Abstract
The title compound, C14H18N2O2, contains two geometrically different molecules in the asymmetric unit: the basal plane of the cyclohexane chair and the N-[pyridin-3-ylmethylidene]methanamine moiety are oriented at dihedral angles of 71.77 (7)° and 83.42 (8)°. In the crystal, the molecules are linked by O—H⋯N hydrogen bonds, generating C(13) head-to-tail chains extending along the base vector [103]. R 2 2(26) ring motifs are formed due to the C—H⋯·O interactions that link neighbouring chains. There also exist π–π interactions [centroid–centroid separation = 3.6925 (12) Å] between the symmetry-related pyridine rings of one of the independent molecules.
Related literature
For related structures, see: Huh & Lee (2007 ▶): Shahzadi et al. (2007 ▶). For graph-set notation, see: Bernstein et al. (1995 ▶).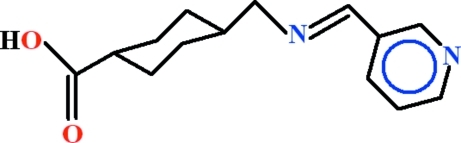
Experimental
Crystal data
C14H18N2O2
M r = 246.31
Monoclinic,

a = 12.7580 (6) Å
b = 11.2504 (6) Å
c = 18.8088 (7) Å
β = 94.720 (2)°
V = 2690.5 (2) Å3
Z = 8
Mo Kα radiation
μ = 0.08 mm−1
T = 296 K
0.34 × 0.25 × 0.22 mm
Data collection
Bruker Kappa APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2005 ▶) T min = 0.975, T max = 0.983
24311 measured reflections
6635 independent reflections
3908 reflections with I > 2σ(I)
R int = 0.034
Refinement
R[F 2 > 2σ(F 2)] = 0.063
wR(F 2) = 0.207
S = 1.05
6635 reflections
327 parameters
H-atom parameters constrained
Δρmax = 0.50 e Å−3
Δρmin = −0.25 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶) and PLATON (Spek, 2009 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶) and PLATON.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811011779/hb5830sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811011779/hb5830Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯N4i | 0.82 | 1.87 | 2.682 (2) | 171 |
| O3—H3⋯N2ii | 0.82 | 1.89 | 2.685 (2) | 164 |
| C11—H11⋯O2iii | 0.93 | 2.56 | 3.478 (3) | 168 |
| C13—H13⋯O2iv | 0.93 | 2.57 | 3.316 (3) | 137 |
| C27—H27⋯O4v | 0.93 | 2.45 | 3.280 (3) | 148 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v) 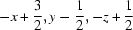 .
.
Acknowledgments
The authors acknowledge the provision of funds for the purchase of the diffractometer and encouragement by Dr Muhammad Akram Chaudhary, Vice Chancellor, University of Sargodha, Pakistan.
supplementary crystallographic information
Comment
The title compound (I, Fig. 1) has been prepared for the study of biological studies and for the synthesis of metallic complexes.
The crystal structure of (II) i.e., 4-(aminomethyl)cyclohexane-1-carboxylic acid (Shahzadi et al., 2007) and (III) i.e., N,N'-bis(pyridin-3-ylmethylene)cyclohexane-trans-1,4-diamine (Huh & Lee, 2007) have been published which are related to the title compound.
The title compound consists of two molecules in the crystallographic asymmetric unit which differ from each other geometrically. In one molecules, the basal plane A (C3/C4/C6/C7) of cyclohexane and N-[pyridin-3-ylmethylidene]methanamine moiety B (C8—C14/N1/N2) are almost planar with r.m.s. deviation of 0.014 and 0.034 Å, respectively. The dihedral angle between A/B is 71.77 (7)°. The carboxylate group C (O1/C1/O2) is of course planar. The dihedral angle between A/C and B/C is 30.07 (15)° and 53.36 (15)°, respectively. In second molecules, the basal plane D (C17/C18/C20/C21) of cyclohexane and N-[pyridin-3-ylmethylidene]methanamine moiety E (C22—C28/N3/N4) are also almost planar with r.m.s. deviation of 0.006 and 0.047 Å, respectively. The dihedral angle between D/E is 83.42 (8)°. The carboxylate group F (O3/C15/O4) makes dihedral angle of 30.03 (26)° and 62.40 (14)° with D and E, respectively.
In the crystal, the molecules are stabilized in the form of infinite C(13) polymeric chains due to O—H···.N H-bonds (Table 1, Fig. 2) extending along the base vector [103]. Due to intermolecular H-bonding of C—H···.O type (Table 1, Fig. 2) ring motifs (Bernstein et al., 1995) R22(26) are formed. The molecules are further stabilized by the π···π interaction between the symmetry related pyridine ring (C24/C25/N4/C26/C27/C28) at a distance of 3.6925 (12) Å.
Experimental
A two-necked reaction flask equipped with a reflux condenser, serum cap and a magnet bar was charged with a methanolic solution (40 ml) of tranexamic acid (0.157 g, 1 mmol) and pyridine-3-carboxaldehyde (0.107 g, 1 mmol) at room temperature under nitrogen atmosphere. An excess amount of triethylamine (1 ml) was dropped into the reaction mixture through a serum cap and subsequently the reaction mixture was refluxed for about 20 h. The disappearance of the starting materials was ascertained by TLC (methanol:chloroform). After completion of the reaction, an equivalent quantity of glacial acetic acid was added to the mixture to ensure neutralization of triethylamine. Later on, the crude mixture was allowed to stand overnight which resulted gradually into crystallized material. The solid was collected by suction filtration, washed with diethyl ether and recrystalized from hot methanol to give colourless prisms of (I).
Refinement
The coordinates of H-atoms of hydroxy groups were refined. The H-atoms were positioned geometrically (O—H = 0.82, C–H = 0.93—0.98 Å) and refined as riding with Uiso(H) = xUeq(C, O), where x = 1.2 for all H-atoms.
Figures
Fig. 1.
View of the title compound with displacement ellipsoids drawn at the 50% probability level. H-atoms are shown as small spheres of arbitrary radius.
Fig. 2.
The partial packing, which shows that the molecules form polymeric chains and ring motifs.
Crystal data
| C14H18N2O2 | F(000) = 1056 |
| Mr = 246.31 | Dx = 1.216 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 3908 reflections |
| a = 12.7580 (6) Å | θ = 1.9–28.3° |
| b = 11.2504 (6) Å | µ = 0.08 mm−1 |
| c = 18.8088 (7) Å | T = 296 K |
| β = 94.720 (2)° | Prism, colourless |
| V = 2690.5 (2) Å3 | 0.34 × 0.25 × 0.22 mm |
| Z = 8 |
Data collection
| Bruker Kappa APEXII CCD diffractometer | 6635 independent reflections |
| Radiation source: fine-focus sealed tube | 3908 reflections with I > 2σ(I) |
| graphite | Rint = 0.034 |
| Detector resolution: 7.50 pixels mm-1 | θmax = 28.3°, θmin = 1.9° |
| ω scans | h = −15→17 |
| Absorption correction: multi-scan (SADABS; Bruker, 2005) | k = −13→14 |
| Tmin = 0.975, Tmax = 0.983 | l = −25→21 |
| 24311 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.063 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.207 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.1099P)2 + 0.2692P] where P = (Fo2 + 2Fc2)/3 |
| 6635 reflections | (Δ/σ)max < 0.001 |
| 327 parameters | Δρmax = 0.50 e Å−3 |
| 0 restraints | Δρmin = −0.25 e Å−3 |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.34542 (12) | 0.20319 (19) | 0.85240 (7) | 0.0774 (7) | |
| O2 | 0.48692 (13) | 0.19021 (19) | 0.79343 (8) | 0.0852 (8) | |
| N1 | 0.18226 (12) | −0.02615 (17) | 0.48003 (7) | 0.0500 (6) | |
| N2 | 0.18135 (14) | 0.04146 (17) | 0.22905 (8) | 0.0552 (6) | |
| C1 | 0.39348 (17) | 0.1868 (2) | 0.79392 (10) | 0.0542 (7) | |
| C2 | 0.31681 (15) | 0.1641 (2) | 0.72955 (9) | 0.0496 (7) | |
| C3 | 0.37171 (17) | 0.1506 (2) | 0.66075 (9) | 0.0607 (8) | |
| C4 | 0.29250 (17) | 0.1273 (2) | 0.59682 (9) | 0.0577 (8) | |
| C5 | 0.22042 (14) | 0.0229 (2) | 0.60795 (8) | 0.0465 (6) | |
| C6 | 0.16868 (15) | 0.0366 (2) | 0.67770 (9) | 0.0558 (7) | |
| C7 | 0.24956 (15) | 0.0556 (2) | 0.74099 (9) | 0.0529 (7) | |
| C8 | 0.13716 (15) | 0.0072 (2) | 0.54600 (9) | 0.0543 (7) | |
| C9 | 0.15528 (15) | 0.0323 (2) | 0.42457 (9) | 0.0482 (6) | |
| C10 | 0.19220 (14) | 0.00271 (18) | 0.35472 (8) | 0.0429 (6) | |
| C11 | 0.15284 (15) | 0.0644 (2) | 0.29463 (9) | 0.0509 (7) | |
| C12 | 0.25168 (16) | −0.0442 (2) | 0.22215 (10) | 0.0552 (7) | |
| C13 | 0.29540 (16) | −0.1099 (2) | 0.27881 (10) | 0.0547 (7) | |
| C14 | 0.26511 (15) | −0.08693 (19) | 0.34589 (9) | 0.0493 (6) | |
| O3 | 0.58371 (14) | 0.35232 (17) | 0.61410 (7) | 0.0740 (7) | |
| O4 | 0.69327 (16) | 0.4490 (2) | 0.55311 (8) | 0.1074 (8) | |
| N3 | 0.50418 (13) | 0.15359 (17) | 0.22618 (8) | 0.0547 (6) | |
| N4 | 0.47454 (15) | 0.19104 (17) | −0.02788 (8) | 0.0584 (6) | |
| C15 | 0.61952 (15) | 0.3860 (2) | 0.55416 (9) | 0.0493 (7) | |
| C16 | 0.55551 (15) | 0.33872 (19) | 0.48915 (9) | 0.0470 (6) | |
| C17 | 0.59449 (19) | 0.3837 (2) | 0.41964 (10) | 0.0636 (8) | |
| C18 | 0.52518 (18) | 0.3364 (2) | 0.35534 (10) | 0.0600 (8) | |
| C19 | 0.52023 (17) | 0.2027 (2) | 0.35542 (9) | 0.0535 (7) | |
| C20 | 0.4829 (2) | 0.1581 (3) | 0.42478 (11) | 0.0726 (9) | |
| C21 | 0.55003 (19) | 0.2050 (2) | 0.48971 (10) | 0.0622 (8) | |
| C22 | 0.45076 (17) | 0.1549 (2) | 0.29154 (9) | 0.0612 (8) | |
| C23 | 0.45152 (16) | 0.17579 (19) | 0.16841 (9) | 0.0485 (6) | |
| C24 | 0.49662 (15) | 0.16380 (18) | 0.09908 (9) | 0.0460 (6) | |
| C25 | 0.44008 (17) | 0.2013 (2) | 0.03678 (9) | 0.0522 (7) | |
| C26 | 0.56779 (19) | 0.1403 (2) | −0.03228 (11) | 0.0623 (8) | |
| C27 | 0.62979 (18) | 0.0993 (2) | 0.02655 (12) | 0.0650 (8) | |
| C28 | 0.59361 (16) | 0.1115 (2) | 0.09252 (10) | 0.0564 (7) | |
| H1 | 0.38897 | 0.20427 | 0.88698 | 0.0929* | |
| H2 | 0.26983 | 0.23300 | 0.72381 | 0.0596* | |
| H3A | 0.41080 | 0.22255 | 0.65240 | 0.0729* | |
| H3B | 0.42131 | 0.08524 | 0.66588 | 0.0729* | |
| H4A | 0.25009 | 0.19799 | 0.58746 | 0.0692* | |
| H4B | 0.33044 | 0.11217 | 0.55512 | 0.0692* | |
| H5 | 0.26354 | −0.04928 | 0.61141 | 0.0557* | |
| H6A | 0.12781 | −0.03406 | 0.68586 | 0.0670* | |
| H6B | 0.12088 | 0.10380 | 0.67389 | 0.0670* | |
| H7A | 0.21368 | 0.06593 | 0.78407 | 0.0634* | |
| H7B | 0.29423 | −0.01402 | 0.74715 | 0.0634* | |
| H8A | 0.08785 | −0.05372 | 0.55814 | 0.0652* | |
| H8B | 0.09846 | 0.08095 | 0.53848 | 0.0652* | |
| H9 | 0.11027 | 0.09675 | 0.42786 | 0.0579* | |
| H11 | 0.10427 | 0.12470 | 0.30007 | 0.0611* | |
| H12 | 0.27229 | −0.06053 | 0.17687 | 0.0663* | |
| H13 | 0.34461 | −0.16888 | 0.27181 | 0.0657* | |
| H14 | 0.29300 | −0.13074 | 0.38490 | 0.0591* | |
| H3 | 0.62309 | 0.37640 | 0.64768 | 0.0888* | |
| H16 | 0.48356 | 0.36827 | 0.49122 | 0.0563* | |
| H17A | 0.66650 | 0.35801 | 0.41633 | 0.0763* | |
| H17B | 0.59343 | 0.46992 | 0.41923 | 0.0763* | |
| H18A | 0.45469 | 0.36834 | 0.35640 | 0.0720* | |
| H18B | 0.55315 | 0.36332 | 0.31170 | 0.0720* | |
| H19 | 0.59168 | 0.17263 | 0.35182 | 0.0642* | |
| H20A | 0.48481 | 0.07191 | 0.42511 | 0.0871* | |
| H20B | 0.41048 | 0.18253 | 0.42792 | 0.0871* | |
| H21A | 0.52030 | 0.17872 | 0.53287 | 0.0747* | |
| H21B | 0.62051 | 0.17260 | 0.48987 | 0.0747* | |
| H22A | 0.42867 | 0.07476 | 0.30202 | 0.0734* | |
| H22B | 0.38817 | 0.20383 | 0.28439 | 0.0734* | |
| H23 | 0.38208 | 0.20049 | 0.16936 | 0.0582* | |
| H25 | 0.37442 | 0.23562 | 0.04046 | 0.0626* | |
| H26 | 0.59272 | 0.13195 | −0.07710 | 0.0748* | |
| H27 | 0.69467 | 0.06412 | 0.02123 | 0.0779* | |
| H28 | 0.63391 | 0.08479 | 0.13282 | 0.0676* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0682 (10) | 0.1258 (17) | 0.0379 (7) | 0.0013 (10) | 0.0026 (7) | −0.0168 (9) |
| O2 | 0.0607 (10) | 0.1432 (19) | 0.0519 (9) | −0.0195 (10) | 0.0063 (7) | −0.0162 (10) |
| N1 | 0.0513 (9) | 0.0658 (12) | 0.0323 (7) | 0.0041 (8) | −0.0004 (6) | −0.0019 (7) |
| N2 | 0.0652 (10) | 0.0674 (13) | 0.0318 (7) | −0.0006 (9) | −0.0030 (7) | 0.0051 (8) |
| C1 | 0.0618 (13) | 0.0623 (15) | 0.0390 (10) | −0.0043 (10) | 0.0075 (9) | −0.0034 (9) |
| C2 | 0.0545 (11) | 0.0606 (14) | 0.0339 (9) | 0.0018 (9) | 0.0042 (8) | −0.0028 (9) |
| C3 | 0.0612 (12) | 0.0862 (18) | 0.0357 (10) | −0.0198 (11) | 0.0095 (9) | −0.0031 (10) |
| C4 | 0.0648 (12) | 0.0790 (17) | 0.0300 (9) | −0.0088 (11) | 0.0088 (8) | 0.0053 (9) |
| C5 | 0.0451 (10) | 0.0635 (14) | 0.0307 (8) | 0.0066 (8) | 0.0026 (7) | 0.0014 (8) |
| C6 | 0.0479 (10) | 0.0848 (17) | 0.0354 (9) | −0.0032 (10) | 0.0077 (8) | 0.0001 (10) |
| C7 | 0.0559 (11) | 0.0744 (16) | 0.0288 (8) | −0.0033 (10) | 0.0064 (8) | 0.0036 (9) |
| C8 | 0.0497 (10) | 0.0801 (16) | 0.0331 (9) | 0.0061 (10) | 0.0035 (8) | −0.0016 (9) |
| C9 | 0.0478 (10) | 0.0583 (13) | 0.0380 (9) | 0.0040 (9) | 0.0005 (7) | −0.0010 (9) |
| C10 | 0.0437 (9) | 0.0510 (12) | 0.0332 (8) | −0.0027 (8) | −0.0016 (7) | 0.0015 (8) |
| C11 | 0.0539 (11) | 0.0581 (14) | 0.0400 (9) | 0.0072 (9) | −0.0009 (8) | 0.0065 (9) |
| C12 | 0.0651 (13) | 0.0657 (15) | 0.0349 (9) | −0.0087 (10) | 0.0041 (9) | −0.0056 (9) |
| C13 | 0.0585 (12) | 0.0562 (14) | 0.0495 (11) | 0.0053 (10) | 0.0044 (9) | −0.0064 (10) |
| C14 | 0.0544 (11) | 0.0548 (13) | 0.0379 (9) | 0.0038 (9) | −0.0010 (8) | 0.0050 (9) |
| O3 | 0.0978 (12) | 0.0927 (14) | 0.0312 (7) | −0.0347 (10) | 0.0032 (7) | −0.0069 (8) |
| O4 | 0.0983 (13) | 0.179 (2) | 0.0455 (9) | −0.0758 (15) | 0.0102 (8) | −0.0169 (11) |
| N3 | 0.0624 (10) | 0.0665 (12) | 0.0341 (8) | −0.0035 (8) | −0.0021 (7) | −0.0072 (8) |
| N4 | 0.0747 (12) | 0.0651 (13) | 0.0346 (8) | −0.0048 (9) | −0.0001 (8) | −0.0044 (8) |
| C15 | 0.0515 (11) | 0.0640 (14) | 0.0327 (9) | −0.0015 (9) | 0.0060 (8) | −0.0063 (9) |
| C16 | 0.0475 (10) | 0.0619 (14) | 0.0316 (8) | −0.0011 (9) | 0.0040 (7) | −0.0061 (8) |
| C17 | 0.0808 (15) | 0.0708 (16) | 0.0391 (10) | −0.0209 (12) | 0.0044 (10) | −0.0015 (10) |
| C18 | 0.0707 (13) | 0.0744 (17) | 0.0346 (9) | −0.0040 (11) | 0.0020 (9) | 0.0045 (10) |
| C19 | 0.0595 (12) | 0.0672 (15) | 0.0336 (9) | −0.0026 (10) | 0.0020 (8) | −0.0062 (9) |
| C20 | 0.1047 (19) | 0.0713 (17) | 0.0401 (10) | −0.0256 (14) | −0.0037 (11) | 0.0018 (10) |
| C21 | 0.0824 (15) | 0.0681 (16) | 0.0353 (9) | −0.0030 (12) | −0.0006 (9) | 0.0057 (10) |
| C22 | 0.0687 (13) | 0.0805 (17) | 0.0337 (9) | −0.0184 (12) | 0.0001 (9) | −0.0061 (10) |
| C23 | 0.0566 (11) | 0.0521 (13) | 0.0364 (9) | 0.0007 (9) | 0.0014 (8) | −0.0043 (8) |
| C24 | 0.0591 (11) | 0.0432 (11) | 0.0350 (9) | −0.0041 (9) | −0.0003 (8) | −0.0056 (8) |
| C25 | 0.0622 (12) | 0.0570 (14) | 0.0367 (9) | 0.0019 (10) | −0.0001 (8) | −0.0033 (9) |
| C26 | 0.0814 (15) | 0.0658 (16) | 0.0414 (10) | −0.0096 (12) | 0.0148 (10) | −0.0126 (10) |
| C27 | 0.0672 (13) | 0.0673 (16) | 0.0612 (13) | 0.0085 (11) | 0.0102 (11) | −0.0137 (11) |
| C28 | 0.0638 (12) | 0.0570 (14) | 0.0471 (11) | 0.0064 (10) | −0.0026 (9) | −0.0032 (9) |
Geometric parameters (Å, °)
| O1—C1 | 1.316 (2) | C8—H8A | 0.9700 |
| O2—C1 | 1.194 (3) | C8—H8B | 0.9700 |
| O1—H1 | 0.8200 | C9—H9 | 0.9300 |
| O3—C15 | 1.307 (2) | C11—H11 | 0.9300 |
| O4—C15 | 1.180 (3) | C12—H12 | 0.9300 |
| O3—H3 | 0.8200 | C13—H13 | 0.9300 |
| N1—C8 | 1.459 (2) | C14—H14 | 0.9300 |
| N1—C9 | 1.257 (2) | C15—C16 | 1.510 (3) |
| N2—C12 | 1.330 (3) | C16—C17 | 1.523 (3) |
| N2—C11 | 1.340 (2) | C16—C21 | 1.506 (3) |
| N3—C22 | 1.454 (2) | C17—C18 | 1.534 (3) |
| N3—C23 | 1.255 (2) | C18—C19 | 1.506 (3) |
| N4—C26 | 1.328 (3) | C19—C20 | 1.511 (3) |
| N4—C25 | 1.332 (2) | C19—C22 | 1.531 (3) |
| C1—C2 | 1.514 (3) | C20—C21 | 1.527 (3) |
| C2—C3 | 1.529 (3) | C23—C24 | 1.474 (2) |
| C2—C7 | 1.518 (3) | C24—C25 | 1.390 (3) |
| C3—C4 | 1.528 (3) | C24—C28 | 1.385 (3) |
| C4—C5 | 1.517 (3) | C26—C27 | 1.385 (3) |
| C5—C6 | 1.524 (2) | C27—C28 | 1.366 (3) |
| C5—C8 | 1.521 (2) | C16—H16 | 0.9800 |
| C6—C7 | 1.525 (3) | C17—H17A | 0.9700 |
| C9—C10 | 1.470 (2) | C17—H17B | 0.9700 |
| C10—C11 | 1.385 (3) | C18—H18A | 0.9700 |
| C10—C14 | 1.391 (3) | C18—H18B | 0.9700 |
| C12—C13 | 1.377 (3) | C19—H19 | 0.9800 |
| C13—C14 | 1.374 (3) | C20—H20A | 0.9700 |
| C2—H2 | 0.9800 | C20—H20B | 0.9700 |
| C3—H3A | 0.9700 | C21—H21A | 0.9700 |
| C3—H3B | 0.9700 | C21—H21B | 0.9700 |
| C4—H4A | 0.9700 | C22—H22A | 0.9700 |
| C4—H4B | 0.9700 | C22—H22B | 0.9700 |
| C5—H5 | 0.9800 | C23—H23 | 0.9300 |
| C6—H6A | 0.9700 | C25—H25 | 0.9300 |
| C6—H6B | 0.9700 | C26—H26 | 0.9300 |
| C7—H7B | 0.9700 | C27—H27 | 0.9300 |
| C7—H7A | 0.9700 | C28—H28 | 0.9300 |
| C1—O1—H1 | 109.00 | C14—C13—H13 | 121.00 |
| C15—O3—H3 | 109.00 | C10—C14—H14 | 120.00 |
| C8—N1—C9 | 118.10 (18) | C13—C14—H14 | 120.00 |
| C11—N2—C12 | 117.77 (17) | O3—C15—C16 | 113.09 (17) |
| C22—N3—C23 | 118.38 (17) | O4—C15—C16 | 125.25 (17) |
| C25—N4—C26 | 117.31 (18) | O3—C15—O4 | 121.64 (18) |
| O1—C1—C2 | 112.17 (18) | C15—C16—C17 | 112.64 (17) |
| O1—C1—O2 | 122.44 (18) | C17—C16—C21 | 110.89 (16) |
| O2—C1—C2 | 125.39 (18) | C15—C16—C21 | 111.62 (16) |
| C3—C2—C7 | 110.11 (17) | C16—C17—C18 | 110.72 (18) |
| C1—C2—C3 | 112.50 (16) | C17—C18—C19 | 111.56 (17) |
| C1—C2—C7 | 110.97 (16) | C18—C19—C22 | 111.83 (17) |
| C2—C3—C4 | 111.33 (17) | C20—C19—C22 | 110.96 (19) |
| C3—C4—C5 | 113.16 (15) | C18—C19—C20 | 110.42 (19) |
| C4—C5—C8 | 112.17 (15) | C19—C20—C21 | 112.3 (2) |
| C4—C5—C6 | 110.54 (16) | C16—C21—C20 | 111.28 (19) |
| C6—C5—C8 | 110.27 (15) | N3—C22—C19 | 112.70 (17) |
| C5—C6—C7 | 111.90 (15) | N3—C23—C24 | 121.85 (18) |
| C2—C7—C6 | 110.87 (16) | C23—C24—C28 | 122.30 (17) |
| N1—C8—C5 | 112.45 (15) | C25—C24—C28 | 117.35 (17) |
| N1—C9—C10 | 122.50 (19) | C23—C24—C25 | 120.31 (18) |
| C11—C10—C14 | 117.84 (15) | N4—C25—C24 | 123.8 (2) |
| C9—C10—C14 | 122.51 (16) | N4—C26—C27 | 123.3 (2) |
| C9—C10—C11 | 119.64 (18) | C26—C27—C28 | 118.6 (2) |
| N2—C11—C10 | 123.18 (19) | C24—C28—C27 | 119.66 (18) |
| N2—C12—C13 | 123.16 (18) | C15—C16—H16 | 107.00 |
| C12—C13—C14 | 118.89 (19) | C17—C16—H16 | 107.00 |
| C10—C14—C13 | 119.16 (17) | C21—C16—H16 | 107.00 |
| C1—C2—H2 | 108.00 | C16—C17—H17A | 110.00 |
| C3—C2—H2 | 108.00 | C16—C17—H17B | 109.00 |
| C7—C2—H2 | 108.00 | C18—C17—H17A | 109.00 |
| C4—C3—H3A | 109.00 | C18—C17—H17B | 110.00 |
| C4—C3—H3B | 109.00 | H17A—C17—H17B | 108.00 |
| C2—C3—H3B | 109.00 | C17—C18—H18A | 109.00 |
| C2—C3—H3A | 109.00 | C17—C18—H18B | 109.00 |
| H3A—C3—H3B | 108.00 | C19—C18—H18A | 109.00 |
| C3—C4—H4A | 109.00 | C19—C18—H18B | 109.00 |
| C3—C4—H4B | 109.00 | H18A—C18—H18B | 108.00 |
| C5—C4—H4B | 109.00 | C18—C19—H19 | 108.00 |
| C5—C4—H4A | 109.00 | C20—C19—H19 | 108.00 |
| H4A—C4—H4B | 108.00 | C22—C19—H19 | 108.00 |
| C8—C5—H5 | 108.00 | C19—C20—H20A | 109.00 |
| C6—C5—H5 | 108.00 | C19—C20—H20B | 109.00 |
| C4—C5—H5 | 108.00 | C21—C20—H20A | 109.00 |
| C7—C6—H6A | 109.00 | C21—C20—H20B | 109.00 |
| C5—C6—H6A | 109.00 | H20A—C20—H20B | 108.00 |
| H6A—C6—H6B | 108.00 | C16—C21—H21A | 109.00 |
| C7—C6—H6B | 109.00 | C16—C21—H21B | 109.00 |
| C5—C6—H6B | 109.00 | C20—C21—H21A | 109.00 |
| C2—C7—H7A | 109.00 | C20—C21—H21B | 109.00 |
| C2—C7—H7B | 109.00 | H21A—C21—H21B | 108.00 |
| C6—C7—H7A | 109.00 | N3—C22—H22A | 109.00 |
| C6—C7—H7B | 109.00 | N3—C22—H22B | 109.00 |
| H7A—C7—H7B | 108.00 | C19—C22—H22A | 109.00 |
| C5—C8—H8B | 109.00 | C19—C22—H22B | 109.00 |
| H8A—C8—H8B | 108.00 | H22A—C22—H22B | 108.00 |
| C5—C8—H8A | 109.00 | N3—C23—H23 | 119.00 |
| N1—C8—H8B | 109.00 | C24—C23—H23 | 119.00 |
| N1—C8—H8A | 109.00 | N4—C25—H25 | 118.00 |
| N1—C9—H9 | 119.00 | C24—C25—H25 | 118.00 |
| C10—C9—H9 | 119.00 | N4—C26—H26 | 118.00 |
| N2—C11—H11 | 118.00 | C27—C26—H26 | 118.00 |
| C10—C11—H11 | 118.00 | C26—C27—H27 | 121.00 |
| N2—C12—H12 | 118.00 | C28—C27—H27 | 121.00 |
| C13—C12—H12 | 118.00 | C24—C28—H28 | 120.00 |
| C12—C13—H13 | 121.00 | C27—C28—H28 | 120.00 |
| C9—N1—C8—C5 | −129.8 (2) | C11—C10—C14—C13 | 0.5 (3) |
| C8—N1—C9—C10 | −177.13 (18) | C9—C10—C14—C13 | 179.63 (19) |
| C12—N2—C11—C10 | −0.7 (3) | N2—C12—C13—C14 | 0.3 (3) |
| C11—N2—C12—C13 | 0.4 (3) | C12—C13—C14—C10 | −0.7 (3) |
| C22—N3—C23—C24 | −173.32 (19) | O3—C15—C16—C17 | 177.02 (19) |
| C23—N3—C22—C19 | −144.5 (2) | O3—C15—C16—C21 | −57.5 (2) |
| C26—N4—C25—C24 | 1.0 (3) | O4—C15—C16—C17 | −1.4 (3) |
| C25—N4—C26—C27 | −0.5 (3) | O4—C15—C16—C21 | 124.1 (3) |
| O1—C1—C2—C3 | 177.38 (19) | C15—C16—C17—C18 | −178.30 (18) |
| O1—C1—C2—C7 | −58.8 (2) | C21—C16—C17—C18 | 55.8 (2) |
| O2—C1—C2—C3 | −2.5 (3) | C15—C16—C21—C20 | 178.56 (17) |
| O2—C1—C2—C7 | 121.4 (2) | C17—C16—C21—C20 | −55.0 (2) |
| C1—C2—C3—C4 | 179.77 (18) | C16—C17—C18—C19 | −56.6 (2) |
| C1—C2—C7—C6 | 177.12 (16) | C17—C18—C19—C20 | 55.7 (2) |
| C3—C2—C7—C6 | −57.7 (2) | C17—C18—C19—C22 | 179.75 (17) |
| C7—C2—C3—C4 | 55.4 (2) | C18—C19—C20—C21 | −55.0 (3) |
| C2—C3—C4—C5 | −53.6 (2) | C22—C19—C20—C21 | −179.5 (2) |
| C3—C4—C5—C6 | 52.2 (2) | C18—C19—C22—N3 | 79.5 (2) |
| C3—C4—C5—C8 | 175.74 (17) | C20—C19—C22—N3 | −156.8 (2) |
| C4—C5—C8—N1 | 66.0 (2) | C19—C20—C21—C16 | 55.1 (3) |
| C4—C5—C6—C7 | −54.0 (2) | N3—C23—C24—C25 | −173.0 (2) |
| C6—C5—C8—N1 | −170.34 (18) | N3—C23—C24—C28 | 9.5 (3) |
| C8—C5—C6—C7 | −178.61 (18) | C23—C24—C25—N4 | −178.5 (2) |
| C5—C6—C7—C2 | 57.7 (2) | C28—C24—C25—N4 | −0.9 (3) |
| N1—C9—C10—C11 | 174.7 (2) | C23—C24—C28—C27 | 177.8 (2) |
| N1—C9—C10—C14 | −4.5 (3) | C25—C24—C28—C27 | 0.3 (3) |
| C9—C10—C11—N2 | −178.96 (19) | N4—C26—C27—C28 | 0.0 (4) |
| C14—C10—C11—N2 | 0.2 (3) | C26—C27—C28—C24 | 0.2 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···N4i | 0.82 | 1.87 | 2.682 (2) | 171 |
| O3—H3···N2ii | 0.82 | 1.89 | 2.685 (2) | 164 |
| C11—H11···O2iii | 0.93 | 2.56 | 3.478 (3) | 168 |
| C13—H13···O2iv | 0.93 | 2.57 | 3.316 (3) | 137 |
| C27—H27···O4v | 0.93 | 2.45 | 3.280 (3) | 148 |
Symmetry codes: (i) x, y, z+1; (ii) x+1/2, −y+1/2, z+1/2; (iii) x−1/2, −y+1/2, z−1/2; (iv) −x+1, −y, −z+1; (v) −x+3/2, y−1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB5830).
References
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Bruker (2005). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2009). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst. 32, 837–838.
- Huh, H. S. & Lee, S. W. (2007). Inorg. Chem. Commun. 10, 1244–1248.
- Shahzadi, S., Ali, S., Parvez, M., Badshah, A., Ahmed, E. & Malik, A. (2007). Russ. J. Inorg. Chem. 52, 386–393.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811011779/hb5830sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811011779/hb5830Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




