Abstract
In the title compound, C30H38O4S2, the centroid of the benzene ring lies on a center of inversion. The thiophene ring is aligned at 49.8 (1)° with respect to the benzene ring. The alkyl chain adopts an extended zigzag conformation.
Related literature
The title compound and its derivatives are used in the preparation of organic semiconductors. For applications of these materials, see: Tian et al. (2010 ▶); Zhang et al. (2010 ▶). For the synthesis of related compounds, see: Fraind & Tovar (2010 ▶); Gurthrie & Tovar (2008 ▶), Hotta (2001 ▶); Kang et al. (1997 ▶); Lois et al. (2007 ▶); Shao & Zhao (2009 ▶); Zhao et al. (2007 ▶). 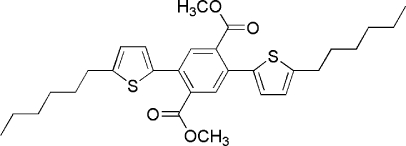
Experimental
Crystal data
C30H38O4S2
M r = 526.72
Monoclinic,

a = 15.617 (6) Å
b = 8.083 (3) Å
c = 11.585 (4) Å
β = 104.470 (4)°
V = 1416.0 (9) Å3
Z = 2
Mo Kα radiation
μ = 0.22 mm−1
T = 293 K
0.35 × 0.32 × 0.19 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2005 ▶) T min = 0.927, T max = 0.959
6083 measured reflections
2490 independent reflections
1865 reflections with I > 2σ(I)
R int = 0.031
Refinement
R[F 2 > 2σ(F 2)] = 0.041
wR(F 2) = 0.104
S = 1.04
2490 reflections
165 parameters
H-atom parameters constrained
Δρmax = 0.16 e Å−3
Δρmin = −0.23 e Å−3
Data collection: APEX2 (Bruker, 2005 ▶); cell refinement: SAINT (Bruker, 2005 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811011718/ng5141sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811011718/ng5141Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors are grateful for financial support from the National Natural Science Foundation of China (NSFC; 20872055, 21073079, J0730425) and the 111 project.
supplementary crystallographic information
Comment
The title compound and its derivatives are important materials for the preparation of various organic semiconductors, which could find applications in the fields of organic light-emitting diodes (OLEDs), organic field-effect transistors (OFETs) or solar cells (Tian et al., 2010; Zhang et al., 2010). The device performance of these organic semiconductors are strongly dependent on the molecular packing in their crystals. This led us to pay attention to the synthesis and crystal structure of the compound. Herein, we report the synthesis and structure of the title compound, namely dimethyl 2,5-bis(5-hexylthiophen-2-yl)benzene-1,4-dioate.
The molecular structure of the title compound is shown in Fig.1. Bond lengths and angles in the molecule are within normal ranges. The bond length of C—H in the thiophene and the benzene rings is 0.93 Å and the angle formed by C—S—C in the thiophene is 92.79°. In this structure, the two thiophene rings and the benzene ring are not in the same plane, which give a dihedral angle of 49.84°.
Experimental
The title compound was synthesized by Suzuki cross-coupling reaction. Briefly, thiophene-2-boronic acid (2.52 g, 10.00 mmol), dimethyl 2,5-dibromobenzene-1,4-dioate (880 mg, 2.50 mmol), Pd(PPh3)4 (115 mg, 0.10 mmol) and NaHCO3 (840 mg, 10.00 mmol) were mixed in dry THF (10 ml) under argon. The mixture was stirred and refluxed for 24 hrs (monitored by TLC) to give the title compound. The product was purified by column chromatography. Colorless crystals were grown via evaporation from n-hexane and dichloromethane (10:1, v:v) mixture solvents at room temperature for X-ray diffraction.
Refinement
All H atoms were placed in geometrically idealized positions, with C—H = 0.93 Å, and constrained to ride on their respective parent atoms, with Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.
The molecular structure of the title compound. Displacement ellipsoids are drawn at the 30% probability level. Symmetry operations: a = -x, 1 - y, -z.
Fig. 2.
Molecular packing of the title compound (I).
Crystal data
| C30H38O4S2 | F(000) = 564 |
| Mr = 526.72 | Dx = 1.235 Mg m−3 |
| Monoclinic, P2/c | Melting point: 362 K |
| Hall symbol: -P 2yc | Mo Kα radiation, λ = 0.71073 Å |
| a = 15.617 (6) Å | Cell parameters from 1616 reflections |
| b = 8.083 (3) Å | θ = 2.5–25.8° |
| c = 11.585 (4) Å | µ = 0.22 mm−1 |
| β = 104.470 (4)° | T = 293 K |
| V = 1416.0 (9) Å3 | Block, colorless |
| Z = 2 | 0.35 × 0.32 × 0.19 mm |
Data collection
| Bruker APEXII CCD diffractometer | 2490 independent reflections |
| Radiation source: fine-focus sealed tube | 1865 reflections with I > 2σ(I) |
| graphite | Rint = 0.031 |
| φ and ω scans | θmax = 25.1°, θmin = 2.5° |
| Absorption correction: multi-scan (SADABS; Bruker, 2005) | h = −16→18 |
| Tmin = 0.927, Tmax = 0.959 | k = −8→9 |
| 6083 measured reflections | l = −13→13 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.041 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.104 | H-atom parameters constrained |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0496P)2 + 0.1877P] where P = (Fo2 + 2Fc2)/3 |
| 2490 reflections | (Δ/σ)max < 0.001 |
| 165 parameters | Δρmax = 0.16 e Å−3 |
| 0 restraints | Δρmin = −0.23 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.07441 (12) | 0.8250 (2) | −0.01209 (17) | 0.0320 (5) | |
| C2 | 0.03976 (12) | 0.6534 (2) | −0.00661 (17) | 0.0301 (4) | |
| C3 | 0.07574 (12) | 0.5472 (2) | 0.08892 (16) | 0.0309 (5) | |
| C4 | 0.15632 (12) | 0.5867 (2) | 0.18357 (17) | 0.0329 (5) | |
| C5 | 0.16918 (13) | 0.5716 (3) | 0.30285 (18) | 0.0411 (5) | |
| H5 | 0.1254 | 0.5364 | 0.3388 | 0.049* | |
| C6 | 0.25579 (14) | 0.6143 (3) | 0.36780 (19) | 0.0457 (6) | |
| H6 | 0.2739 | 0.6121 | 0.4506 | 0.055* | |
| C7 | 0.30953 (12) | 0.6585 (2) | 0.29820 (18) | 0.0364 (5) | |
| C8 | 0.40559 (13) | 0.7071 (3) | 0.3327 (2) | 0.0480 (6) | |
| H8A | 0.4122 | 0.8115 | 0.2946 | 0.058* | |
| H8B | 0.4393 | 0.6246 | 0.3020 | 0.058* | |
| C9 | 0.44453 (13) | 0.7245 (3) | 0.4653 (2) | 0.0472 (6) | |
| H9A | 0.4376 | 0.6205 | 0.5037 | 0.057* | |
| H9B | 0.4115 | 0.8080 | 0.4960 | 0.057* | |
| C10 | 0.54203 (14) | 0.7720 (3) | 0.4984 (2) | 0.0504 (6) | |
| H10A | 0.5750 | 0.6890 | 0.4672 | 0.060* | |
| H10B | 0.5489 | 0.8764 | 0.4604 | 0.060* | |
| C11 | 0.58113 (14) | 0.7884 (3) | 0.6310 (2) | 0.0539 (6) | |
| H11A | 0.5466 | 0.8688 | 0.6621 | 0.065* | |
| H11B | 0.5752 | 0.6830 | 0.6682 | 0.065* | |
| C12 | 0.67691 (15) | 0.8399 (3) | 0.6673 (2) | 0.0581 (7) | |
| H12A | 0.7115 | 0.7618 | 0.6342 | 0.070* | |
| H12B | 0.6827 | 0.9475 | 0.6330 | 0.070* | |
| C13 | 0.71464 (18) | 0.8491 (3) | 0.7999 (2) | 0.0792 (9) | |
| H13A | 0.6816 | 0.9277 | 0.8334 | 0.119* | |
| H13B | 0.7754 | 0.8833 | 0.8164 | 0.119* | |
| H13C | 0.7111 | 0.7422 | 0.8345 | 0.119* | |
| C14 | 0.03448 (12) | 0.3957 (2) | 0.09348 (17) | 0.0327 (5) | |
| H14 | 0.0574 | 0.3248 | 0.1569 | 0.039* | |
| C15 | 0.11166 (16) | 1.0295 (3) | −0.1356 (2) | 0.0549 (6) | |
| H15A | 0.0682 | 1.1060 | −0.1218 | 0.082* | |
| H15B | 0.1181 | 1.0434 | −0.2153 | 0.082* | |
| H15C | 0.1673 | 1.0503 | −0.0797 | 0.082* | |
| O1 | 0.08339 (9) | 0.86154 (16) | −0.12051 (12) | 0.0441 (4) | |
| O2 | 0.09033 (9) | 0.91985 (17) | 0.07033 (12) | 0.0444 (4) | |
| S1 | 0.25288 (3) | 0.65154 (7) | 0.15037 (5) | 0.0447 (2) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0258 (10) | 0.0378 (11) | 0.0301 (12) | −0.0019 (8) | 0.0026 (9) | −0.0002 (9) |
| C2 | 0.0278 (10) | 0.0345 (10) | 0.0280 (11) | −0.0041 (8) | 0.0071 (8) | −0.0029 (9) |
| C3 | 0.0264 (10) | 0.0364 (11) | 0.0283 (12) | −0.0025 (8) | 0.0039 (8) | −0.0035 (9) |
| C4 | 0.0285 (10) | 0.0340 (10) | 0.0337 (12) | −0.0047 (8) | 0.0031 (9) | −0.0004 (9) |
| C5 | 0.0347 (12) | 0.0546 (13) | 0.0321 (13) | −0.0116 (10) | 0.0048 (9) | 0.0031 (10) |
| C6 | 0.0422 (13) | 0.0577 (14) | 0.0311 (13) | −0.0109 (10) | −0.0025 (10) | 0.0009 (10) |
| C7 | 0.0282 (10) | 0.0383 (11) | 0.0379 (13) | −0.0035 (9) | −0.0010 (9) | −0.0023 (9) |
| C8 | 0.0298 (11) | 0.0531 (13) | 0.0562 (16) | −0.0086 (10) | 0.0017 (11) | −0.0064 (11) |
| C9 | 0.0319 (12) | 0.0486 (13) | 0.0535 (15) | −0.0053 (10) | −0.0032 (11) | −0.0030 (11) |
| C10 | 0.0336 (12) | 0.0491 (13) | 0.0603 (17) | −0.0045 (10) | −0.0037 (11) | −0.0043 (12) |
| C11 | 0.0404 (13) | 0.0528 (14) | 0.0580 (17) | −0.0087 (11) | −0.0072 (12) | 0.0015 (12) |
| C12 | 0.0387 (13) | 0.0582 (15) | 0.0659 (18) | −0.0052 (11) | −0.0086 (12) | −0.0039 (13) |
| C13 | 0.0651 (18) | 0.085 (2) | 0.066 (2) | −0.0177 (15) | −0.0225 (15) | 0.0089 (16) |
| C14 | 0.0298 (10) | 0.0379 (11) | 0.0279 (11) | −0.0003 (8) | 0.0023 (9) | 0.0011 (9) |
| C15 | 0.0672 (16) | 0.0483 (14) | 0.0546 (16) | −0.0134 (12) | 0.0257 (13) | 0.0083 (11) |
| O1 | 0.0577 (10) | 0.0423 (9) | 0.0349 (9) | −0.0128 (7) | 0.0162 (7) | −0.0015 (6) |
| O2 | 0.0542 (9) | 0.0409 (8) | 0.0345 (9) | −0.0097 (7) | 0.0039 (7) | −0.0071 (7) |
| S1 | 0.0315 (3) | 0.0660 (4) | 0.0355 (4) | −0.0110 (3) | 0.0062 (2) | −0.0034 (3) |
Geometric parameters (Å, °)
| C1—O2 | 1.201 (2) | C9—H9B | 0.9700 |
| C1—O1 | 1.331 (2) | C10—C11 | 1.510 (3) |
| C1—C2 | 1.496 (3) | C10—H10A | 0.9700 |
| C2—C14i | 1.390 (3) | C10—H10B | 0.9700 |
| C2—C3 | 1.403 (3) | C11—C12 | 1.508 (3) |
| C3—C14 | 1.391 (3) | C11—H11A | 0.9700 |
| C3—C4 | 1.483 (2) | C11—H11B | 0.9700 |
| C4—C5 | 1.351 (3) | C12—C13 | 1.503 (3) |
| C4—S1 | 1.728 (2) | C12—H12A | 0.9700 |
| C5—C6 | 1.416 (3) | C12—H12B | 0.9700 |
| C5—H5 | 0.9300 | C13—H13A | 0.9600 |
| C6—C7 | 1.349 (3) | C13—H13B | 0.9600 |
| C6—H6 | 0.9300 | C13—H13C | 0.9600 |
| C7—C8 | 1.505 (3) | C14—C2i | 1.390 (3) |
| C7—S1 | 1.721 (2) | C14—H14 | 0.9300 |
| C8—C9 | 1.511 (3) | C15—O1 | 1.452 (2) |
| C8—H8A | 0.9700 | C15—H15A | 0.9600 |
| C8—H8B | 0.9700 | C15—H15B | 0.9600 |
| C9—C10 | 1.523 (3) | C15—H15C | 0.9600 |
| C9—H9A | 0.9700 | ||
| O2—C1—O1 | 123.96 (18) | C11—C10—H10A | 108.8 |
| O2—C1—C2 | 124.28 (18) | C9—C10—H10A | 108.8 |
| O1—C1—C2 | 111.74 (16) | C11—C10—H10B | 108.8 |
| C14i—C2—C3 | 119.57 (16) | C9—C10—H10B | 108.8 |
| C14i—C2—C1 | 118.65 (16) | H10A—C10—H10B | 107.7 |
| C3—C2—C1 | 121.56 (16) | C12—C11—C10 | 115.4 (2) |
| C14—C3—C2 | 118.10 (17) | C12—C11—H11A | 108.4 |
| C14—C3—C4 | 118.50 (17) | C10—C11—H11A | 108.4 |
| C2—C3—C4 | 123.39 (17) | C12—C11—H11B | 108.4 |
| C5—C4—C3 | 128.22 (18) | C10—C11—H11B | 108.4 |
| C5—C4—S1 | 109.88 (14) | H11A—C11—H11B | 107.5 |
| C3—C4—S1 | 121.82 (14) | C13—C12—C11 | 114.0 (2) |
| C4—C5—C6 | 113.54 (19) | C13—C12—H12A | 108.7 |
| C4—C5—H5 | 123.2 | C11—C12—H12A | 108.7 |
| C6—C5—H5 | 123.2 | C13—C12—H12B | 108.7 |
| C7—C6—C5 | 113.65 (19) | C11—C12—H12B | 108.7 |
| C7—C6—H6 | 123.2 | H12A—C12—H12B | 107.6 |
| C5—C6—H6 | 123.2 | C12—C13—H13A | 109.5 |
| C6—C7—C8 | 129.7 (2) | C12—C13—H13B | 109.5 |
| C6—C7—S1 | 110.14 (15) | H13A—C13—H13B | 109.5 |
| C8—C7—S1 | 120.20 (16) | C12—C13—H13C | 109.5 |
| C7—C8—C9 | 114.55 (19) | H13A—C13—H13C | 109.5 |
| C7—C8—H8A | 108.6 | H13B—C13—H13C | 109.5 |
| C9—C8—H8A | 108.6 | C2i—C14—C3 | 122.33 (18) |
| C7—C8—H8B | 108.6 | C2i—C14—H14 | 118.8 |
| C9—C8—H8B | 108.6 | C3—C14—H14 | 118.8 |
| H8A—C8—H8B | 107.6 | O1—C15—H15A | 109.5 |
| C8—C9—C10 | 113.74 (19) | O1—C15—H15B | 109.5 |
| C8—C9—H9A | 108.8 | H15A—C15—H15B | 109.5 |
| C10—C9—H9A | 108.8 | O1—C15—H15C | 109.5 |
| C8—C9—H9B | 108.8 | H15A—C15—H15C | 109.5 |
| C10—C9—H9B | 108.8 | H15B—C15—H15C | 109.5 |
| H9A—C9—H9B | 107.7 | C1—O1—C15 | 115.35 (16) |
| C11—C10—C9 | 113.8 (2) | C7—S1—C4 | 92.78 (10) |
| O2—C1—C2—C14i | 129.1 (2) | C5—C6—C7—S1 | 1.1 (2) |
| O1—C1—C2—C14i | −49.3 (2) | C6—C7—C8—C9 | −6.6 (3) |
| O2—C1—C2—C3 | −45.5 (3) | S1—C7—C8—C9 | 174.20 (15) |
| O1—C1—C2—C3 | 136.11 (18) | C7—C8—C9—C10 | 179.42 (19) |
| C14i—C2—C3—C14 | −0.7 (3) | C8—C9—C10—C11 | −179.62 (19) |
| C1—C2—C3—C14 | 173.85 (17) | C9—C10—C11—C12 | −178.4 (2) |
| C14i—C2—C3—C4 | 178.01 (17) | C10—C11—C12—C13 | −177.9 (2) |
| C1—C2—C3—C4 | −7.4 (3) | C2—C3—C14—C2i | 0.7 (3) |
| C14—C3—C4—C5 | −48.0 (3) | C4—C3—C14—C2i | −178.05 (17) |
| C2—C3—C4—C5 | 133.3 (2) | O2—C1—O1—C15 | −2.0 (3) |
| C14—C3—C4—S1 | 128.25 (17) | C2—C1—O1—C15 | 176.42 (16) |
| C2—C3—C4—S1 | −50.5 (2) | C6—C7—S1—C4 | −0.48 (17) |
| C3—C4—C5—C6 | 177.57 (18) | C8—C7—S1—C4 | 178.84 (17) |
| S1—C4—C5—C6 | 0.9 (2) | C5—C4—S1—C7 | −0.28 (16) |
| C4—C5—C6—C7 | −1.4 (3) | C3—C4—S1—C7 | −177.16 (16) |
| C5—C6—C7—C8 | −178.1 (2) |
Symmetry codes: (i) −x, −y+1, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: NG5141).
References
- Bruker (2005). SMART, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Fraind, A. M. & Tovar, J. D. (2010). J. Phys. Chem. B, 114, 3104–3116. [DOI] [PubMed]
- Gurthrie, D. A. & Tovar, J. D. (2008). Org. Lett. 10, 4323–4326. [DOI] [PubMed]
- Hotta, S. (2001). J. Heterocycl. Chem. 38, 923–927.
- Kang, S.-K., Kim, J.-S. & Choi, S.-C. (1997). J. Org. Chem. 62, 4208–4209. [DOI] [PubMed]
- Lois, S., Florès, J.-C., Lère-Porte, J.-P., Serein-Spirau, F., Moreau, J. J. E., Miqueu, K., Sotiropoulos, J.-M., Baylère, P., Tillard, M. & Belin, C. (2007). Eur. J. Org. Chem. 4019–4031.
- Shao, M. & Zhao, Y. (2009). Tetrahedron Lett. 50, 6897–6900.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Tian, H., Deng, Y., Pan, F., Huang, L., Yan, D., Geng, Y. & Wang, F. (2010). J. Mater. Chem. 20, 7998-8004.
- Zhang, W., Smith, J., Watkins, S. E., Gysel, R., McGehee, M., Salleo, A., Kirkpatrick, J., Ashraf, S., Anthopoulos, T., Heeney, M. & McCulloch, I. (2010). J. Am. Chem. Soc. 132, 11437–11439. [DOI] [PubMed]
- Zhao, C., Zhang, Y. & Ng, M.-K. (2007). J. Org. Chem. 72, 6364–6371. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811011718/ng5141sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811011718/ng5141Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




