Abstract
In the crystal structure of the title compound, C14H12O4, the molecules form classical O—H⋯O hydrogen-bonded carboxylic acid dimers. These dimers are linked by C—H⋯pi; interactions into a three-dimensional network. The benzene rings are oriented at a dihedral angle of 69.6 (3)°.
Related literature
For applications of the title compound, see: Jackson et al. (1993) ▶; Gapinski et al. (1990 ▶). For related structures, see: Shi et al. (2011 ▶); Raghunathan et al. (1982 ▶). For the synthesis of the title compound, see: Pellón et al. (1995 ▶). For bond-length data, see: Allen et al. (1987 ▶).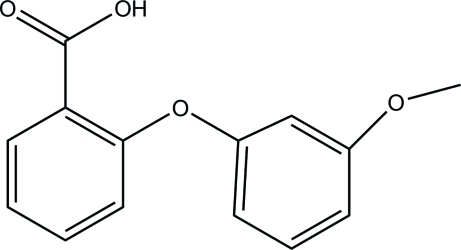
Experimental
Crystal data
C14H12O4
M r = 244.24
Orthorhombic,

a = 14.309 (3) Å
b = 8.5330 (17) Å
c = 19.432 (4) Å
V = 2372.6 (8) Å3
Z = 8
Mo Kα radiation
μ = 0.10 mm−1
T = 295 K
0.30 × 0.10 × 0.05 mm
Data collection
Enraf–Nonius CAD-4 diffractometer
Absorption correction: ψ scan (North et al., 1968 ▶) T min = 0.970, T max = 0.995
4273 measured reflections
2175 independent reflections
1056 reflections with I > 2σ(I)
R int = 0.093
3 standard reflections every 200 reflections intensity decay: 1%
Refinement
R[F 2 > 2σ(F 2)] = 0.062
wR(F 2) = 0.133
S = 1.00
2175 reflections
163 parameters
H-atom parameters constrained
Δρmax = 0.16 e Å−3
Δρmin = −0.16 e Å−3
Data collection: CAD-4 Software (Enraf–Nonius, 1985 ▶); cell refinement: CAD-4 Software; data reduction: XCAD4 (Harms & Wocadlo, 1995 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811011019/hg5013sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811011019/hg5013Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 is the centroid of the C2–C7 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O4—H4A⋯O3i | 0.82 | 1.82 | 2.633 (3) | 173 |
| C1—H1B⋯Cg1ii | 0.96 | 2.89 | 3.784 (4) | 155 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
The author gratefully acknowledges financial support from the Natural Science Foundation of the Education Department of Shaanxi Provincial Government (09JK844) and is grateful for support provided by the key industry problem plan of Yulin (gygg200807) and the special research projects of Yulin University (08YK17).
supplementary crystallographic information
Comment
The title compound, 2-(3-methoxyphenoxy)benzoic acid is an important intermediate of xanthone dicarboxylic acids (Jackson et al., 1972). Xanthone dicarboxylic acid, such as LY223982, which inhibited the binding of LTB4 to receptors on intact human neutrophils nearly as well as nonradioactive LTB4 (Gapinski et al., 1990). Knowledge of the crystal structure of such benzoicacid derivatives gives us not only information about nuclearity of the complex molecule, but is important in understanding the behaviour of these compounds with respect to the mechanisms of pharmacological activities and physiological activities. Therefore, we have synthesized the title compound, (I), and report its crystal structure here.
The molecular structure of (I) is shown in Fig. 1, and the intermolecular O—H···O hydrogen bond (Table 1) results in the formation of carboxylic acid dimers (Fig. 2.). The bond lengths are within normal ranges (Allen et al., 1987). Similar crystal structure of some compounds have been reported (Shi et al., 2011; Raghunathan et al., 1982).
In the molecule of (I), the dihedral angle of the rings(C3—C6) and (C8—C13) is 69.6 (3)°, the molecules were connected together via O—H···O intermolecular hydrogen bonds to form dimers. These dimers are linked by C—H···π and weak C—H···O interactions to give a three-dimensional network, which seems to be very effective in the stabilization of the crystal structure.
Experimental
The title compound (I) was prepared by the method of Ullmann condensation reaction reported in literature (Pellón et al., 1995). A mixture of 2-chlorobenzoic acid (6.26 g, 0.04 mol), 3-methoxyphenol (9.93 g, 0.08 mol), anhydrous K2CO3 (11.04 g' 0.08 mol), pyridine (1.58 g' 0.02 mol), Cu powder (0.2 g) and cuprous iodide (0.2 g) in 25 ml water was kept at reflux for two hours. The mixture was then basified with Na2CO3 solution and extracted with diethyl ether. The aqueous solution was acidified with HCl, the precipitated solid was filtered off and disolved in NaOH; the basic solution was filtered (charcoal) and acidified with acetic acid. The 2-(3-methoxyphenoxy)benzoic acid was crystalized from the mixture.
Refinement
H atoms were positioned geometrically and refined as riding groups, with O—H = 0.82 and C—H = 0.93 Å for aromatic H, and constrained to ride on their parent atoms, with Uiso(H) = xUeq(C), where x = 1.2 for aromatic H, and x = 1.5 for other H.
Figures
Fig. 1.
The molecular structure of (I) (thermal ellipsoids are shown at 30% probability levels).
Fig. 2.
The structure of a dimer of (I).
Crystal data
| C14H12O4 | F(000) = 1024 |
| Mr = 244.24 | Dx = 1.367 Mg m−3 |
| Orthorhombic, Pbca | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 25 reflections |
| a = 14.309 (3) Å | θ = 9–12° |
| b = 8.5330 (17) Å | µ = 0.10 mm−1 |
| c = 19.432 (4) Å | T = 295 K |
| V = 2372.6 (8) Å3 | Block, colourless |
| Z = 8 | 0.30 × 0.10 × 0.05 mm |
Data collection
| Enraf–Nonius CAD-4 diffractometer | 1056 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.093 |
| graphite | θmax = 25.4°, θmin = 2.1° |
| ω/2θ scans | h = 0→17 |
| Absorption correction: ψ scan (North et al., 1968) | k = 0→10 |
| Tmin = 0.970, Tmax = 0.995 | l = −23→23 |
| 4273 measured reflections | 3 standard reflections every 200 reflections |
| 2175 independent reflections | intensity decay: 1% |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.062 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.133 | H-atom parameters constrained |
| S = 1.00 | w = 1/[σ2(Fo2) + (0.039P)2] where P = (Fo2 + 2Fc2)/3 |
| 2175 reflections | (Δ/σ)max < 0.001 |
| 163 parameters | Δρmax = 0.16 e Å−3 |
| 0 restraints | Δρmin = −0.16 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.60412 (16) | 0.2617 (3) | 0.77635 (10) | 0.0641 (7) | |
| C1 | 0.6361 (3) | 0.4135 (4) | 0.79576 (18) | 0.0738 (11) | |
| H1A | 0.6239 | 0.4863 | 0.7592 | 0.111* | |
| H1B | 0.7021 | 0.4098 | 0.8046 | 0.111* | |
| H1C | 0.6040 | 0.4468 | 0.8366 | 0.111* | |
| O2 | 0.70190 (16) | 0.0652 (2) | 0.99264 (12) | 0.0668 (7) | |
| C2 | 0.6162 (2) | 0.1419 (4) | 0.82240 (16) | 0.0510 (8) | |
| O3 | 0.88741 (15) | 0.0003 (3) | 0.97959 (10) | 0.0654 (7) | |
| C3 | 0.6528 (2) | 0.1601 (4) | 0.88657 (16) | 0.0495 (8) | |
| H3A | 0.6720 | 0.2584 | 0.9016 | 0.059* | |
| O4 | 0.95145 (16) | −0.0699 (3) | 1.07827 (11) | 0.0783 (8) | |
| H4A | 0.9992 | −0.0465 | 1.0573 | 0.117* | |
| C4 | 0.6614 (2) | 0.0306 (4) | 0.92945 (16) | 0.0472 (8) | |
| C5 | 0.6323 (2) | −0.1143 (4) | 0.90941 (18) | 0.0570 (9) | |
| H5A | 0.6371 | −0.2000 | 0.9388 | 0.068* | |
| C6 | 0.5953 (3) | −0.1298 (4) | 0.84420 (18) | 0.0660 (10) | |
| H6A | 0.5751 | −0.2279 | 0.8296 | 0.079* | |
| C7 | 0.5878 (2) | −0.0056 (5) | 0.80078 (18) | 0.0638 (10) | |
| H7A | 0.5636 | −0.0193 | 0.7568 | 0.077* | |
| C8 | 0.7034 (2) | −0.0465 (4) | 1.04428 (17) | 0.0533 (8) | |
| C9 | 0.7889 (2) | −0.0922 (3) | 1.06981 (15) | 0.0454 (7) | |
| C10 | 0.7896 (2) | −0.1884 (4) | 1.12863 (16) | 0.0535 (9) | |
| H10A | 0.8463 | −0.2195 | 1.1477 | 0.064* | |
| C11 | 0.7077 (3) | −0.2363 (4) | 1.15792 (17) | 0.0598 (9) | |
| H11A | 0.7090 | −0.3012 | 1.1964 | 0.072* | |
| C12 | 0.6238 (3) | −0.1898 (5) | 1.1312 (2) | 0.0666 (10) | |
| H12A | 0.5683 | −0.2236 | 1.1512 | 0.080* | |
| C13 | 0.6218 (2) | −0.0919 (5) | 1.0743 (2) | 0.0699 (10) | |
| H13A | 0.5650 | −0.0575 | 1.0567 | 0.084* | |
| C14 | 0.8796 (2) | −0.0495 (4) | 1.03863 (15) | 0.0488 (8) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0741 (17) | 0.0676 (16) | 0.0506 (13) | 0.0054 (14) | −0.0066 (12) | 0.0027 (13) |
| C1 | 0.091 (3) | 0.067 (3) | 0.064 (2) | 0.008 (2) | −0.008 (2) | 0.010 (2) |
| O2 | 0.0799 (17) | 0.0490 (13) | 0.0716 (15) | −0.0051 (12) | −0.0318 (14) | 0.0080 (12) |
| C2 | 0.049 (2) | 0.055 (2) | 0.0488 (19) | 0.0094 (18) | 0.0042 (15) | 0.0037 (18) |
| O3 | 0.0670 (15) | 0.0839 (17) | 0.0452 (12) | −0.0097 (13) | −0.0051 (11) | 0.0177 (14) |
| C3 | 0.046 (2) | 0.0420 (19) | 0.061 (2) | −0.0060 (16) | −0.0011 (16) | −0.0044 (16) |
| O4 | 0.0547 (15) | 0.115 (2) | 0.0654 (16) | −0.0057 (16) | −0.0086 (13) | 0.0296 (15) |
| C4 | 0.0391 (18) | 0.046 (2) | 0.0565 (19) | 0.0019 (14) | −0.0081 (16) | −0.0041 (18) |
| C5 | 0.062 (2) | 0.045 (2) | 0.064 (2) | 0.0033 (19) | 0.0009 (19) | −0.0012 (18) |
| C6 | 0.081 (3) | 0.043 (2) | 0.074 (2) | −0.0001 (19) | 0.002 (2) | −0.014 (2) |
| C7 | 0.064 (2) | 0.070 (2) | 0.057 (2) | 0.001 (2) | −0.0037 (17) | −0.020 (2) |
| C8 | 0.062 (2) | 0.0448 (19) | 0.0530 (19) | 0.0070 (18) | −0.0061 (17) | 0.0012 (16) |
| C9 | 0.0518 (18) | 0.0384 (17) | 0.0461 (16) | −0.0025 (15) | 0.0024 (16) | −0.0004 (14) |
| C10 | 0.058 (2) | 0.059 (2) | 0.0439 (19) | 0.0086 (18) | −0.0046 (18) | −0.0023 (15) |
| C11 | 0.070 (2) | 0.061 (2) | 0.0485 (19) | −0.002 (2) | 0.0065 (19) | 0.0034 (17) |
| C12 | 0.053 (2) | 0.075 (3) | 0.072 (3) | 0.003 (2) | 0.015 (2) | −0.002 (2) |
| C13 | 0.048 (2) | 0.079 (3) | 0.083 (3) | 0.011 (2) | 0.001 (2) | −0.006 (2) |
| C14 | 0.057 (2) | 0.0475 (18) | 0.0423 (18) | −0.0035 (17) | −0.0138 (16) | 0.0029 (15) |
Geometric parameters (Å, °)
| O1—C2 | 1.369 (3) | C5—H5A | 0.9300 |
| O1—C1 | 1.425 (4) | C6—C7 | 1.360 (5) |
| C1—H1A | 0.9600 | C6—H6A | 0.9300 |
| C1—H1B | 0.9600 | C7—H7A | 0.9300 |
| C1—H1C | 0.9600 | C8—C13 | 1.362 (4) |
| O2—C8 | 1.384 (3) | C8—C9 | 1.377 (4) |
| O2—C4 | 1.389 (3) | C9—C10 | 1.407 (4) |
| C2—C3 | 1.362 (4) | C9—C14 | 1.477 (4) |
| C2—C7 | 1.388 (4) | C10—C11 | 1.366 (4) |
| O3—C14 | 1.229 (3) | C10—H10A | 0.9300 |
| C3—C4 | 1.389 (4) | C11—C12 | 1.366 (4) |
| C3—H3A | 0.9300 | C11—H11A | 0.9300 |
| O4—C14 | 1.296 (3) | C12—C13 | 1.386 (5) |
| O4—H4A | 0.8200 | C12—H12A | 0.9300 |
| C4—C5 | 1.362 (4) | C13—H13A | 0.9300 |
| C5—C6 | 1.379 (4) | ||
| C2—O1—C1 | 117.7 (2) | C6—C7—C2 | 119.7 (3) |
| O1—C1—H1A | 109.5 | C6—C7—H7A | 120.1 |
| O1—C1—H1B | 109.5 | C2—C7—H7A | 120.1 |
| H1A—C1—H1B | 109.5 | C13—C8—C9 | 121.8 (3) |
| O1—C1—H1C | 109.5 | C13—C8—O2 | 119.6 (3) |
| H1A—C1—H1C | 109.5 | C9—C8—O2 | 118.1 (3) |
| H1B—C1—H1C | 109.5 | C8—C9—C10 | 117.7 (3) |
| C8—O2—C4 | 120.0 (2) | C8—C9—C14 | 124.2 (3) |
| O1—C2—C3 | 124.2 (3) | C10—C9—C14 | 118.1 (3) |
| O1—C2—C7 | 116.2 (3) | C11—C10—C9 | 120.5 (3) |
| C3—C2—C7 | 119.6 (3) | C11—C10—H10A | 119.8 |
| C2—C3—C4 | 119.5 (3) | C9—C10—H10A | 119.8 |
| C2—C3—H3A | 120.2 | C10—C11—C12 | 120.6 (3) |
| C4—C3—H3A | 120.2 | C10—C11—H11A | 119.7 |
| C14—O4—H4A | 109.5 | C12—C11—H11A | 119.7 |
| C5—C4—O2 | 125.0 (3) | C11—C12—C13 | 119.8 (3) |
| C5—C4—C3 | 121.5 (3) | C11—C12—H12A | 120.1 |
| O2—C4—C3 | 113.4 (3) | C13—C12—H12A | 120.1 |
| C4—C5—C6 | 117.9 (3) | C8—C13—C12 | 119.7 (4) |
| C4—C5—H5A | 121.1 | C8—C13—H13A | 120.2 |
| C6—C5—H5A | 121.1 | C12—C13—H13A | 120.2 |
| C7—C6—C5 | 121.7 (3) | O3—C14—O4 | 122.0 (3) |
| C7—C6—H6A | 119.1 | O3—C14—C9 | 123.2 (3) |
| C5—C6—H6A | 119.1 | O4—C14—C9 | 114.8 (3) |
| C1—O1—C2—C3 | 3.1 (5) | C13—C8—C9—C10 | −0.1 (5) |
| C1—O1—C2—C7 | −177.4 (3) | O2—C8—C9—C10 | 171.1 (3) |
| O1—C2—C3—C4 | 179.6 (3) | C13—C8—C9—C14 | 178.6 (3) |
| C7—C2—C3—C4 | 0.1 (5) | O2—C8—C9—C14 | −10.2 (5) |
| C8—O2—C4—C5 | −9.0 (5) | C8—C9—C10—C11 | 1.3 (4) |
| C8—O2—C4—C3 | 171.1 (3) | C14—C9—C10—C11 | −177.5 (3) |
| C2—C3—C4—C5 | −1.3 (5) | C9—C10—C11—C12 | −1.0 (5) |
| C2—C3—C4—O2 | 178.6 (3) | C10—C11—C12—C13 | −0.6 (6) |
| O2—C4—C5—C6 | −178.6 (3) | C9—C8—C13—C12 | −1.5 (5) |
| C3—C4—C5—C6 | 1.3 (5) | O2—C8—C13—C12 | −172.5 (3) |
| C4—C5—C6—C7 | −0.1 (5) | C11—C12—C13—C8 | 1.8 (6) |
| C5—C6—C7—C2 | −1.1 (6) | C8—C9—C14—O3 | −16.7 (5) |
| O1—C2—C7—C6 | −178.4 (3) | C10—C9—C14—O3 | 162.0 (3) |
| C3—C2—C7—C6 | 1.1 (5) | C8—C9—C14—O4 | 163.9 (3) |
| C4—O2—C8—C13 | −67.7 (4) | C10—C9—C14—O4 | −17.4 (4) |
| C4—O2—C8—C9 | 121.0 (3) |
Hydrogen-bond geometry (Å, °)
| Cg1 is the centroid of the C2–C7 ring. |
| D—H···A | D—H | H···A | D···A | D—H···A |
| O4—H4A···O3i | 0.82 | 1.82 | 2.633 (3) | 173 |
| C1—H1B···Cg1ii | 0.96 | 2.89 | 3.784 (4) | 155 |
Symmetry codes: (i) −x+2, −y, −z+2; (ii) −x+3/2, y+1/2, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HG5013).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc Perkin Trans.2, pp. S1–19.
- Enraf–Nonius (1985). CAD-4 Software Enraf–Nonius, Delft, The Netherlands.
- Gapinski, D. M., Mallett, B. E., Froelich, L. L. & Jackson, W. T. (1990). J. Med. Chem. 33, 2798-2807. [DOI] [PubMed]
- Harms, K. & Wocadlo, S. (1995). XCAD4 University of Marburg, Germany.
- Jackson, W. T., Boyd, R. J., Froelich, L. L., Gapinski, D. M., Mallett, B. E. & Sawyer, J. S. (1993). J. Med. Chem. 36, 1726–1734. [DOI] [PubMed]
- North, A. C. T., Phillips, D. C. & Mathews, F. S. (1968). Acta Cryst. A24, 351–359.
- Pellón, R. F., Carrasco, R., Milián, V.& Rodes, L. (1995). Synth. Commun. 25, 1077–1083.
- Raghunathan, S., Chandrasekhar, K. & Pattabhi, V. (1982). Acta Cryst. B38, 2536–2538.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shi, L., Zhang, Q., Xiao, Q., Wu, T. & Zhu, H.-J. (2011). Acta Cryst. E67, o748. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811011019/hg5013sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811011019/hg5013Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




