title compound with the atom numbering scheme. Displacement ellipsoids are drawn at the 30% probability level. H atoms are presented as a small spheres of arbitrary radius.
Fig. 2.
The molecular packing structure of the title compound, viewed down the c axis showing the O—H···N hydrogen bonds as dashed lines.
Crystal data
| C10H13NO2S | F(000) = 448 |
| Mr = 211.27 | Dx = 1.339 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 4205 reflections |
| a = 7.3672 (9) Å | θ = 2.5–26.1° |
| b = 8.8534 (11) Å | µ = 0.28 mm−1 |
| c = 16.0632 (19) Å | T = 296 K |
| V = 1047.7 (2) Å3 | Block, silver |
| Z = 4 | 0.39 × 0.20 × 0.11 mm |
Data collection
| Bruker APEXII CCD diffractometer | 2619 independent reflections |
| Radiation source: fine-focus sealed tube | 2096 reflections with I > 2σ(I) |
| graphite | Rint = 0.025 |
| φ and ω scans | θmax = 28.4°, θmin = 2.5° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −6→9 |
| Tmin = 0.898, Tmax = 0.970 | k = −11→11 |
| 11038 measured reflections | l = −21→21 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.047 | H-atom parameters constrained |
| wR(F2) = 0.149 | w = 1/[σ2(Fo2) + (0.086P)2 + 0.1644P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.08 | (Δ/σ)max < 0.001 |
| 2619 reflections | Δρmax = 0.47 e Å−3 |
| 127 parameters | Δρmin = −0.36 e Å−3 |
| 0 restraints | Absolute structure: Flack (1983), 1087 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: 0.02 (14) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | −0.00757 (12) | 0.90745 (7) | 0.64150 (4) | 0.0503 (2) | |
| C2 | −0.0074 (4) | 0.8815 (3) | 0.53306 (13) | 0.0390 (4) | |
| C3 | −0.0070 (5) | 0.7321 (3) | 0.51314 (15) | 0.0446 (5) | |
| C4 | −0.0075 (4) | 0.6395 (3) | 0.58618 (17) | 0.0525 (6) | |
| H4A | −0.0080 | 0.5345 | 0.5841 | 0.063* | |
| C5 | −0.0071 (5) | 0.7169 (3) | 0.65749 (17) | 0.0568 (6) | |
| H5 | −0.0067 | 0.6721 | 0.7099 | 0.068* | |
| C6 | −0.0104 (4) | 1.0139 (2) | 0.47984 (12) | 0.0368 (4) | |
| N7 | −0.0045 (4) | 1.1462 (2) | 0.51234 (11) | 0.0448 (4) | |
| O8 | −0.0178 (4) | 1.25711 (18) | 0.45051 (10) | 0.0490 (5) | |
| C9 | −0.0104 (4) | 1.1857 (3) | 0.36792 (13) | 0.0405 (5) | |
| C10 | −0.0244 (4) | 1.0174 (3) | 0.38670 (13) | 0.0423 (6) | |
| H10A | −0.1394 | 0.9763 | 0.3679 | 0.051* | |
| H10B | 0.0740 | 0.9615 | 0.3610 | 0.051* | |
| C11 | 0.1691 (4) | 1.2299 (4) | 0.3280 (2) | 0.0630 (9) | |
| H11A | 0.1707 | 1.3369 | 0.3182 | 0.094* | |
| H11B | 0.1828 | 1.1774 | 0.2760 | 0.094* | |
| H11C | 0.2672 | 1.2033 | 0.3645 | 0.094* | |
| C12 | −0.1672 (4) | 1.2500 (4) | 0.31872 (17) | 0.0452 (6) | |
| H12A | −0.1627 | 1.3593 | 0.3219 | 0.054* | |
| H12B | −0.1527 | 1.2219 | 0.2607 | 0.054* | |
| O13 | −0.3406 (2) | 1.2007 (2) | 0.34646 (11) | 0.0489 (5) | |
| H13 | −0.3554 | 1.2261 | 0.3951 | 0.073* | |
| C14 | −0.0073 (5) | 0.6707 (3) | 0.42966 (17) | 0.0507 (6) | |
| H14A | −0.0080 | 0.7518 | 0.3900 | 0.076* | |
| H14B | −0.1134 | 0.6093 | 0.4220 | 0.076* | |
| H14C | 0.0994 | 0.6101 | 0.4216 | 0.076* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0500 (4) | 0.0629 (4) | 0.0381 (3) | 0.0022 (4) | −0.0002 (4) | 0.0035 (2) |
| C2 | 0.0289 (10) | 0.0484 (11) | 0.0399 (9) | 0.0021 (13) | −0.0002 (12) | 0.0035 (8) |
| C3 | 0.0314 (11) | 0.0483 (12) | 0.0540 (13) | 0.0039 (14) | 0.0010 (14) | 0.0021 (9) |
| C4 | 0.0406 (12) | 0.0495 (13) | 0.0673 (16) | 0.0052 (15) | −0.0003 (17) | 0.0198 (11) |
| C5 | 0.0458 (13) | 0.0715 (17) | 0.0529 (13) | 0.0036 (18) | 0.0057 (16) | 0.0222 (12) |
| C6 | 0.0320 (10) | 0.0401 (10) | 0.0383 (10) | −0.0011 (13) | −0.0029 (12) | −0.0017 (8) |
| N7 | 0.0555 (12) | 0.0430 (9) | 0.0361 (8) | 0.0010 (14) | −0.0045 (13) | 0.0004 (7) |
| O8 | 0.0711 (14) | 0.0379 (8) | 0.0380 (8) | 0.0026 (11) | −0.0086 (11) | −0.0009 (6) |
| C9 | 0.0412 (11) | 0.0465 (11) | 0.0340 (9) | −0.0041 (14) | −0.0004 (13) | 0.0005 (8) |
| C10 | 0.0525 (15) | 0.0389 (10) | 0.0355 (9) | 0.0021 (12) | −0.0024 (12) | −0.0035 (8) |
| C11 | 0.0416 (15) | 0.079 (2) | 0.069 (2) | −0.0069 (16) | 0.0063 (15) | 0.0075 (18) |
| C12 | 0.0408 (14) | 0.0556 (15) | 0.0391 (12) | 0.0012 (13) | 0.0003 (12) | 0.0045 (11) |
| O13 | 0.0397 (9) | 0.0606 (11) | 0.0462 (10) | −0.0008 (9) | 0.0025 (8) | −0.0063 (9) |
| C14 | 0.0455 (12) | 0.0429 (12) | 0.0637 (14) | 0.0026 (15) | 0.0004 (16) | 0.0016 (10) |
Geometric parameters (Å, °)
| S1—C5 | 1.706 (3) | C9—C11 | 1.522 (4) |
| S1—C2 | 1.757 (2) | C9—C10 | 1.524 (3) |
| C2—C3 | 1.361 (3) | C10—H10A | 0.9700 |
| C2—C6 | 1.451 (3) | C10—H10B | 0.9700 |
| C3—C4 | 1.431 (3) | C11—H11A | 0.9600 |
| C3—C14 | 1.447 (3) | C11—H11B | 0.9600 |
| C4—C5 | 1.335 (4) | C11—H11C | 0.9600 |
| C4—H4A | 0.9300 | C12—O13 | 1.422 (3) |
| C5—H5 | 0.9300 | C12—H12A | 0.9700 |
| C6—N7 | 1.283 (3) | C12—H12B | 0.9700 |
| C6—C10 | 1.500 (3) | O13—H13 | 0.8200 |
| N7—O8 | 1.400 (2) | C14—H14A | 0.9600 |
| O8—C9 | 1.470 (3) | C14—H14B | 0.9600 |
| C9—C12 | 1.511 (4) | C14—H14C | 0.9600 |
| C5—S1—C2 | 91.14 (12) | C6—C10—H10A | 111.3 |
| C3—C2—C6 | 130.3 (2) | C9—C10—H10A | 111.3 |
| C3—C2—S1 | 111.12 (17) | C6—C10—H10B | 111.3 |
| C6—C2—S1 | 118.57 (17) | C9—C10—H10B | 111.3 |
| C2—C3—C4 | 111.4 (2) | H10A—C10—H10B | 109.2 |
| C2—C3—C14 | 125.7 (2) | C9—C11—H11A | 109.5 |
| C4—C3—C14 | 123.0 (2) | C9—C11—H11B | 109.5 |
| C5—C4—C3 | 114.1 (2) | H11A—C11—H11B | 109.5 |
| C5—C4—H4A | 122.9 | C9—C11—H11C | 109.5 |
| C3—C4—H4A | 122.9 | H11A—C11—H11C | 109.5 |
| C4—C5—S1 | 112.24 (19) | H11B—C11—H11C | 109.5 |
| C4—C5—H5 | 123.9 | O13—C12—C9 | 114.0 (2) |
| S1—C5—H5 | 123.9 | O13—C12—H12A | 108.7 |
| N7—C6—C2 | 119.83 (19) | C9—C12—H12A | 108.7 |
| N7—C6—C10 | 112.94 (18) | O13—C12—H12B | 108.7 |
| C2—C6—C10 | 127.22 (19) | C9—C12—H12B | 108.7 |
| C6—N7—O8 | 110.42 (17) | H12A—C12—H12B | 107.6 |
| N7—O8—C9 | 109.65 (16) | C12—O13—H13 | 109.5 |
| O8—C9—C12 | 106.4 (2) | C3—C14—H14A | 109.5 |
| O8—C9—C11 | 107.6 (2) | C3—C14—H14B | 109.5 |
| C12—C9—C11 | 110.33 (19) | H14A—C14—H14B | 109.5 |
| O8—C9—C10 | 103.84 (16) | C3—C14—H14C | 109.5 |
| C12—C9—C10 | 114.8 (2) | H14A—C14—H14C | 109.5 |
| C11—C9—C10 | 113.2 (3) | H14B—C14—H14C | 109.5 |
| C6—C10—C9 | 102.27 (17) | ||
| C5—S1—C2—C3 | 0.0 (3) | C2—C6—N7—O8 | −177.1 (3) |
| C5—S1—C2—C6 | 179.1 (2) | C10—C6—N7—O8 | 1.6 (4) |
| C6—C2—C3—C4 | −178.8 (3) | C6—N7—O8—C9 | −7.3 (3) |
| S1—C2—C3—C4 | 0.2 (4) | N7—O8—C9—C12 | 131.1 (2) |
| C6—C2—C3—C14 | 0.8 (6) | N7—O8—C9—C11 | −110.7 (3) |
| S1—C2—C3—C14 | 179.8 (3) | N7—O8—C9—C10 | 9.5 (3) |
| C2—C3—C4—C5 | −0.4 (4) | N7—C6—C10—C9 | 4.4 (4) |
| C14—C3—C4—C5 | −180.0 (3) | C2—C6—C10—C9 | −177.1 (3) |
| C3—C4—C5—S1 | 0.4 (4) | O8—C9—C10—C6 | −8.0 (3) |
| C2—S1—C5—C4 | −0.2 (3) | C12—C9—C10—C6 | −123.7 (2) |
| C3—C2—C6—N7 | −177.8 (4) | C11—C9—C10—C6 | 108.3 (3) |
| S1—C2—C6—N7 | 3.3 (4) | O8—C9—C12—O13 | −70.1 (3) |
| C3—C2—C6—C10 | 3.7 (5) | C11—C9—C12—O13 | 173.5 (3) |
| S1—C2—C6—C10 | −175.2 (3) | C10—C9—C12—O13 | 44.2 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O13—H13···N7i | 0.82 | 2.17 | 2.905 (3) | 150 |
Symmetry codes: (i) x−1/2, −y+5/2, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: TK2733).
References
- Boger, P., Wakabayashi, K. & Hirai, K. (2002). Herbicide Classeses in Development (mode of action, targets, genetic engineering, chemistry), edited by P. Boger, K. Wakabayashi & K. Hirai. Berlin, Heidelberg: Springer-Verlag.
- Bruker (2009). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Bryant, R. & Bite, M. (2010). Ag Chem New Compound Review, Vol 28, p. 59. Orpington: Agranova.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Hwang, I. T., Kim, H. R., Jeon, D. J., Hong, K. S., Song, J. H. & Cho, K. Y. (2005). J. Agric. Food Chem. 53, 8639–8643. [DOI] [PubMed]
- Koo, S. J. & Hwang, K. H. (2008). Korea Patent 0814420.
- Koo, S. J., Hwang, K. H., Hwang, I. T., Jeon, D. J. & Kim, H. R. (2007). Proceedings of the 21th Asian Pacific Weed Science Society Conference, pp. 591–601.
- Ryu, E. K., Kim, H. R., Jeon, D. J., Song, J. H., Kim, K. M., Lee, J. N., Kim, H. C. & Hong, K. S. (2005). US Patent 6838416.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811011639/tk2733sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811011639/tk2733Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




