Abstract
Neurospora comprises a primary model system for the study of fungal genetics and biology. In spite of this, little is known about genome evolution in Neurospora. For example, the evolution of synonymous codon usage is largely unknown in this genus. In the present investigation, we conducted a comprehensive analysis of synonymous codon usage and its relationship to gene expression and gene length (GL) in Neurospora tetrasperma and Neurospora discreta. For our analysis, we examined codon usage among 2,079 genes per organism and assessed gene expression using large-scale expressed sequenced tag (EST) data sets (279,323 and 453,559 ESTs for N. tetrasperma and N. discreta, respectively). Data on relative synonymous codon usage revealed 24 codons (and two putative codons) that are more frequently used in genes with high than with low expression and thus were defined as optimal codons. Although codon-usage bias was highly correlated with gene expression, it was independent of selectively neutral base composition (introns); thus demonstrating that translational selection drives synonymous codon usage in these genomes. We also report that GL (coding sequences [CDS]) was inversely associated with optimal codon usage at each gene expression level, with highly expressed short genes having the greatest frequency of optimal codons. Optimal codon frequency was moderately higher in N. tetrasperma than in N. discreta, which might be due to variation in selective pressures and/or mating systems.
Keywords: Neurospora tetrasperma, Neurospora discreta, optimal codons, gene expression, bias, gene length
Introduction
The Neurospora genus contains organisms that are central to research in fungal genomics, biochemistry, and evolution. At present, however, much remains unknown about patterns and factors driving genome evolution in this taxonomic group, particularly for the emerging model organisms Neurospora tetrasperma and Neurospora discreta.
Although N. tetrasperma and N. discreta have been less extensively characterized than their close relative N. crassa (Davis and Perkins 2002; Borkovich et al. 2004), available data has revealed key life history and genomic traits for these taxa. Specifically, N. tetrasperma has been shown to be a highly self-fertile (pseudohomothallic) organism assumed to have evolved from a self-incompatible (heterothallic) ancestor (Raju and Perkins 1994; Dettman et al. 2003). In contrast, N. discreta is heterothallic (Perkins and Raju 1986). In both organisms, similar to most filamentous ascomycetes, sexual attraction and reproduction is regulated by the mating-type (mat) locus on the mating-type chromosomes (Shiu and Glass 2000; Casselton 2008). For N. tetrasperma, self-fertilization is promoted by the fact that nuclei carrying the two opposite mating types are contained in each ascospore (Shear and Dodge 1927; Raju and Perkins 1994; Jacobson 1995), whereas in N. discreta, mating occurs between two haploid partners containing nuclei of opposite type (Perkins and Raju 1986). Comparative phylogenetic analyses suggest that N. tetrasperma and N. discreta are not discrete species per se but rather are each comprised of distinct closely related phylogenetic lineages, that is with high levels of genetic diversity among lineages and low-genetic diversity within lineages (Dettman et al. 2006; Menkis et al. 2009). The haploid chromosome number in each taxon is 7, containing approximately 43 MB DNA (Dodge et al. 1950; Perkins and Raju 1986; http://genome.jgi-psf.org/). Currently, few data are available about the genomic traits and evolution of N. tetrasperma and N. discreta, including the evolution of codon usage in gene coding DNA.
Synonymous codons are not used randomly. Biases in synonymous codon usage may result from mutational pressure (Osawa et al. 1988; Sueoka 1988; Kano et al. 1991; Sharp et al. 1995) or from selective pressure for more efficient and accurate translation (Duret and Mouchiroud 1999; Duret 2000; Stoletzki and Eyre-Walker 2006). The hypothesis that codon usage is driven by selection has been supported by findings that codon usage biases are correlated to tRNA abundance (Ikemura 1982, 1985; Duret 2000). In addition, codon usage bias has been positively correlated with gene expression in numerous taxonomic groups (e.g., Escherichia coli, Saccharomyces, Caenorhabditis, Drosophila, Arabidopsis, Silene, Populus), findings indicative of selection for efficient translation (Sharp 1987; Duret and Mouchiroud 1999; Cutter et al. 2006, 2008; Ingvarsson 2008; Qiu et al. 2011). A key indicator of translational selection within genomes is the enhanced usage of a specific set of codons (i.e., optimal codons) in highly expressed genes as compared with lowly expressed genes. In this regard, an evaluation of the presence/absence of optimal codons and factors associated with their usage provides a means to test whether selection for translational efficiency and/or accuracy plays a major role in genome evolution (Sharp 1987; Stenico et al. 1994; Duret and Mouchiroud 1999; Cutter et al. 2008; Ingvarsson 2008).
Several factors in addition to gene expression have been associated with codon usage. In particular, gene length (GL) is believed to be inversely associated with bias in codon usage. For example, it has been reported that shorter genes (shorter coding sequences [CDS] regions) tend to have greater bias in codon usage in certain animals and plants (e.g., Drosophila, Arabidopsis, Silene, species of Caenorhabditis, Duret and Mouchiroud 1999; Cutter et al. 2008; Qiu et al. 2011). However, this correlation has not been found in certain other taxa (e.g., species of Caenorhabditis, Populus, Cutter et al. 2008; Ingvarsson 2008). At present, the role of GL, and particularly the relationship between gene expression and GL, remains largely unknown for most organisms. Another factor that may alter codon usage is mating system. Theory predicts that inbreeding species should have lower effective recombination rates (due to recombination among identical or nearly identical genomes) and reduced effective population size as compared with their outcrossing counterparts, each of which act to relax selective pressure for genomic traits such as codon usage (see Charlesworth and Wright 2001). Altogether, it is evident that a further understanding of the role of translational selection in Neurospora genomes requires an assessment of gene expression level, combined with evaluation of the possible roles of GL and mating systems.
One effective means to study gene expression, and its relationship to codon usage, in newly emerging model species such as N. tetrasperma and N. discreta, is through the examination of expressed sequence tag (EST) data. In particular, the study of ESTs have proven to be a highly effective method to quantify gene expression as the extent of redundancy in ESTs reflects the abundance of mRNA in tissues/cells (Duret and Mouchiroud 1999; Akashi 2001; Wright et al. 2002; Subramanian and Kumar 2004; Cutter et al. 2008; Ingvarsson 2008). In this regard, the recent availability of comprehensive EST data sets for N. tetrasperma and N. discreta allows for an examination of gene expression in these organisms and its association with codon usage.
The objective of the present investigation was to study synonymous codon usage within the N. tetrasperma and N. discreta genomes. For our analysis, we conducted a thorough assessment of codon usage within each of these taxa relative to gene expression. In addition, we assessed the codon usage relative to GL and the substitution rates at synonymous (dS) sites. As a complementary analysis, we assessed the role of base composition on codon usage, based on comparisons of introns and gene coding DNA. In combination, these analyses were aimed toward revealing the key factors driving the evolution of synonymous codon usage in these Neurospora genomes.
Materials and Methods
In this study, we investigated synonymous codon usage in the two Neurospora taxa for which both large-scale genomic and EST data sets are currently available: N. tetrasperma and N. discreta. For our analysis, we utilized the unannotated genome sequences for these two taxa available at the Joint Genome Institute (http://www.jgi.doe.gov/; The sequence data were produced by the US Department of Energy Joint Genome Institute in collaboration with the user community). For N. tetrasperma, the data are available from the haploid mat A strain with Fungal Genomics Stock Center (FGSC) ID 2508, and for N. discreta, the data are available from the haploid mat A strain FGSC 8579. Annotated gene sequences for N. crassa, which were used as reference taxon for gene identification in N. tetrasperma and N. discreta, were obtained from the N. crassa database (Annotation version 4, available June 2010; FGSC 2489; http://www.broad.mit.edu/annotation/genome/neurospora/). Among these three taxa, previous phylogenetic analyses have shown that N. tetrasperma and N. discreta form a clade relative to N. discreta; and the phylogentic tree is ([NT, NC], ND) (Dettman et al. 2003). For gene expression analysis in N. tetrasperma and N. discreta, we used EST data sets available at the Joint Genome Institute, which contain 279,323 and 453,559 ESTs, respectively (table 1). EST data sets were generated slightly differently for each taxon (e.g., tissues were grown and harvested at the vegetative stages, with evidence of emerging sexual tissue in N. tetrasperma but not N. discreta; Takao Kasuga, personal communication). Thus, independent analyses were always conducted for N. tetrasperma and for N. discreta.
Table 1.
Summary of Gene and EST Data Used in the Present Analysis
| Taxon | Genes |
ESTs |
|||
| Number of Genes | Number of Highly Expressed Genes | Number of Lowly Expressed Genes | Number of ESTs Examined | Number of ESTs Matching Genes | |
| Neurospora tetrasperma | 2,079 | 688 | 1,391 | 279,323 | 105,003 |
| N. discreta | 2,079 | 702 | 1,377 | 453,559 | 161,242 |
Gene Identification
Genes (defined herein as CDS regions) for N. tetrasperma and N. discreta were identified by comparison of the assembled raw genomic sequence for these taxa to the annotated CDS data set for N. crassa using BLAT software (Kent 2002). Genes showing quality alignments, including a start and stop codon and spanning the complete CDS region for N. crassa, were identified, yielding a total of 2,151 genes. Genes were automatically aligned and gaps removed using the BLAT procedure. The identified gene regions in N. tetrasperma and N. discreta were assigned the NCU gene identifier of the matching gene in N. crassa. We removed sequences from gene families with highly similar (>90% identity) paralogous sequences, based on a comparison of the gene list against itself using MEGABLAST (http://www.ncbi.nlm.nih.gov/blast/megablast.shtm). The final gene list contained a total of 2,079 genes that were found in both N. tetrasperma and in N. discreta (table 1) and which were used in our gene expression and codon usage analysis. The NCU identifier for each of the 2,079 genes examined herein is provided in supplementary data file 1 (Supplementary Material online).
Introns were identified in both N. tetrasperma and N. discreta. For this, we extracted the genomic regions between exons of genes in N. tetrasperma and separately for N. discreta. In total, we identified introns for 1,752 of the 2,079 genes from N. tetrasperma and N. discreta. For each gene, introns were concatenated prior to analysis.
Quantification of Gene Expression
For the quantification of gene expression levels, each of the 2,079 genes for N. tetrasperma and the 2,079 homologous genes for N. discreta were compared against their corresponding taxon-specific EST data set (table 1) using MEGABLAST. ESTs having more than 95% sequence identity were considered a match; this level of identity is sufficient for correct EST matches among closely related genes and allows for minor variation due to sequencing errors (Subramanian and Kumar 2004). It is worthwhile to note that we examined a single haploid strain of N. tetrasperma and of N. discreta such that there is no allelic variation per gene and that we a priori removed genes having >90% identity, which ensures accurate matches between ESTs and genes herein. For N. tetrasperma, we found that a total of 105,003 ESTs (of 279,323 available ESTs) matched its 2,079 genes, whereas 161,242 ESTs (of 453,559 ESTs) matched the corresponding gene set in N. discreta (table 1). Using these data, we calculated the frequency of ESTs for each of the 2,079 genes for each of the two Neurospora taxa as follows: ESTs per 100,000 = the number of EST matches per gene/total number of ESTs per taxon-specific data set × 100,000. Subsequently, each of the 2,079 genes was categorized as either lowly (<10 ESTs per 100,000, including genes showing no evidence of expression) or highly expressed (≥10 ESTs per 100,000) for N. tetrasperma and for N. discreta, respectively. Approximately, 1/3 of genes were above the cutoff of ten ESTs per 100,000, whereas 2/3 of genes were below this level for each taxon. A summary of the number of genes within each category of gene expression for N. tetrasperma and for N. discreta is provided in table 1.
Codon Usage Analyses
Using the gene sequences and gene expression data sets generated above, we assessed the association between codon usage and gene expression within N. tetrasperma and within N. discreta. For this, we examined the effective number of codons (ENCs); a broad measure of how far the codon usage of a gene departs from equal usage of synonymous codons. ENC can have a lowermost value of 20, in the case of extreme bias where one codon is solely used for each amino acid, and an uppermost value of 61 when all alternative synonymous codons are used equally. Thus, lower ENC values represent greater bias in codon usage (Wright 1990). We also examined the GC content of third nucleotide positions of codons (GC3). GC3 and ENC were determined in CodonW (J. Peden, http://codonw.sourceforge.net).
The preferential usage of codons has been described by various terms in the literature. For example, “preferred” and “major” may refer to those most frequent in the genome, those most frequent in genes showing biased codon usage and/or those associated with high gene expression (e.g., Akashi 2001; Vicario et al. 2007). For clarity, for our purposes, we have used the term “optimal” codons herein to denote those codons specifically associated with elevated gene expression (Duret and Mouchiroud 1999; Cutter et al. 2008; Ingvarsson 2008). In order to determine whether optimal codons are inherent to N. tetrasperma and N. discreta, we determined the relative synonymous codon usage (RSCU) for each codon per synonymous codon family, for each of the genes examined in this study. RSCU measures the observed frequency of a particular codon relative to the expected frequency if all synonymous codons were used equally. RSCU values greater than 1 indicate preferential usage and higher values among codons within a synonymous codon family denote increased usage (Sharp et al. 1986). The RCSU values were determined by using CAIcal software (Puigbo et al. 2008), and the analyses were performed independently in the two taxa. Using these data, we determined the value of ▵RSCU = Mean RSCUHighlyExpressed Genes – Mean RSCULowly Expressed Genes for each codon per synonymous codon family. Optimal codons were defined as those codons having statistically significant and positive ▵RSCU values (Duret and Mouchiroud 1999; Cutter et al. 2008; Ingvarsson 2008). Statistical significance was assessed using t-tests of RSCU values among highly and lowly expressed genes (P < 0.05). A Bonferroni correction was applied to all P values.
Based on the optimal codon list defined using the above methodology, we determined the frequency of optimal codons (Fop) per gene for N. tetrasperma and for N. discreta using CodonW (J. Peden, http://codonw.sourceforge.net). Subsequently, we conducted two-way analysis of variance (ANOVA) for each taxon with Fop as the dependent variable and gene expression and GL as categorical factors (Cutter et al. 2006). GLs were assumed to equal the values from N. crassa, as the complete and/or annotated genomic DNA or protein sequences are not yet available for N. tetrasperma and N. discreta. This approach has also been utilized in other organisms (Cutter et al. 2008; Ingvarsson 2008) as protein lengths tend to be highly conserved among eukaryotes (Wang et al. 2004). For the ANOVA's, gene expression was categorized as low or high (<10 ESTs per 100,000 or ≥10 ESTs per 100,000, respectively) and GLs were subdivided into three categories (short, GL < 250 codons; medium, ≤ 250 GL > 500; and long, GL ≥ 500 codons). Each taxon was analyzed independently in the ANOVA's. Post hoc pair-wise analyses of our ANOVA data were conducted using the Holm-Sidak method. All statistical analyses, including ANOVAs and t-tests, were conducted using Sigmastat v3 (http://www.systat.com).
Analysis of Synonymous and Nonsynonymous Substitutions
Measurements of dS for N. tetrasperma (NT) and for N. discreta (ND) were conducted on concatenated sequences for the lowly expressed genes and separately for highly expressed genes for each taxon. The corresponding genes from N. crassa (NC) were identified for each of these gene sets and used to measure dS values. dS was determined using the PAML package yn00 (Yang 2007).
Results
Based on the analysis of 2,079 genes and comprehensive EST data sets, we assessed the relationship between codon usage and gene expression in N. tetrasperma and N. discreta. Lowly expressed genes were defined as those with less than 10 ESTs hits per 100,000 (including genes showing no evidence of expression), whereas highly expressed genes have equal to or greater than 10 ESTs per 100,000 (table 1). We first assessed the ENCs and the GC content of third nucleotide positions (GC3) of codons relative to gene expression. Our data reveal that highly expressed genes have statistically significantly lower ENC values than lowly expressed gene (P < 0.05; fig. 1). This is consistent with bias toward the preferential usage of a specific subset of codons in more highly expressed genes. In addition, we found that GC3 was positively associated with gene expression (fig. 1), suggesting that the observed codon usage bias is associated with greater usage of GC ending codons in highly expressed genes; this parallels an association between GC3 and bias in codon usage reported in certain other organisms (Akashi 2001).
FIG. 1.—
The relationship between gene expression level and codon usage in Neurospora species. (A) The mean ENCs for lowly (<10 ESTs per 100,000) and highly (≥10ESTs per 100,000) expressed genes. (B) The mean GC3 values for lowly and highly expressed genes. Error bars represent standard errors. Different letters (a or b) above bars indicate statistically significant differences (P < 0.05).
Optimal Codons
We identified optimal codons based on a statistically significantly higher RSCU within genes having high expression as compared with those with low expression, that is a positive ▵RSCU (Duret and Mouchiroud 1999; Cutter et al. 2008; Ingvarsson 2008). Our analysis of ▵RSCU across all 2,079 genes for N. tetrasperma and for N. discreta reveal a list of optimal codons that are identical for the two taxa: 24 codons are statistically significantly more frequent in highly expressed than in lowly expressed genes (P < 0.05; in all t-tests after Bonferroni correction, table 2; note that standard errors are provided in supplementary table 1, Supplementary Material online). Two putative optimal codons were also identified for the amino acids aspartic acid and proline. For these two amino acids, one codon was utilized more often in highly expressed genes, although this difference was not statistically significant. In totality, our data provide marked evidence that elevated gene expression is associated with usage of an optimal set of codons in Neurospora taxa, a finding consistent with selection on codon usage in their genomes.
Table 2.
The Difference in Relative Synonymous Codon Usage (▵RSCU) in Highly Versus Lowly Expressed Genes in Neurospora tetrasperma and N. discreta
|
N. tetrasperma |
N. discreta |
||||||||
| Mean RSCU |
▵RSCU | P Value | Mean RSCU |
▵RSCU | P Value | ||||
| Codon | Amino Acid | High Exp | Low Exp | High Exp | Low Exp | ||||
| GCC | Ala | 2.2219 | 1.7797 | +0.4422 | ** | 2.1579 | 1.7937 | +0.3642 | ** |
| GCT | Ala | 1.0598 | 0.8731 | +0.1867 | ** | 1.1142 | 0.8552 | +0.2589 | ** |
| GCA | Ala | 0.1956 | 0.4777 | −0.2821 | ** | 0.2250 | 0.4806 | −0.2556 | ** |
| GCG | Ala | 0.5228 | 0.8695 | −0.3467 | ** | 0.5029 | 0.8704 | −0.3675 | ** |
| CGC | Arg | 2.8901 | 2.0840 | +0.8061 | ** | 2.7968 | 2.0820 | +0.7148 | ** |
| CGT | Arg | 1.3149 | 0.8111 | +0.5038 | ** | 1.3491 | 0.8175 | +0.5316 | ** |
| AGA | Arg | 0.4472 | 0.6415 | −0.1943 | ** | 0.4670 | 0.6282 | −0.1612 | ** |
| CGA | Arg | 0.2029 | 0.5418 | −0.3390 | ** | 0.2552 | 0.5433 | −0.2881 | ** |
| CGG | Arg | 0.4235 | 0.7713 | −0.3478 | ** | 0.4070 | 0.7761 | −0.3691 | ** |
| AGG | Arg | 0.6866 | 1.1374 | −0.4508 | ** | 0.6823 | 1.1442 | −0.4619 | ** |
| AAC | Asn | 1.7438 | 1.5007 | +0.2431 | ** | 1.7106 | 1.4822 | +0.2284 | ** |
| AAT | Asn | 0.2504 | 0.4893 | −0.2388 | ** | 0.2837 | 0.5047 | −0.2211 | ** |
| GACa | Asp | 1.2528 | 1.1994 | +0.0534 | 1.2209 | 1.1920 | +0.0289 | ||
| GAT | Asp | 0.7297 | 0.7949 | −0.0651 | * | 0.7563 | 0.8036 | −0.0473 | |
| TGC | Cys | 1.4708 | 1.3093 | +0.1615 | * | 1.4880 | 1.3006 | +0.1874 | ** |
| TGT | Cys | 0.2152 | 0.4578 | −0.2426 | ** | 0.2157 | 0.4583 | −0.2426 | ** |
| CAG | Gln | 1.6003 | 1.2852 | +0.3151 | ** | 1.5706 | 1.2830 | +0.2876 | ** |
| CAA | Gln | 0.3881 | 0.7019 | −0.3138 | ** | 0.4180 | 0.7097 | −0.2917 | ** |
| GAG | Glu | 1.6666 | 1.3836 | +0.2830 | ** | 1.6307 | 1.3854 | +0.2452 | ** |
| GAA | Glu | 0.3305 | 0.6150 | −0.2845 | ** | 0.3636 | 0.6117 | −0.2480 | ** |
| GGT | Gly | 1.4238 | 0.9472 | +0.4766 | ** | 1.4477 | 0.9362 | +0.5115 | ** |
| GGC | Gly | 2.0348 | 1.8612 | +0.1736 | ** | 1.9888 | 1.8626 | +0.1262 | ** |
| GGA | Gly | 0.3594 | 0.6709 | −0.3115 | ** | 0.3850 | 0.6634 | −0.2784 | ** |
| GGG | Gly | 0.1820 | 0.5178 | −0.3358 | ** | 0.1785 | 0.5349 | −0.3564 | ** |
| CAC | His | 1.5375 | 1.2801 | +0.2574 | ** | 1.5154 | 1.2620 | +0.2534 | ** |
| CAT | His | 0.4014 | 0.6610 | −0.2595 | ** | 0.4219 | 0.6799 | −0.2579 | ** |
| ATC | Ile | 2.0657 | 1.8729 | +0.1928 | ** | 2.0337 | 1.8630 | +0.1708 | ** |
| ATT | Ile | 0.8619 | 0.9150 | −0.0531 | 0.8770 | 0.9196 | −0.0426 | ||
| ATA | Ile | 0.0549 | 0.2078 | −0.1529 | ** | 0.0637 | 0.2153 | −0.1516 | ** |
| CTC | Leu | 2.9099 | 2.0755 | +0.8344 | ** | 2.8151 | 2.0548 | +0.7603 | ** |
| CTT | Leu | 1.1064 | 0.9779 | +0.1285 | * | 1.1957 | 0.9677 | +0.2280 | ** |
| TTA | Leu | 0.0391 | 0.1267 | −0.0876 | ** | 0.0474 | 0.1303 | −0.0829 | ** |
| CTA | Leu | 0.1668 | 0.3896 | −0.2228 | ** | 0.1693 | 0.3999 | −0.2306 | ** |
| TTG | Leu | 0.7561 | 1.0437 | −0.2876 | ** | 0.7508 | 1.0453 | −0.2944 | ** |
| CTG | Leu | 1.0216 | 1.3779 | −0.3563 | ** | 1.0046 | 1.4020 | −0.3974 | ** |
| AAG | Lys | 1.8633 | 1.6207 | +0.2427 | ** | 1.8431 | 1.6130 | +0.2302 | ** |
| AAA | Lys | 0.1338 | 0.3765 | −0.2427 | ** | 0.1569 | 0.3841 | −0.2273 | ** |
| TTC | Phe | 1.5947 | 1.3220 | +0.2727 | ** | 1.5631 | 1.3051 | +0.2580 | ** |
| TTT | Phe | 0.3908 | 0.6737 | −0.2829 | ** | 0.4283 | 0.6905 | −0.2622 | ** |
| CCC | Pro | 2.3242 | 1.6341 | +0.6901 | ** | 2.2490 | 1.6394 | +0.6097 | ** |
| CCTa | Pro | 0.9147 | 0.8604 | +0.0543 | 0.9765 | 0.8508 | +0.1256 | * | |
| CCA | Pro | 0.2523 | 0.5846 | −0.3322 | ** | 0.2679 | 0.5970 | −0.3291 | ** |
| CCG | Pro | 0.4855 | 0.9180 | −0.4325 | ** | 0.4838 | 0.9099 | −0.4260 | ** |
| TCC | Ser | 2.1815 | 1.5580 | +0.6235 | ** | 2.0904 | 1.5761 | +0.5143 | ** |
| TCT | Ser | 0.9250 | 0.7278 | +0.1972 | ** | 0.9664 | 0.7151 | +0.2513 | ** |
| AGC | Ser | 1.3687 | 1.4512 | −0.0825 | 1.3735 | 1.4404 | −0.0668 | ||
| TCG | Ser | 1.0090 | 1.1733 | −0.1642 | * | 0.9901 | 1.1778 | −0.1877 | * |
| AGT | Ser | 0.2827 | 0.5621 | −0.2794 | ** | 0.3103 | 0.5628 | −0.2525 | ** |
| TCA | Ser | 0.2156 | 0.5276 | −0.3120 | ** | 0.2523 | 0.5278 | −0.2756 | ** |
| ACC | Thr | 2.3811 | 1.7518 | +0.6293 | ** | 2.3053 | 1.7700 | +0.5353 | ** |
| ACT | Thr | 0.7616 | 0.6536 | +0.1079 | * | 0.7898 | 0.6393 | +0.1504 | ** |
| ACA | Thr | 0.3168 | 0.6412 | −0.3243 | ** | 0.3561 | 0.6244 | −0.2683 | ** |
| ACG | Thr | 0.5405 | 0.9533 | −0.4128 | ** | 0.5431 | 0.9662 | −0.4231 | ** |
| TAC | Tyr | 1.5653 | 1.3606 | +0.2047 | ** | 1.5366 | 1.3610 | +0.1756 | ** |
| TAT | Tyr | 0.4027 | 0.5991 | −0.1964 | ** | 0.4179 | 0.6056 | −0.1877 | ** |
| GTC | Val | 2.2600 | 1.7407 | +0.5193 | ** | 2.2085 | 1.7176 | +0.4909 | ** |
| GTT | Val | 0.9744 | 0.8771 | +0.0973 | * | 1.0108 | 0.8908 | +0.1199 | ** |
| GTA | Val | 0.1475 | 0.3198 | −0.1723 | ** | 0.1673 | 0.3160 | −0.1488 | ** |
| GTG | Val | 0.6124 | 1.0538 | −0.4414 | ** | 0.6021 | 1.0696 | −0.4676 | ** |
Note.—Pair-wise t-tests were conducted for each codon across all highly expressed versus all lowly expressed genes. P values are shown and have been adjusted for Bonferroni correction (*indicates 0.05< P > 0.00001; **indicates P ≤ 0.00001). Codons in bold have been assigned as optional codons (N = 26). Underlined codons are the primary optimal codon per synonymous codon family that is largest positive ▵RSCU. The standard errors for mean RSCU values are provided in supplementary table 1 (Supplementary Material online).
This codon was designated as a putative optimal codon for this amino acid as RSCU has a greater value in the highly expressed genes, even though comparison is not statistically significant.
Several trends were detected in our analyses of optimal codon usage in these two Neurospora genomes. Specifically, we found that the vast majority of optimal codons contain C at the third nucleotide position (table 2). For amino acids encoded by four or more codons (e.g., leucine, valine, serine, proline, threonine, alanine, arginine, and glycine), we found that the primary optimal codon, defined as the codon with the largest positive ▵RSCU values, always contained C at the third codon position. The secondary optimal codon (with the second highest ▵RSCU) tended to be terminated by T. Our data also show that there is a strong preference for the primary optimal codon as compared with the secondary optimal codon. For example, for leucine, the ▵RSCU for the primary optimal codon (CTC) is +0.8344, whereas this value is only +0.1285 for the secondary optimal codon (CTT) in N. tetrasperma. For those amino acids that do not have any synonymous codons with C at the third nucleotide position, for example, glutamine (encoded by two codons, CAG and CAA) and lysine (encoded by AAG and AAT), the optimal codon tended to end in G. The data demonstrates that these Neurospora genomes contain optimal codons spanning the 18 amino acids with synonymous codons and particularly favors those codons ending in C or G.
Fop Relative to Gene Expression and GL
Based on the optimal codon list defined above, we determined the Fop per gene for N. tetrasperma and for N. discreta. Subsequently, in order to assess the relative roles of gene expression and GL, we conducted two-way ANOVA analyses for each taxon with Fop as the dependent variable and gene expression and GL as categorical factors. Gene expression was categorized as low or high (as described above) and GLs subdivided into three categories (short, GL < 250 codons; medium, ≤ 250 GL > 500; and long, GL ≥ 500 codons). An independent analysis was conducted for N. tetrasperma and for N. discreta. The ANOVA results show that gene expression level explains a major component of variation in optimal codon usage in N. tetrasperma and in N. discreta, with values exceeding > 31.9% (table 3). The data also reveal that the proportion of Fop variation that is explained by gene expression is more than 6-fold higher than that explained by GL (e.g., 32%/4% = 8 for N. tetrasperma and 31.9%/5.18% = 6.2 for N. discreta, table 3). Statistically significant interaction was detected between gene expression and GL (P < 0.05) but was a relatively minor factor determining variation in codon usage (<1%; table 3). Notably, nearly identical results were obtained when three categories of gene expression: low, high, and no expression, were used in the ANOVA's (data not shown). In sum, these data indicate that gene expression level is a key factor driving codon usage, whereas GL plays a statistically significant, but relatively minor, role in these genomes.
Table 3.
Two-Way ANOVA Results for the Fop with Gene Expression and GL as Factors
|
Neurospora tetrasperma |
N. discreta |
|||||||||||
| DOFa | Sum of Squares | Mean Square | F | P Value | Proportion of Variation Explained | DOFa | Sum of Squares | Mean Square | F | P Value | Proportion of Variation Explained | |
| Gene expression | 1 | 9.95 | 9.95 | 1038.92 | <10−10 | 32.0% | 1 | 9.77 | 9.77 | 1026.74 | <10−10 | 31.9% |
| GL | 2 | 1.24 | 0.62 | 63.75 | <10−10 | 4.00% | 2 | 1.59 | 0.80 | 83.81 | <10−10 | 5.18% |
| Gene expression × GL | 2 | 0.07 | 0.03 | 3.49 | 0.03 | 0.22% | 2 | 0.30 | 0.15 | 15.93 | 3.0 × <10−7 | 0.98% |
| Residual | 2,073 | 19.85 | 0.01 | 2,073 | 19.72 | 0.01 | ||||||
| Total | 2,078 | 31.07 | 0.02 | 2,078 | 30.64 | 0.02 | ||||||
Three categories of GLs (short, medium, and long) and two categories (low and high) of gene expression were utilized. The N values for each of the six categories of gene expression and GL were as follows: N. tetrasperma NHigh_Short = 200, NHigh_Medium = 251, NHigh_Long = 237, NLow_Short = 398, NLow_Medium = 575, NLow_Long = 418; N. discreta, NHigh_Short = 186, NHigh_Medium = 261, NHigh_Long = 255, NLow_Short = 412, NLow_Medium = 565 NLow_Long = 400.
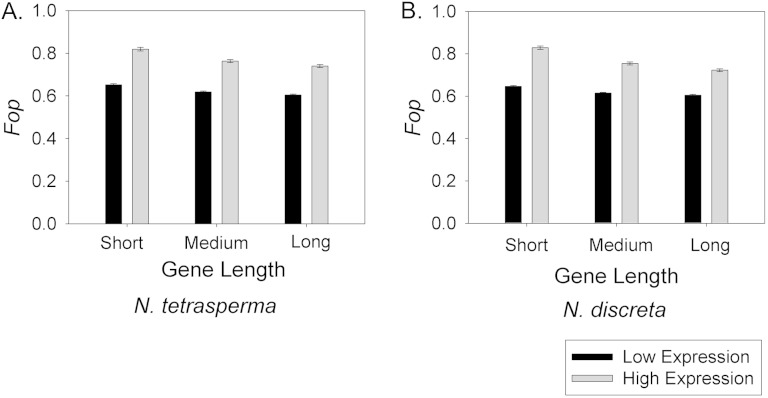
Post hoc pair-wise analyses of gene expression (within each GL category) and GL (within each gene expression category) from our ANOVA data using the Holm-Sidak method further reveal the patterns of optimal codon usage N. tetrasperma and in N. discreta. For example, these comparisons reveal that Fop is inversely correlated with GL, with a statistically significantly higher Fop for shorter genes than longer genes for each gene expression category. In addition, highly expressed genes have statistically significantly higher Fop than lowly expressed genes at all three GLs (fig. 2). Among all possible gene categories (i.e., among the six possible combinations of GL and gene expression level), the results show that the highest Fop values occur in highly expressed short genes; this trend suggests that these traits may be associated with the most efficient translation. It is particularly notable that highly expressed long genes have higher Fop values than lowly expressed medium length or short genes (fig. 2); this is consistent with the notion that gene expression plays a greater role in driving codon usage bias than GL. In totality, these findings support the conclusion that gene expression is a primary factor driving optimal codon usage in these Neurospora genomes.
FIG. 2.—
The Fop value for genes from different GL categories and gene expression levels. (A) Neurospora tetrasperma and (B) N. discreta. All comparisons among GLs (per expression level) and among gene expression levels (per GL) within each figure are statistically significantly different (P < 0.05) after post hoc analysis of ANOVAs and correction for multiple tests. Error bars represent standard errors.
As a supplemental analysis, we assessed the synonymous substitutions (dS) for concatenated highly expressed genes and concatenated lowly expressed genes in N. tetrasperma and separately in N. discreta (relative to N. crassa). We found that dS values for highly expressed genes were markedly reduced (by between 5% and 22% depending on the model) as compared with lowly expressed genes for N. tetrasperma and for N. discreta (see supplementary table 2, Supplementary Material online). The findings of reduced dS for highly expressed genes were consistent across several substitution models. More than a 20% decline in dS was detected in highly expressed genes using models by Nei and Gojobori (1986) and Li (1993). These models do not account for codon usage bias; thus, we may infer that selection on codon usage likely drives the decline in dS. The model of Yang and Nielsen (2000), which accounts for codon usage bias, also showed reduced dS for highly expressed genes. In this case, the decline in dS in highly expressed genes relative to lowly expressed genes was between 5% and 7.8% (for N. tetrasperma and N. discreta, respectively), which is much lower than the other models and is consistent with a correction for codon usage in the estimation of dS. Nonetheless, the fact that a decline was still observed despite the correction for codon frequencies, suggests that the selection on codon usage in these Neurospora taxa may exceed the level that can be accounted for by the model. In sum, all the substitution models support reduced dS in highly expressed genes. This suggests that highly expressed genes, which have elevated bias in codon usage, evolve at a lower rate at synonymous sites; this finding further supports the role of translational selection in these Neurospora genomes (McVean and Vieira 2001; Ingvarsson 2008).
Introns
Mutational bias is one factor that could underlie variation in codon usage (Akashi 2001; Plotkin and Kudla 2010). This is an unlikely explanation in our study as bias in codon usage was highly correlated with gene expression (fig. 2 and tables 2 and 3). Nonetheless, it is conceivable that mutational biases might be associated with gene expression level and thus could explain our findings reported herein. In order to assess this possibility, we examined DNA sequences assumed to be selectively neutral, for example, introns, in order to further discern whether mutation or selection drives codon usage. In contrast to studies where gene sequences were derived from ESTs and thus introns were not available (e.g., Cutter et al. 2008; Ingvarsson 2008), our gene data set were derived from genomic DNA and thus allow separate analyses of intronic and exonic sequences. Introns were identified for 1,752 of the 2,079 genes from N. tetrasperma and N. discreta.
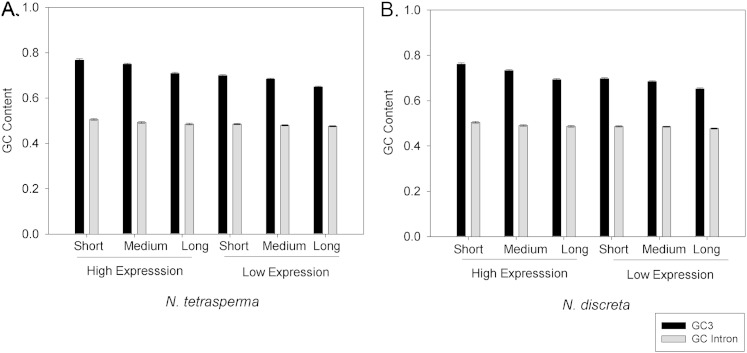
As described above, most of the optimal codons identified herein end in G or C nucleotides (table 2). Thus, comparison of GC3 and the GC content of introns (GCI) may be used to assess whether biases in codon usage are associated with mutational pressure (Wolfe et al. 1989; Smith and Eyre-Walker 2001; Vicario et al. 2007). As shown in figure 3, our data show that GC3 values are not associated with GCI for either N. tetrasperma or for N. discreta. For example, for highly expressed short genes in N. tetrasperma, we found that the mean GC3 value was 0.768 (±0.005), whereas the mean GCI value was only 0.524 (±0.004; P value of t-test <10−16). In fact, we found that the GC3 values were statistically significantly higher than GCI for genes within each of the six possible combinations of gene expression level (high, low) and GL (short, medium, long), with P < 10−16 in each of these comparisons (after Bonferroni correction). These findings indicate that the GC3 content is not evolving neutrally and that mutational biases cannot explain our findings. Thus, the marked association between gene expression and optimal codon usage in N. tetrasperma and in N. discreta are best explained by selection for translational efficiency and/or accuracy in these taxa.
FIG. 3.—
The mean GC content at third nucleotide positions of codons (GC3) and for introns of genes (GCI) from each combination of gene expression level (High, Low) and GL (Short, Medium, Long). (A) Neurospora tetrasperma (B) N. discreta. All comparisons among GC3 and GCI are statistically significantly different within each of the six combinations of gene expression and GL per taxon (t-tests P < 0.05 after Bonferroni correction). Error bars represent standard errors.
Differences in Codon Usage among Neurospora Taxa
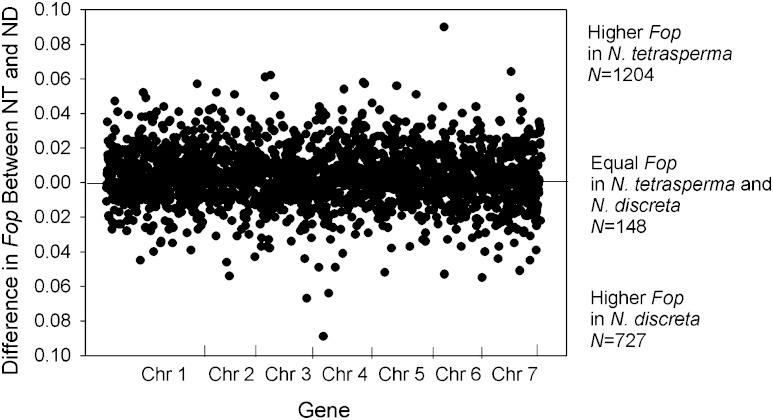
In order to further understand the evolutionary dynamics driving optimal codon usage in N. tetrasperma and N. discreta, we determined the difference in the Fop among these two taxa for each of the 2,079 genes examined herein. Our results indicate that 92.9% (1,931 of the 2,079 genes) of the genes examined show differences in Fop among these taxa, with only 148 having identical Fop values (fig. 4). Notably, a statistically significantly greater number of genes have a higher Fop value in N. tetrasperma (N = 1,204) than those that have a higher value in N. discreta (N = 727; chi-square test, P = 0.013), suggesting that selection for optimal codon usage is stronger in N. tetrasperma. We also compared the ▵RSCU values for the optimal codons among these two organisms. For this, we examined only the primary optimal codon per synonymous codon family, which is the optimal codon with the largest positive ▵RSCU. The data show that the ▵RSCU value was higher in N. tetrasperma than for N. discreta for 16 of the 18 primary optimal codons (table 2). This is consistent with greater selective pressure on optimal codon usage in N. tetrasperma than for N. discreta across the vast majority of synonymous codon families.
FIG. 4.—
The difference in the Fop among Neurospora tetrasperma and N. discreta for each for each of the 2,079 genes examined herein. Genes are provided on the x axis in the order they occur on the chromosomes (Chr).
In totality, the data suggest that there have been mild shifts in optimal codon usage among genes in N. discreta as compared with N. tetrasperma (fig. 4), with more genes having a higher Fop in N. tetrasperma. Thus, adaptive codon usage is greater in N. tetrasperma. It may be speculated that this results from a generally greater level of purifying selection in the N. tetrasperma lineage but could also result from positive selection events. Despite this variation, however, our data show that most aspects of codon usage are highly conserved for N. discreta and N. tetrasperma, with these taxa having identical optimal codon lists (table 2) whose frequency is largely dependent on gene expression level and to a lesser extent GL (figs. 1 and 2 and tables 2 and 3).
Discussion
Although mutational biases or gene conversion may be associated with codon usage bias in certain organisms (D'Onofrio et al. 1991; Wright et al. 2007; Haudry et al. 2008; Plotkin and Kudla 2010), the marked codon bias reported herein for N. discreta and for N. tetrasperma is best explained by selection for translational efficiency and/or accuracy. This is demonstrated by several findings. First and most importantly, our data show unequivocally that optimal codons are associated with gene expression level in N. tetrasperma and in N. discreta (table 2 and fig. 2). In addition, a very high fraction of variation in Fop is explained by expression level (table 3), a level equal to, or beyond that, reported in other species such as Populus and Caenorhabditis (Cutter et al. 2008; Ingvarsson 2008). Secondly, the effect of gene expression is prevalent across all three GL categories, and thus GL cannot explain these findings (fig. 2). Thirdly, dS is markedly lower in highly expressed genes, a finding which suggests that selection has altered synonymous substitution rates among taxa (supplementary table 2, Supplementary Material online; McVean and Vieira 2001). Such an effect is only expected under strong selection for codon usage (McVean and Vieira 2001). Finally, we found markedly higher GC content at third nucleotide positions of codons than for the associated introns for the genes examined herein (fig. 3); such findings demonstrate that mutational or gene conversion biases cannot explain our results (Wolfe et al. 1989; Eyre-Walker 1999; Smith and Eyre-Walker 2001; Vicario et al. 2007). Thus, it is evident that codon usage in N. tetrasperma and N. discreta is shaped by selection for translational efficiency/accuracy.
The fact that the 26 optimal codons identified herein (including two putative optimal codons) are identical across N. tetrasperma and N. discreta (table 2), suggests that optimal codon usage has been highly conserved among these divergent Neurospora taxa. The ▵RSCU values, which is an indicator of the strength of selection for optimal codons (Cutter et al. 2008; Ingvarsson 2008), are notably higher than those reported for Populus and Arabidopsis, wherein almost all values for optimal codons were less than 0.30 (Duret and Mouchiroud 1999; Ingvarsson 2008), for example, nine of the optimal codons for N. tetrasperma and N. discreta had ▵RSCU values higher than 0.30 and the highest value was 0.8344 (for CTC for leucine in N. tetrasperma; table 2). However, the uppermost ▵RSCU values reported herein for Neurospora are in a similar range as found in Drosophila and Caenorhabditis, whose values often exceeded 0.30 (Duret and Mouchiroud 1999). Thus, these data suggest that the Neurospora genomes examined here have among the highest selective pressures for optimal codons usage, relative to these other organisms. It is worthwhile to note that prior research using tentative models for selection pressure on codon usage has suggested that selection coefficients might be greater in N. crassa than in other eukaryotes such as Arabidopsis and equal or greater than species of Drosophila and Caenorhabditis (dos Reis and Wernisch 2008). Our present data is consistent with marked selective pressure on codon usage in N. tetrasperma and N. discreta.
Fop and GL
Our data demonstrate that GL contributes to optimal codon usage in N. tetrasperma and N. discreta but to a lesser extent than gene expression (fig. 2 and table 3). The gene-length effects may result from background selection and/or Hill-Robertson effects, each of which interfere with the effectiveness of selection and has a greater effect for longer genes (Comeron et al. 1999; Comeron and Guthrie 2005; Loewe and Charlesworth 2007) that is such processes may interfere with the fixation of optimal codons in longer genes. Accordingly, shorter genes may have more efficient selection for optimal codons.
The gene-length effects reported herein are consistent with reports of an inverse association between GL and optimal codon frequency in certain other multicellular organisms (e.g., Drosophila, Caenorhabditis, and Silene, Duret and Mouchiroud 1999; Cutter et al. 2008; Qiu et al. 2011). Our findings, however, differ from recent results in Caenorhabditis, wherein GL effects on optimal codon usage were inferred in some taxa but not in others (Cutter et al. 2008). Similarly, no effect of GL on optimal codon usage was found in various species of Populus (Ingvarsson 2008). In these studies, it has been proposed that the lack of GL effects (or lack of a consistent effect in Caenorhabditis) might be due to the fact that GLs were assumed to be equal to those from a reference taxon (Cutter et al. 2008; Ingvarsson 2008). Thus, there might be unknown differences in GLs among species that impeded the detection of GL effects. This explanation seems unlikely, as protein lengths (and thus CDS regions) tend to be highly conserved among eukaryotes (Wang et al. 2004). Furthermore, we estimated GLs for N. tetrasperma and in N. discreta, using the reference taxon N. crassa, which did not obscure the GL effects reported in present analysis (fig. 2 and table 3). Another proposed explanation for the lack of GL effects in certain species of Caenorhabditis and in Populus is that different sets of genes were used for the various species examined in each of these studies (Cutter et al. 2008; Ingvarsson 2008). This was not an issue in our analyses, as we utilized identical gene sets (2,079 genes) for both N. tetrasperma and N. discreta, which likely enhanced our ability to consistently detect GL effects on the Fop across taxa. Overall, our present findings point toward the conclusion that GL plays a significant role in determining the Fop per gene in N. tetrasperma and N. discreta.
The findings of the highest Fop values for short highly expressed genes for each of the Neurospora taxa examined herein (fig. 2) corresponds with results reported for other multicellular organisms such as species of Drosophila, Caenorhabditis, and Silene (Duret and Mouchiroud 1999; Qiu et al. 2011). Such findings suggest that in combination these traits promote highly efficient translation. Highly expressed genes have elevated fitness costs due to the energy and cellular resources required for synthesis and correcting synthesis errors (mRNA and protein; Akashi 2003; Drummond and Wilke 2009). Longer proteins are believed to have greater phenotypic costs for protein-synthesis errors (and accumulate more errors, Drummond and Wilke 2009) and have greater energy costs for synthesis than shorter proteins (Akashi 2003). Thus, natural selection may favor evolution of highly expressed longer proteins into shorter proteins in order to minimize fitness costs (Akashi 2003); accordingly, such a phenomenon might partially contribute toward the elevated optimal codon usage observed within short genes as compared with longer genes in Neurospora.
Comparison among Neurospora Taxa
Although the optimal codon list is highly conserved among N. tetrasperma and N. discreta (table 2), it is worthwhile to note that small but nontrivial differences in codon usage were evident among these two taxa. For example, the findings that N. tetrasperma has a higher Fop value for a large majority of the 2,079 genes examined herein (fig. 4) and has an elevated ▵RSCU value for 16 of 18 of the primary optimal codons than its relative N. discreta, suggest there is greater selective pressure on codon usage in this taxon. This finding is surprising as N. tetrasperma is a self-compatible and primarily inbreeding taxon with enforced intratetrad mating (Raju and Perkins 1994), whereas N. discreta is a self-incompatible taxon. Theory predicts that inbreeding species should have relaxed selective pressure for genomic traits such as codon usage due to lower effective recombination rates giving rise to reduced effective population size as compared with their outcrossing counterparts (see Charlesworth and Wright 2001). This notion has been supported by findings from species of Caenorhabditis, which have shown that the transition from outbreeding to selfing correlates with a decrease in codon usage bias (Cutter et al. 2008). In addition, data from inbreeding species of Lycopersicon have been found to have more transposable elements than outcrossers, consistent with less selective pressure for inbreeders (Young et al. 1994; Charlesworth D and Charlesworth B 1995). However, our present data do not support this trend and suggests that the inbreeding species N. tetrasperma has greater selective pressure for codon usage than the self-incompatible N. discreta.
Several plausible factors might give rise to differences in codon usage among N. tetrasperma and N. discreta. For example, N. tetrasperma might have a higher effective population size than N. discreta; this is not expected given that N. tetrasperma is a primarily inbreeding pseudohomothallic species, whereas N. discreta is heterothallic (and thus likely subjected to much less inbreeding, Perkins and Raju 1986). However, N. tetrasperma has been shown to be capable of occasional outcrossing events and its nine independent phylogenetic lineages are not perfectly reproductively isolated (Menkis et al. 2009), traits which may enhance its effective population size. Furthermore, although N. discreta is one of the most extensively distributed Neurospora species, for example, accounting for more than 95% of isolates collected in western North America (Jacobson et al. 2004), it consists of multiple phylogenetic lineages of unknown population size (and thus might consist of small populations; Dettman et al. 2006). Another factor that could play a significant role is the time of origin of the mating systems. For example, it has been suggested that lack of codon usage differences in certain species of Caenorhabditis might result from the fact that self-fertilization arose less than 4 Ma (Cutter et al. 2008). Recent data has suggested that inbreeding is also the derived state in N. tetrasperma originating less than 6 Ma (Menkis et al. 2008; Nygren et al. 2011); accordingly, it may be inferred that selfing might have existed for too short of a time period to alter codon usage in this taxon. In this regard, it is plausible that effects of effective population sizes and/or a recent origin of reinforced inbreeding may explain why, contrary to theoretical predictions, selective pressure is higher for the highly inbreeding taxon N. tetrasperma.
Conclusions
The present investigation reveals that gene expression level is a primary factor driving codon usage within the filamentous ascomycetes N. tetrasperma and in N. discreta. The selective pressure equals or exceeds that reported in the limited other taxa with comparable data available to data (e.g., Caenorhabditis, Populus, Drosophila, and Arabidopsis, Duret and Mouchiroud 1999; Cutter et al. 2008; Ingvarsson 2008). Moreover, in contrast to certain other organisms, our data also show that GL contributes substantially to shaping codon usage in Neurospora. Further studies will be needed to ascertain whether such optimal codon usage is inherent to other species of Neurospora, including N. crassa and other genera of filamentous ascomycetes and fungi.
Supplementary Material
Supplementary tables S1–S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors gratefully acknowledge research funding from Royal Swedish Academy of Sciences (Hierta-Retzius Research Grant) (to C.A.W.), The Royal Physiographic Society in Lund (to C.A.W.), The Lars Hierta Minne Foundation (to C.A.W.), the Wenner Gren Foundation (to C.A.W.), The Magn Bergvall Foundation (to H.J.) and The Swedish Research Council (to H.J.). In addition, valuable comments regarding our manuscript by the two anonymous reviewers and the Associate Editor are greatly appreciated.
References
- Akashi H. Gene expression and molecular evolution. Curr Opin Genet Dev. 2001;11:660–666. doi: 10.1016/s0959-437x(00)00250-1. [DOI] [PubMed] [Google Scholar]
- Akashi H. Translational selection and yeast proteome evolution. Genetics. 2003;164:1291–1303. doi: 10.1093/genetics/164.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich KA, et al. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev. 2004;68:1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casselton LA. Fungal sex genes-searching for the ancestors. Bioessays. 2008;30:711–714. doi: 10.1002/bies.20782. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. Transposable elements in inbreeding and outbreeding populations. Genetics. 1995;140:415–417. doi: 10.1093/genetics/140.1.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Wright SI. Breeding systems and genome evolution. Curr Opin Genet Dev. 2001;11:685–690. doi: 10.1016/s0959-437x(00)00254-9. [DOI] [PubMed] [Google Scholar]
- Comeron JM, Guthrie TB. Intragenic Hill-Robertson interference influences selection intensity on synonymous mutations in Drosophila. Mol Biol Evol. 2005;22:2519–2530. doi: 10.1093/molbev/msi246. [DOI] [PubMed] [Google Scholar]
- Comeron JM, Kreitman M, Aguadéb M. Natural selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics. 1999;151:239–249. doi: 10.1093/genetics/151.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Wasmurth JD, Blaxter ML. The evolution of biased codon and amino acid usage in Nematode genomes. Mol Biol Evol. 2006;23:2303–2315. doi: 10.1093/molbev/msl097. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Wasmuth JD, Washington NL. Patterns of molecular evolution in Caenorhabditis preclude ancient origins of selfing. Genetics. 2008;178:2093–2104. doi: 10.1534/genetics.107.085787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RH, Perkins DD. Neurospora: a model of model microbes. Nature Rev Genet. 2002:397–403. doi: 10.1038/nrg797. [DOI] [PubMed] [Google Scholar]
- Dettman JR, Jacobon DJ, Taylor JW. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution. 2003;57:2703–2720. doi: 10.1111/j.0014-3820.2003.tb01514.x. [DOI] [PubMed] [Google Scholar]
- Dettman JR, Jacobson DJ, Taylor JW. Multilocus sequence data reveal extensive phylogenetic species diversity in the Neurospora discreta complex. Mycologia. 2006;98:436–446. doi: 10.3852/mycologia.98.3.436. [DOI] [PubMed] [Google Scholar]
- Dodge BO, Singleton JR, Rolnick A. Studies on lethal E gene in Neurospora tetrasperma, including chromosome counts in races of N. Sitophila. Proc Am Phil Soc. 1950;94:38–52. [Google Scholar]
- D'Onofrio G, Mouchiroud D, Aïssani B, Gautier C, Bernardi G. Correlations between the compositional properties of human genes, codon usage, and amino acid composition of proteins. J Mol Evol. 1991;32:504–510. doi: 10.1007/BF02102652. [DOI] [PubMed] [Google Scholar]
- dos Reis M, Wernisch L. Estimating translational selection in eukaryotic genomes. Mol Biol Evol. 2008;26:451–461. doi: 10.1093/molbev/msn272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet. 2009;10:715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L. tRNA gene number and codon usage in the C. elegans genome are coadapted for optimal translation of highly expressed genes. Trends Genet. 2000;16:287–289. doi: 10.1016/s0168-9525(00)02041-2. [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci U S A. 1999;96:4482–4487. doi: 10.1073/pnas.96.8.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A. Evidence of selection on silent site base composition in mammals: potential implications for the evolution of isochores and junk DNA. Genetics. 1999;152:675–683. doi: 10.1093/genetics/152.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudry A, et al. Mating system and recombination affect molecular evolution in four Triticeae species. Genet Res. 2008;90:97–109. doi: 10.1017/S0016672307009032. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. J Mol Biol. 1982;158:573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and transfer-RNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Ingvarsson PD. Molecular evolution of synonymous codon usage in Populus. BMC Evol Biol. 2008;8:307. doi: 10.1186/1471-2148-8-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson DJ. Sexual dysfunction associated with outcrossing in Neurospora tetrasperma, a pseudohomothallic ascomycete. Mycologia. 1995;87:604–617. [Google Scholar]
- Jacobson DJ, Powell AJ, Dettman JR, Saenz GS. Neurospora in temperate forests of western North America. Mycologia. 2004;96:66–74. [PubMed] [Google Scholar]
- Kano A, Andachi Y, Ohama T, Osawa S. Novel anticodon composition of transfer RNAs in Micrococcus luteus, a bacterium with a high genomic G + C content: correlation with codon usage. J Mol Biol. 1991;221:387–401. doi: 10.1016/0022-2836(91)80061-x. [DOI] [PubMed] [Google Scholar]
- Kent WJ. Blast-the BLAST alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- Loewe L, Charlesworth B. Background selection in single genes may explain patterns of codon bias. Genetics. 2007;175:1381–1393. doi: 10.1534/genetics.106.065557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean GA, Vieira J. Inferring parameters of mutation, selection and demography from patterns of synonymous site evolution in Drosophila. Genetics. 2001;157:245–257. doi: 10.1093/genetics/157.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkis A, Bastiaans E, Jacobson DJ, Johannesson H. Phylogenetic and biological species diversity within the Neurospora tetrasperma complex. J Evol Biol. 2009;22:1923–1936. doi: 10.1111/j.1420-9101.2009.01801.x. [DOI] [PubMed] [Google Scholar]
- Menkis A, Jacobson DJ, Gustafsoon T, Johannesson H. The mating-type chromosome in the filamentous asycomycete Neurospora tetrasperma represents a model for early evolution of sex chromosomes. PLoS Genet. 2008;4:e1000030. doi: 10.1371/journal.pgen.1000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nygren K, et al. A comprehensive phylogeny of the genus Neurospora (Ascomycota) reveals a link between reproductive mode and molecular evolution in fungi. Mol Phylogenet Evol. 2011 doi: 10.1016/j.ympev.2011.03.023. doi:10.1016/j.ympev.2011.03.023. [DOI] [PubMed] [Google Scholar]
- Osawa S, et al. Directional mutation pressure and transfer RNA in choice of the third nucleotide of synonymous two-codon sets. Proc Natl Acad Sci U S A. 1988;85:1124–1128. doi: 10.1073/pnas.85.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DD, Raju NB. Neurospora discreta, a new heterothallic species defined by its crossing behavior. Exp Mycol. 1986;10:323–338. [Google Scholar]
- Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nature Rev Genet. 2010 doi: 10.1038/nrg2899. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbo P, Bravo IG, Garcia-Vallve S. CAIcal: a combined set of tools to assess codon usage adaptation. Biol Direct. 2008;3:38. doi: 10.1186/1745-6150-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S, Bergero R, Zeng K, Charlesworth D. Patterns of codon usage bias in Silene latifolia. Mol Biol Evol. 2011;178:2093–2104. doi: 10.1093/molbev/msq251. [DOI] [PubMed] [Google Scholar]
- Raju NB, Perkins DD. Diverse programs of ascus development in pseudohomothallic species of Neurospora, Gelasinospora and Podospora. Dev Genetics. 1994;15:104–118. doi: 10.1002/dvg.1020150111. [DOI] [PubMed] [Google Scholar]
- Sharp PM. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Averof M, Lloyd AT, Matassi G, Peden JF. DNA sequence evolution, the sounds of silence. Philos Trans R Soc Lond B Biol Sci. 1995;349:241–247. doi: 10.1098/rstb.1995.0108. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Tuohy TMF, Mosurski KR. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986;14:5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shear CL, Dodge BO. Life histories and heterothallism of the red bread-mold fungi of the Monilia sitophila group. J Agric Res. 1927;34:1019–1041. [Google Scholar]
- Shiu PKT, Glass NL. Cell and nuclear recognition mechanisms mediated by mating type in filamentous ascomycetes. Curr Opin Microbiol. 2000;3:183–188. doi: 10.1016/s1369-5274(00)00073-4. [DOI] [PubMed] [Google Scholar]
- Smith N, Eyre-Walker A. Synonymous codon bias is not caused by mutation bias in G + C-rich genes in humans. Mol Biol Evol. 2001;18:982–998. doi: 10.1093/oxfordjournals.molbev.a003899. [DOI] [PubMed] [Google Scholar]
- Stenico M, Lloyd AT, Sharp PM. Codon usage in Caenorhabditis elegans: delineation of translational selection and mutational biases. Nucleic Acids Res. 1994;22:2437–2446. doi: 10.1093/nar/22.13.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletzki N, Eyre-Walker A. Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol Biol Evol. 2006;24:374–381. doi: 10.1093/molbev/msl166. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics. 2004;168:373–381. doi: 10.1534/genetics.104.028944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. Directional mutation pressure and neutral molecular evolution. Proc Natl Acad Sci U S A. 1988;85:2653–2657. doi: 10.1073/pnas.85.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicario S, Moriyama EN, Powell JR. Codon usage in twelve species of Drosophila. BMC Evol Biol. 2007;7:226. doi: 10.1186/1471-2148-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Hsieh M, Li WH. A general tendency for conservation of protein lengths across eukaryotic kingdoms. Mol Biol Evol. 2004;22:142–147. doi: 10.1093/molbev/msh263. [DOI] [PubMed] [Google Scholar]
- Wolfe K, Sharp HPM, Li WH. Mutation rates differ among regions of the mammalian genome. Nature. 1989;337:283–285. doi: 10.1038/337283a0. [DOI] [PubMed] [Google Scholar]
- Wright SI. The “effective number of codons” used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Wright SI, Lauga B, Charlesworth D. Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Mol Biol Evol. 2002;19:1407–1420. doi: 10.1093/oxfordjournals.molbev.a004204. [DOI] [PubMed] [Google Scholar]
- Wright SE, Iorgovan G, Misra S, Mokhtari M. Neutral evolution of synonymous base composition in the Brassicaceae. J Mol Evol. 2007;64:136–141. doi: 10.1007/s00239-005-0274-1. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Young RJ, Francis DM, St Clair DA, Taylor BH. A dispersed gamily of repetitive DNA sequences exhibits characteristics of a transposable element in the genus Lycopersicon. Genetics. 1994;137:581–588. doi: 10.1093/genetics/137.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]