Abstract
A 19-kb DNA region containing genes involved in the biosynthesis of the capsule of Haemophilus influenzae serotype f (Hif) has been cloned and characterized. The Hif cap locus organization is typical of group II capsule biosynthetic loci found in other H. influenzae serotype b bacteria and other gram-negative bacteria. However, the Hif cap locus was not associated with an IS1016 element. Three new open reading frames, Fcs1, Fcs2, and Fcs3, were identified in the Hif capsule-specific region II. The chromosomal location of the Hif cap locus and the organization of the flanking sequences differed significantly from previously described division I H. influenzae serotypes.
Many bacterial species produce an extracellular polysaccharide capsule (CPS), which contributes to the pathogenicity of these organisms. The capsule of Haemophilus influenzae, a human pathogen causing disease in both adults and children, is important in establishing and maintaining invasive disease such as bacteremia and meningitis by conferring bacterial resistance to complement-mediated phagocytosis (28). H. influenzae is serotyped based on the presence of six antigenically distinct CPS (types a to f) (31). H. influenzae strains that lack CPS are classified as nontypeable or nonencapsulated. Each of the six capsule types can be identified by both serologic (31) and molecular methods (5). Nonencapsulated strains fail to react specifically with antisera against capsular serotypes a through f or to hybridize with capsule-specific genes. Encapsulated strains of H. influenzae can be divided further into two phylogenetic divisions, I and II, based on multilocus enzyme electrophoresis typing (30).
Prior to routine immunization of infants with conjugate H. influenzae serotype b (Hib) vaccine, Hib was the most common H. influenzae serotype causing invasive disease (>90%), and H. influenzae serotype f (Hif), although rare, was the second most common (44). With the postvaccine reduction in Hib disease, the proportion of invasive H. influenzae disease caused by Hif rose from 1% in 1989 to 17% in 1994 (40). Invasive Hif disease now contributes to a substantial proportion of all invasive H. influenzae disease (39, 43) and has been associated with infections such as bacteremia, endocarditis, and a mycotic aneurysm (1, 8, 35). Recently, Hif was isolated from a case of rapidly fatal sepsis in an otherwise healthy child (45).
Strong homology of the genes involved in capsule biosynthesis from a number of gram-negative bacteria suggests a common molecular origin. Although the genetic elements of H. influenzae capsule biosynthesis have been most extensively characterized in Hib (13, 14, 19, 34), the H. influenzae cap locus for all six serotypes contains the same three functionally unique regions, I, II, and III (18, 20). Regions I and III are common to all six capsular types and contain genes involved in the exportation and processing of the capsular material. Region I genes (bexDCBA) code for an ATP-driven capsule export apparatus (16). Region II contains serotype-specific biosynthesis genes that appear to be unique to each of the six capsule types (34, 41). Region III genes, hcsA and hcsB, recently identified in Hib, appear to be involved in postpolymerization steps (10, 25, 34). In order to further understand the role of the Hif capsule, we identified and characterized the genetic locus for the capsule biosynthetic genes of H. influenzae serotype f.
Cosmid sequencing and assembly of the Hif cap locus.
A cosmid library of the American Type Culture Collection (Manassas, Va.) H. influenzae serotype f strain 700222 (40) Sau3A-partially digested chromosomal DNA was constructed in Escherichia coli XL1-Blue MR cells using the GigaPack III XL packaging system (Stratagene, La Jolla, Calif.) as described by the manufacturer. Cosmids pTS66 and pTS67 were found to contain DNA specific to region II of the Hif cap locus by PCR for the presence of Hif cap-specific region II DNA using oligonucleotide primers f1 and f2 as previously described (5) (Table 1). pTS66 and pTS67 were further characterized by Southern hybridization analysis (33) using a Hif cap-specific region II probe (f1 to f2) as well as with probes generated by PCR from the Hib cap locus in conserved regions I (Hi-1 to Hi-2) (5) and III (ORF6 [24] to Reg3b; 5′-GCAATGGCACATCATGCAC-3′).
TABLE 1.
Oligonucleotide primer pairs used in RT-PCR of Fcs1, Fcs2, and Fcs3, analysis of the adjacent Hif cap DNA, and as probes in Southern blot hybridization
| Oligonucleotide name | Sequence (5′-3′) | Gene(s) (size [bp]) | Reference |
|---|---|---|---|
| f1 | GCTACTATCCAAGTCCAAATC | Hif cap specific (450) | 5 |
| f2 | CGCAATTATGGAAGAAAGCT | 5 | |
| HI-1 | CGTTTGTATGATGTTGATCCAGAC | bexA (343) | 5 |
| HI-2 | TGTCCATGTCTTCAAAATGATG | 5 | |
| ORF6 | GTTATTACTTGCGTGATCGT | hcsB (311) | 24 |
| Reg3b | GCAATGGCACATCATGCAC | This study | |
| Fcs1-a | CGTGACGCATTGAATAATAAATAC | fcs1 (720) | This study |
| Fcs1-b | CTGTGCCTTAGCATTTGCGGAG | This study | |
| Fcs2-a | GTTAATGCAAAAGATCAAG | fcs2 (1,149) | This study |
| Fcs2-b | CATAGTTATACCATGCTGTAACC | This study | |
| Fcs3-a | CCGGGAATCCATAGAGTTCA | fcs3 (992) | This study |
| Fcs3-b | GAGCATTCTATAATATGCAATGG | This study | |
| Fcs1-2-a | CGCTCCGCAAATGCTAA | fcs1-fcs2 (323) | This study |
| Fcs1-2-b | TTCTTCACTGTTATCAGGGCTAC | This study | |
| Fcs2-3-a | CGATGAATTATCTCTATCTTACAAGG | fcs2-fcs3 (417) | This study |
| Fcs2-3-b | CTGATGGTGTATGAACTCTATG | This study | |
| capfSodC | CATGCGCATTTTCCACGCCAGC | bexA-sodC (875) | This study |
| capfBexA | GGGATTGAGGCGCAATGATTCG | This study | |
| capfMerT | CCTTTTGGGTTGCCATAGCC | merT-sodC (774) | This study |
| SodC 3970-2951 | CACTCAGATCATCCTGCTCC | This study | |
| capf region III | CAGTACGAGTGGTATTTCAG | hcsB-HI1637 (700) | This study |
| HI1637 | AAATTTCCATTATGGGAAACG | This study |
Cosmid pTS66 was sent to Lark Technologies, Houston, Tex., for sequencing using randomly sheared cosmid DNA in a pUC19 shotgun library (4). DNA sequencing was performed using a BigDye Terminator Cycle Sequencing Ready Reaction kit, with AmpliTaq DNA polymerase FS (Applied Biosystems, Foster City, Calif.) and analyzed using an ABI377 apparatus from Lark Technologies. Sequencing reads provided approximately 8.8-fold coverage of the cosmid clone. Oligonucleotide primers were created to close gaps using PCR and sequencing directly from a second cosmid, pTS67, using primers f1 and f2 (5) and primer walking as needed.
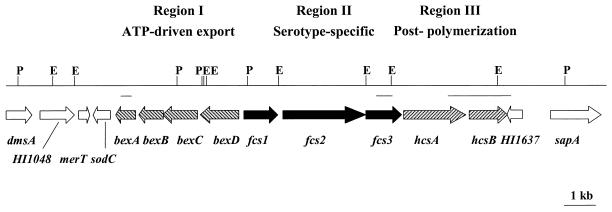
Sequencing and computer assembly using the Phred-Phrap package (CodonCode Corporation, Dedham, Mass.) and LaserGene version 5.03 (DNASTAR, Inc., Madison, Wis.) of pTS66 and pTS67 resulted in a contiguous region of 19,576 bp from the Hif chromosome. Nucleotide and amino acid homology comparisons using GenBank DNA and protein sequence databases of the National Center for Biotechnology Information BLAST network server (3) and TBLASTX analysis (3) identified a total of 15 open reading frames (ORFs), 9 of which appear to be specifically associated with the Hif cap locus (Fig. 1).
FIG. 1.
Genetic organization of the Hif capsule locus and adjacent DNA. Arrows indicate genes and ORFs. Region I contains four genes, on the opposite strand, homologous to those found in Hia and Hib, bexDCBA (left-hatched arrows). Region II is comprised of three serotype-specific genes designated fcs1, fcs2, and fcs3 (black arrows). Region III has two genes, hcsA and hcsB, also found in the cap locus of Hib (right-hatched arrows). Six additional ORFs outside of the Hif cap locus are noted with white arrows. Vertical lines show cleavage sites for restriction endonucleases EcoRI (E) and PstI (P). Horizontal lines below the restriction map show the locations and sizes of the probes used to identify cosmid clones with the Hif cap locus.
From analysis of the sequence data and ORFs, it is apparent that the Hif cap locus is composed of three distinct regions (I, II, and III), a genetic organization common to the Hib cap locus as well as other group II capsule biosynthetic clusters (Fig. 1). However, the capsule locus of the serotype f capsule includes genes that are not found in previously reported capsule loci of H. influenzae serotypes a and b (7, 34, 41), and it is not associated with an IS1016 element. The DNA that flanks the Hif cap locus does not appear to be directly involved in capsule production, although functions associated with the capsule cannot be excluded.
Sequence analysis of Hif cap locus regions I and III.
Region I contains four ORFs with homology to Hib, bexA, bexB, bexC, and bexD (Fig. 1), as well as other region I capsule genes from E. coli, Neisseria meningitidis, Actinobacillus pleuropneumoniae, and Mannheimia haemolytica (34). Because the encoded polypeptides from each of these four genes were nearly identical (89 to 94% identity) to the region I gene products from Hib, they were named bexDCBA. The BexD protein in Hif appeared to be slightly less conserved than BexA, BexB, and BexC, with only 89% identity to the Hib counterpart.
The nucleotide sequence in region III contained two genes, hcsA and hcsB (for Haemophilus capsule synthesis), recently sequenced from the Hib capsule locus (34). HcsA and HcsB are highly conserved between the Hib and Hif capsule loci at 97 and 91% similarity, respectively (Fig. 1).
Sequence analysis of Hif cap locus region II.
Region II, the central, serotype-specific region of the Hif cap locus, included three ORFs. The G+C content of the DNA in region II was 26%, significantly lower than the 38 and 39.7% G+C content of regions I and III and the overall G+C content of H. influenzae DNA. This may indicate that region II DNA was more recently acquired by the bacterium. The three ORFs had no homology with Hib Bcs1 to Bsc4 and were designated Fcs1, Fcs2, and Fcs3 for f capsule-specific gene products (Fig. 1).
The deduced amino acid sequence of Fcs1 was compared to those of other known sequences using BLAST (2), and homology with other capsule polysaccharide genes was detected. Fcs1 is 68% similar to Cps1A, capsular polysaccharide synthesis A from A. pleuropneumoniae (GenBank accession no. AAM69355), 67% similar to LcbA, a putative N. meningitidis serogroup L region D capsule protein (GenBank accession no. AAF21950), and 54% similar to the N. meningitidis serogroup X strain M7575 putative capsule biosynthesis protein XcbA (39a). Less similarity (49%) was seen between Fcs1 and the C-terminal half of SacB, an N. meningitidis serogroup A capsule biosynthesis gene (GenBank accession no. AAC38286) (36).
Sequence homology search with Fcs2 for known proteins detected two putative conserved domains. The N-terminal amino acids 8 to 178 of Fcs2 were 98.8% identical to the consensus sequence from the conserved domain database (26, 27) protein family glycosyltransferase 2, Pfam00535. This is a highly diverse family of proteins with the capacity to transfer sugar from UDP-glucose, UDP-N-acetylgalactosamine, GDP-mannose, and CDP-abequose to a range of substrates that includes cellulose, dolichol phosphate, and teichoic acids. C-terminal amino acids 516 to 887 were 94.1% identical to the consensus sequence from COG1887, a cluster of orthologous groups (COG) (37, 38) of putative glycosl-glycerophosphate transferases involved in teichoic acid biosynthesis.
CDART (conserved domain architecture retrieval tool) (11) identified six proteins with similar domain architecture. Fcs2 is 54% similar to Cps1B and 45% similar to Cps1C, capsular polysaccharide synthesis-B and -C from A. pleuropneumoniae (GenBank accession no. AAM69356 and AAM69358, respectively). Fcs2 is 46% similar to EpsJ, a Lactococcus lactis subsp. cremoris glycosyltransferase involved in synthesis of the polysaccharide backbone (GenBank accession no. NP_053024) (42), 48% similar to GgaB, a Bacillus subtilis minor teichoic acids biosynthesis protein (GenBank accession no. P46918) (21), and 40% similar to LcbB, a putative bifunctional polymerase from N. meningitidis serogroup L (GenBank accession no. AAF21951). Fcs2 is also 41% similar to a hypothetical protein (Acs3), the third of four ORFs found in region II of H. influenzae serotype a strain RM107 (GenBank accession no. S49240). Finally, no sequence homology was found for the third ORF, Fcs3, with known proteins as detected by BLAST (2). Functional characterization of Fcs1, Fcs2, and Fcs3 remains to be established.
Region II fcs1, fcs2, and fcs3 transcription.
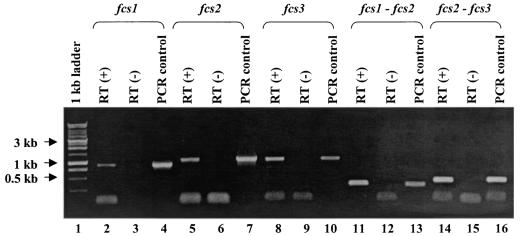
To determine whether fcs1, fcs2, and fcs3 were transcribed and organized as an operon, we performed reverse transcription-PCR (RT-PCR) experiments with whole-cell RNA obtained from exponential broth cultures of Hif 700222 with RNeasy (Qiagen, Valencia, Calif.) and SuperScript II reverse transcriptase (Invitrogen, Carlsbad, Calif.) and PCR reagents from Applied Biosystems at an annealing temperature of 55°C. Transcription of fcs1 (Fig. 2, lane 2), fcs2 (Fig. 2, lane 5), and fcs3 (Fig. 2, lane 8) was confirmed by RT-PCR using primers designed from their coding sequences (Table 1). An RT-PCR product of 323 bp was amplified between fcs1 and fcs2 using a primer in the coding sequence of fcs2 (Fcs1-2-b) and a second primer in fcs1 (Fcs1-2-a) during the amplification of the cDNA (Fig. 2, lane 11). An RT-PCR product of 417 bp was amplified between fcs2 and fcs3 using a primer in the coding sequence of fcs3 (Fcs2-3-b) and a second in fcs2 (Fcs2-3-a) during the amplification of the cDNA (Fig. 2, lane 14). These results indicate that the three region II genes, fcs1, fcs2, and fcs3, are cotranscribed on the same mRNA.
FIG. 2.
fcs1, fcs2, and fcs3 expression detected in log-phase Hif 700222 cells and shown to be cotranscribed by RT-PCR. Lane 1, 1-kb standard marker (Promega Corp., Madison, Wis.); lanes 3, 6, 9, 12, and 15, RT negative controls; lanes 4, 7, 10, 13, and 16, positive controls with chromosomal DNA from Hif 700222 as template; lane 2, Fcs1-a and Fcs1-b RT (+); lane 5, Fcs2-a and Fcs2-b RT (+); lane 8, Fcs3-a and Fcs3-b RT (+); lane 11, Fcs1-2-a and Fcs1-2-b RT (+); lane 12, Fcs1-2-a and Fcs1-2-b RT (-); lane 13, Fcs1-2-a and Fcs1-2-b as PCR control; lane 14, Fcs2-3-a and Fcs2-3-b RT (+). Primers and product sizes are listed in Table 1.
Chromosomal location of Hif cap locus.
Sequence analysis of the DNA adjacent to the Hif cap locus within cosmid pTS66 identified six additional ORFs, five highly homologous to previously identified ORFs from H. influenzae Rd (GenBank accession no. NC_000907): merT, sapA, HI1048, HI1637, and an incomplete copy of dmsA. An additional gene, sodC, not found in the Rd genome, is located immediately adjacent to bexA and the 5′ junction of the Hif cap locus. The layout of the flanking sequences with respect to the Hif cap locus is shown in Fig. 1. HI1048 and HI1637 are hypothetical proteins (GenBank accession no. AAC22710 and AAC23282, respectively); dmsA (HI1047) encodes an anaerobic dimethyl sulfoxide reductase, chain A protein (GenBank accession no. AAC22706); merT (HI1049) encodes a mercuric ion transport protein (GenBank accession no. AAC22707); sapA (HI1638) encodes a peptide ABC transporter periplasmic protein (GenBank accession no. AAC23285); and sodC (GenBank accession no. M84012) encodes a copper-zinc superoxide dismutase (Cu,Zn-SOD).
Unlike the DNA flanking the Hib cap locus, which is contiguous in the H. influenzae Rd genome, the DNA outside of the Hif cap locus is found in two widely separated sections of the Rd genome. dmsA (HI1047), HI1048, and merT (HI1049) are found in section 100 of the Rd genome, while HI1637 and sapA (HI1638) are in sections 151 and 152, respectively (sodC is not present in the Rd genome). Sections 100 and 151 to 152 are separated by 589,419 bp in Rd (GenBank accession no. NC_000907) (6). This finding was not necessarily unexpected. Rd, formerly an encapsulated H. influenzae serotype d strain, is a division I H. influenzae strain, and all H. influenzae serotype f strains belong to division II. Divisions I and II have been demonstrated to be widely separated phylogenetic divisions (30). Furthermore, sodC is present only in division II and not present in division I strains with the exception of serotype e, which shares features of both divisions I and II (15, 22).
To determine if the layout of the Hif cap locus and flanking sequences found on the cosmid clones was identical in the type f chromosome, PCR analysis of the end junctions of the Hif cap locus was performed using primers specific to region I, sodC, merT, region III, and HI1637 (Table 1) and chromosomal DNA from Hif strain 700222. The same layout of genes was observed in Hif strain 700222 and the sequenced cosmid pTS66 (data not shown).
We looked further at a collection of 69 Hif strains isolated from patients with invasive H. influenzae disease. Isolates were confirmed to be H. influenzae serotype f strains by serologic and molecular methods (5). All 69 Hif isolates were examined by one-colony PCR (32) using primer pairs as described above and were found to have the Hif cap locus in the identical location as that of Hif 700222 and associated with the same flanking genes (data not shown). This suggests that the chromosomal location of the Hif cap locus is highly conserved and is as presented in Fig. 1.
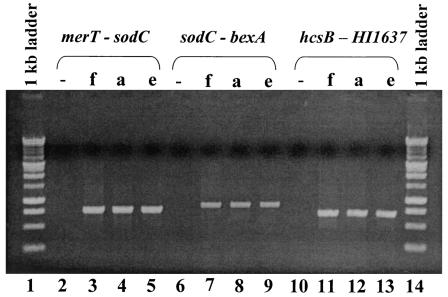
It has been suggested that all division I H. influenzae strains have the same chromosomal cap locus location, between direct repeats of IS1016 (34). It is possible that all cap loci in H. influenzae division II strains are located between sodC and HI1637. Previously, we noted that H. influenzae serotype a strain ATCC 9006 and H. influenzae serotype e strain 8142 did not have the same cap locus chromosomal location shared by most division I H. influenzae strains (34). Although most Hia belong to division I, some have been classified as division II. Hie, while classified as division I, has the greatest genetic distance from all other division I H. influenzae (29) and is the only division I H. influenzae reported to have sodC (22). We also examined the end junctions of Hia strain 9006 and Hie strain 8142 using primers specific to region I, sodC, merT, region III, and HI1637 (Table 1). Both the Hia and the Hie strains had the same PCR profile as the Hif strains, suggesting the identical chromosomal location of the cap loci in these strains (Fig. 3). Further studies will be necessary to determine whether the cap locus location is uniformly conserved among all H. influenzae division II strains (all Hif, and a minority of Hia and Hib) and division I Hie strains.
FIG. 3.
PCR analysis of the end junctions and outside DNA of the Hif cap locus and H. influenzae serotypes a and e. Lanes 1 and 14, 1-kb standard marker (Promega Corp.). Hif 700222 chromosomal DNA was amplified with primer pairs capfMerT and SodC 3970-2951 (lane 3), capfSodC and capfBexA (lane7), and capf region III and HI1637 (lane 11). Hia ATCC 9006 chromosomal DNA was amplified with primer pairs capfMerT and SodC 3970-2951 (lane 4), capfSodC and capfBexA (lane 8), and capf region III and HI1637 (lane 12). Hie ATCC 8142 chromosomal DNA was amplified with primer pairs capfMerT and SodC 3970-2951 (lane 5), capfSodC and capfBexA (lane 9), and capf region III and HI1637 (lane 13). Lanes 2, 6, and 10 are the no-template controls. Primers and product sizes are listed in Table 1.
The location of sodC adjacent to bexA and the capsule locus of H. influenzae NCTC8468, a division II type b strain, have been previously reported (15). Hif sodC is 98.4% identical to the Hib sodC described in detail by Kroll et al. (15) and also located adjacent to bexA. The identical palindromic pairs that match the 11-bp Haemophilus DNA uptake motif seen 19 nucleotides downstream from the stop codon of Hib sodC (GenBank accession no. M84012) (17) are found at the same location downstream from the termination codon in the Hif sodC. The sodC gene encodes for Cu,Zn-SOD, a metallo-enzyme catalyzing the conversion of superoxide radicals into hydrogen peroxide and oxygen (9, 12). Cu,Zn-SOD has been found in the periplasm of a wide range of gram-negative commensal and pathogenic bacteria, including Haemophilus species (22), and its role in virulence has been suggested for pathogenic bacteria (23). Unlike Fe-SODs and Mn-SODs, which are located in the cytosol and have a primary role of minimizing the effects from anaerobic respiration, Cu,Zn-SODs are exported from the cytosol to the periplasm or beyond. Since superoxide generated within the cytosol cannot cross the cytoplasmic membrane, this suggests that Cu,Zn-SODs provide protection from extracellular superoxide production by phagocytic host cells (23).
The possession of a polysaccharide capsule greatly enhances the virulence of H. influenzae and its ability to cause invasive disease. Although the Hib conjugate vaccine has been highly successful in dramatically reducing Hib invasive disease, occurrence of invasive disease due to other capsule serotypes as well as nontypeable H. influenzae has persisted and may be increasing in some populations. The apparent increase of Hif disease in the post-Hib vaccine era suggests the need for further characterization of H. influenzae serotype f. We report the genetic analysis of the Hif cap locus and its surrounding DNA. Future studies to better understand the functional activities of the capsule-specific genes fcs1, fcs2, and fcs3 are warranted.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been deposited in GenBank under the accession no. AF549211.
Editor: J. N. Weiser
REFERENCES
- 1.Adlakha, A., S. H. Yale, R. Patel, R. S. Edson, H. J. Schultz, and A. W. Stanson. 1994. Haemophilus influenzae serotype f: an unusual cause of a mycotic aneurysm in an adult. Mayo Clin. Proc. 69:467-468. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Falla, T. J., D. W. M. Crook, L. N. Brophy, D. Maskell, J. S. Kroll, and E. R. Moxon. 1994. PCR for capsular typing of Haemophilus influenzae. J. Clin. Microbiol. 32:2382-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 7.Follens, A., M. Veiga-da-Cunha, R. Merckx, E. van Schaftingen, and J. van Eldere. 1999. acs1 of Haemophilus influenzae type a capsulation locus region II encodes a bifunctional ribulose 5-phosphate reductase-CDP-ribitol pyrophosphorylase. J. Bacteriol. 181:2001-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frayha, H. H., A. K. Kalloghlian, and M. M. deMoor. 1996. Endocarditis due to Haemophilus influenzae serotype f. Clin. Infect. Dis. 23:401-402. [DOI] [PubMed] [Google Scholar]
- 9.Fridovich, I. 1986. Superoxide dismutases. Adv. Enzymol. Rel. Areas Mol. Biol. 58:61-97. [DOI] [PubMed] [Google Scholar]
- 10.Frosch, M., and A. Muller. 1993. Phospholipid substitution of capsular polysaccharides and mechanisms of capsule formation in Neisseria meningitidis. Mol. Microbiol. 8:483-493. [DOI] [PubMed] [Google Scholar]
- 11.Geer, L. Y., M. Domrachev, D. J. Lipman, and S. H. Bryant. 2002. CDART: protein homology by domain architecture. Genome Res. 12:1619-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassan, H. M. 1989. Microbial superoxide dismutases. Adv. Genet. 26:65-97. [DOI] [PubMed] [Google Scholar]
- 13.Hoiseth, S. K., C. J. Connelly, and E. R. Moxon. 1985. Genetics of spontaneous, high-frequency loss of b capsule expression in Haemophilus influenzae. Infect. Immun. 49:389-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoiseth, S. K., E. R. Moxon, and R. P. Silver. 1986. Genes involved in Haemophilus influenzae type b capsule expression are part of an 18-kilobase tandem duplication. Proc. Natl. Acad. Sci. USA 83:1106-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroll, J. S., P. R. Langford, and B. M. Loynds. 1991. Copper-zinc superoxide dismutase of Haemophilus influenzae and H. parainfluenzae. J. Bacteriol. 173:7449-7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kroll, J. S., B. Loynds, L. N. Brophy, and E. R. Moxon. 1990. The bex locus in encapsulated Haemophilus influenzae: a chromosomal region involved in capsule polysaccharide export. Mol. Microbiol. 4:1853-1862. [DOI] [PubMed] [Google Scholar]
- 17.Kroll, J. S., B. M. Loynds, and P. R. Langford. 1992. Palindromic Haemophilus DNA uptake sequences in presumed transcriptional terminators from H. influenzae and H. parainfluenzae. Gene 114:151-152. [DOI] [PubMed] [Google Scholar]
- 18.Kroll, J. S., B. M. Loynds, and E. R. Moxon. 1991. The Haemophilus influenzae capsulation gene cluster: a compound transposon. Mol. Microbiol. 5:1549-1560. [DOI] [PubMed] [Google Scholar]
- 19.Kroll, J. S., and E. R. Moxon. 1988. Capsulation and gene copy number at the cap locus of Haemophilus influenzae type b. J. Bacteriol. 170:859-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroll, J. S., S. Zamze, B. Loynds, and E. R. Moxon. 1989. Common organization of chromosomal loci for production of different capsular polysaccharides in Haemophilus influenzae. J. Bacteriol. 171:3343-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 22.Langford, P. R., B. M. Loynds, and J. S. Kroll. 1992. Copper-zinc superoxide dismutase in Haemophilus species. J. Gen. Microbiol. 138:517-522. [DOI] [PubMed] [Google Scholar]
- 23.Langford, P. R., B. J. Sheehan, T. Shaikh, and J. S. Kroll. 2002. Active copper- and zinc-containing superoxide dismutase in the cryptic genospecies of Haemophilus causing urogenital and neonatal infections discriminates them from Haemophilus influenzae sensu stricto. J. Clin. Microbiol. 40:268-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leaves, N. I., T. J. Falla, and D. W. M. Crook. 1995. The elucidation of novel capsular genotypes of Haemophilus influenzae type b with the polymerase chain reaction. J. Med. Microbiol. 43:120-124. [DOI] [PubMed] [Google Scholar]
- 25.Lo, R. Y., L. J. McKerral, T. L. Hills, and M. Kostrzynska. 2001. Analysis of the capsule biosynthetic locus of Mannheimia (Pasteurella) haemolytica A1 and proposal of a nomenclature system. Infect. Immun. 69:4458-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchler-Bauer, A., A. R. Panchenko, B. A. Shoemaker, P. A. Thiessen, L. Y. Geer, and S. H. Bryant. 2002. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 30:281-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moxon, E. R., and J. S. Kroll. 1990. The role of bacterial polysaccharide capsules as virulence factors. Curr. Top. Microbiol. Immunol. 150:65-85. [DOI] [PubMed] [Google Scholar]
- 29.Musser, J. M., J. S. Kroll, D. M. Granoff, E. R. Moxon, B. R. Brodeur, J. Campos, H. Dabernat, W. Frederiksen, J. Hamel, G. Hammond, E. A. Hoiby, K. E. Jonsdottir, M. Kabeer, I. Kallings., W. N. Khan, M. Kilian, K. Knowles, H. J. Koornhof, B. Law, K. I. Li, J. Montgomery, P. E. Pattison, J.-C. Piffaretti, A. K. Takala, M. L. Thong, R. A. Wall, J. I. Ward, and R. K. Selander. 1990. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev. Infect. Dis. 12:75-111. [DOI] [PubMed] [Google Scholar]
- 30.Musser, J. M., J. S. Kroll, E. R. Moxon, and R. K. Selander. 1988. Evolutionary genetics of the encapsulated strains of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 85:7758-7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pittman, M. 1931. Variation and type specificity in the bacterial species Haemophilus influenzae. J. Exp. Med. 53:471-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Read, T. D., S. W. Satola, J. A. Opdyke, and M. M. Farley. 1998. Copy number of pilus gene clusters in Haemophilus influenzae and variation in the hifE pilin gene. Infect. Immun. 66:1622-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsh, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Satola, S. W., P. L. Schirmer, and M. M. Farley. 2003. Complete sequence of the cap locus of Haemophilus influenzae serotype b and nonencapsulated b capsule-negative variants. Infect. Immun. 71:3639-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slater, L. N., J. Guarnaccia, S. Makintubee, and G. R. Istre. 1990. Bacteremic disease due to Haemophilus influenzae capsular type f in adults: report of five cases and review. Rev. Infect. Dis. 12:628-635. [DOI] [PubMed] [Google Scholar]
- 36.Swartley, J. S., L. J. Liu, Y. K. Miller, L. E. Martin, S. Edupuganti, and D. S. Stephens. 1998. Characterization of the gene cassette required for biosynthesis of the (α1→6)-linked N-acetyl-d-mannosamine-1-phosphate capsule of serogroup A Neisseria meningitidis. J. Bacteriol. 180:1533-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 38.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonin. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trollfors, B., B. Claesson, T. Lagergard, and T. Sandberg. 1984. Incidence, predisposing factors and manifestations of invasive Haemophilus influenzae infections in adults. Eur. J. Clin. Microbiol. 3:180-184. [DOI] [PubMed] [Google Scholar]
- 39a.Tzeng, Y.-L., C. Noble, and D. S. Stephens. 2003. Genetic basics for biosynthesis of the (α1->4)-linked N-acetyl-d-glucosamine 1-phosphate capsule of Neisseria meningitidis serogroup X. Infect. Immun. 71:6712-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urwin, G., J. A. Krohn, K. Deaver-Robinson, J. D. Wenger, M. M. Farley, et al. 1996. Invasive disease due to Haemophilus influenzae serotype f: clinical and epidemiologic characteristics in the H. influenzae serotype b vaccine era. Clin. Infect. Dis. 22:1069-1076. [DOI] [PubMed] [Google Scholar]
- 41.van Eldere, J., L. Brophy, B. Loynds, P. Celis, I. Hancock, S. Carman, J. S. Kroll, and E. R. Moxon. 1995. Region II of the Haemophilus influenzae type b capsulation locus is involved in serotype-specific polysaccharide synthesis. Mol. Microbiol. 15:107-118. [DOI] [PubMed] [Google Scholar]
- 42.van Kranenburg, R., I. I. van Swam, J. D. Marugg, M. Kleerebezem, and W. M. de Vos. 1999. Exopolysaccharide biosynthesis in Lactococcus lactis NIZO B40: functional analysis of the glycosyltransferase genes involved in synthesis of the polysaccharide backbone. J. Bacteriol. 181:338-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallace, R. J., Jr., D. M. Musher, E. J. Septimus, J. E. McGowan, Jr., F. J. Quinones, K. Wiss, P. H. Vance, and P. A. Trier. 1981. Haemophilus influenzae infections in adults: characterization of strains by serotypes, biotypes, and beta-lactamase production. J. Infect. Dis. 144:101-106. [DOI] [PubMed] [Google Scholar]
- 44.Wenger, J. D., R. Pierce, K. Deaver, R. Franklin, G. Bosley, N. Pigott, and C. V. Broome. 1992. Invasive Haemophilus influenzae disease: a population-based evaluation of the role of capsular polysaccharide serotype. J. Infect. Dis. 165(Suppl. 1):S34-S35. [DOI] [PubMed] [Google Scholar]
- 45.Zacharisen, M. C., S. K. Watters, and J. Edwards. 2003. Rapidly fatal Haemophilus influenzae serotype f sepsis in a healthy child. J. Infect. 46:194-196. [DOI] [PubMed] [Google Scholar]