Abstract
NspA is a conserved membrane protein that elicits protective antibody responses in mice against Neisseria meningitidis. A recent crystallographic study showed that NspA adopts an eight-stranded β-barrel structure when reconstituted in detergent. In order to define the segments of NspA-containing epitopes recognized by protective murine anti-NspA antibodies, we studied the binding of two bactericidal and protective anti-NspA monoclonal antibodies (MAbs), AL12 and 14C7. Neither MAb binds to overlapping synthetic peptides (10-mers, 12-mers, and cyclic 12-mers) corresponding to the entire mature sequence of NspA, or to denatured recombinant NspA (rNspA), although binding to the protein can be restored by refolding in liposomes. Based on the ability of the two MAbs to bind to Escherichia coli microvesicles prepared from a set of rNspA variants created by site-specific mutagenesis, the most important contacts between the MAbs and NspA appear to be located within the LGG segment of loop 3. The conformation of loop 2 also appears to be an important determinant, as particular combinations of residues in this segment resulted in loss of antibody binding. Thus, the two anti-NspA MAbs recognize discontinuous conformational epitopes that result from the close proximity of loops 2 and 3 in the three-dimensional structure of NspA. The data suggest that optimally immunogenic vaccines using rNspA will require formulations that permit proper folding of the protein.
Capsular group B strains of Neisseria meningitidis cause ∼30% of meningococcal disease in the United States (18, 19) and up to 80% in northern Europe (4). Although capsular polysaccharide-based vaccines have been developed for prevention of disease caused by strains with capsular groups A, C, Y, and W135, this approach has been problematic for group B (9, 16). The group B polysaccharide capsule [α(2→8) N-acetyl neuraminic acid] is identical to human polysialic acid (6, 8) and is poorly immunogenic, possibly as a result of immune tolerance. Further, there are safety issues that are difficult to resolve for a vaccine that has the potential to elicit autoreactive antibodies. Considerable effort, therefore, has focused on the identification of noncapsular antigens that might provide broad protection against group B strains (reviewed in references 9 and 16).
Neisserial surface protein A (NspA) is an 18.6-kDa membrane protein of unknown function that was first described by Martin, Brodeur, and colleagues (11). Unlike other neisserial surface proteins, such as PorA and Opc that also have been shown to elicit bactericidal protective antibodies, NspA is conserved and expressed by all N. meningitidis strains tested to date (11, 15). Immunization of mice with recombinant NspA (rNspA) also conferred protection against bacteremia in animals challenged with a group B strain (11). In subsequent studies, our investigators showed that both polyclonal antibodies and monoclonal antibodies (MAbs) elicited in mice by immunization with rNspA were bactericidal against ∼50% of group B strains tested and passively protected against meningococcal bacteremia in an infant rat challenge model (13, 15). Additionally, a MAb (14C7) elicited by immunization with native NspA in outer membrane vesicle preparations was bactericidal against strains that were resistant to complement-mediated bacteriolysis by the most active MAb (AL12) that we had prepared by immunization with rNspA produced in Escherichia coli (14). Taken together, the results show that NspA is a promising vaccine candidate for prevention of meningococcal disease. Little is known, however, about the NspA epitopes that are surface exposed on the bacteria and capable of eliciting bactericidal antibody. The identification of the surface-exposed epitopes of NspA that interact with protective antibodies may allow for rational design of improved rNspA vaccines.
In order to define the segments of NspA containing epitopes recognized by protective anti-NspA antibodies, we have studied the binding of the two anti-NspA MAbs, AL12 and 14C7, described above, to a set of rNspA variants created by site-specific mutagenesis. The mutants have substitutions in surface-exposed loops 2 and 3. In this report we provide evidence that conformational epitopes defined by both loops 2 and 3 are targets of these two bactericidal anti-NspA MAbs.
MATERIALS AND METHODS
MAbs AL12 and 14C7.
Two anti-NspA MAbs (AL12 and 14C7) were used to investigate the effect of amino acid substitutions on epitope structure. MAb AL12 (immunoglobulin G2a [IgG2a]) was produced by immunizing mice with rNspA expressed in outer membrane vesicles that were blebbed from E. coli strain BL21(DE3) that had been transformed with the plasmid pGMS1.0. The plasmid contains nspA cloned from group B strain 8047. MAb AL12 has complement-mediated bactericidal activity against approximately 50% of genetically diverse group B strains tested and also confers passive protection against meningococcal bacteremia in infant rats challenged with group B strains 8047 and BZ232 but not M986 (15). MAb 14C7 (IgG3) was made against native NspA by sequentially immunizing a mouse with outer membrane vesicle preparations from three heterologous neisserial strains (14). The NspA proteins expressed by each of the vaccine strains are heterologous to each other and include one amino acid difference in surface-exposed loop 3. Both MAb AL12 and 14C7 bind to NspA expressed by all three vaccine strains (data not shown). For AL12-susceptible strains, MAb 14C7 is bactericidal in the presence of complement at lower antibody concentrations than those for MAb AL12. 14C7 also has activity against some strains that are resistant to AL12-mediated bacteriolysis, and 14C7 passively protects against bacterial challenge in the infant rat model against some group B strains, such as M986 (14), which is resistant to passive protection by MAb AL12 (15).
ELISA.
The whole-cell enzyme-linked immunosorbent assay (ELISA) was performed as described by Abdillahi and Poolman (1). Briefly, bacterial cells grown overnight at 37°C in 5% CO2 on chocolate agar plates were resuspended in sterile phosphate-buffered saline (PBS) buffer. The cells were inactivated by heating to 56°C in a water bath for 30 min. The suspension was adjusted to an optical density at 620 nm (OD620) of 0.1, and 100-μl aliquots of the suspension were added to wells of flat-bottom 96-well microtiter plates (Nalge Nunc International, Rochester, N.Y.). The liquid in the wells was allowed to evaporate at ambient temperature in a fume hood. Before addition of antibodies, the plates were washed once with wash buffer (0.1% [wt/vol] Tween 20 in PBS), blocked by adding blocking buffer (2% [wt/vol] nonfat milk in PBS), and incubated at 37°C for 1 h. After removal of the blocking buffer, test antibodies diluted in blocking buffer were added to the wells and incubated at 4°C overnight. The plates were washed five times with wash buffer followed by the addition of rabbit anti-mouse IgG-, IgA-, and IgM-alkaline phosphatase-conjugated polyclonal antibody (Zymed, South San Francisco, Calif.) diluted in wash buffer containing 1% (wt/vol) bovine serum albumin. After 1 h of incubation at ambient temperature, the plates were washed five times with wash buffer and developed with p-nitrophenylphosphate substrate (1 mg/ml; Sigma, St. Louis, Mo.) in 1 M diethanolamine (pH 9.8) containing 0.5 mM magnesium chloride. The OD405 was measured using a microtiter plate reader (Bio-Rad, Richmond, Calif.).
Cloning and mutagenesis of rNspA in E. coli.
The NspA gene from N. meningitidis strain 8047 was cloned (pGMS 1.0) as previously described (13). The same strategy was employed for cloning the NspA gene from N. meningitidis strain MCH88. The following primers were used for the amplification of the NspA gene: 5′-ACAGCAGGATCCTTTAACGGATTC-3′ and 5′-GTGGATGAAGCTTTGGACATTTC-3′. These primers also contain cleavage sites for the restriction endonucleases BamHI and HindIII at the 5′ and 3′ ends, respectively, of the NspA gene. The primers were used in the PCR using a Peltier thermal cycler (MJ Research, Inc., South San Francisco, Calif.) to amplify a 746-bp DNA segment from the genome of MCH88. The settings included an initial hold step at 95°C for 3 min followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and elongation at 72°C for 1 min. An additional hold step at 72°C for 5 min was used at the end of the amplification cycle to extend incomplete ends. Genomic DNA for MCH88 was isolated by using a commercial kit (Qiagen, Valencia, Calif.) according to the directions of the manufacturer for the preparation of genomic DNA from bacteria. Strain MCH88 was kindly provided by Denis Martin (Unité de recherche en vaccinologie, Entre Hospitalier Universitaire de Québec, Quebec, Canada). The fragment, which includes the wild-type promoter region, was subsequently cloned into the multicopy plasmid, pKS(+) (Stratagene, La Jolla, Calif.), generating plasmid pMCH88. Note that pGMS 1.0 is cloned into an identical vector pSK(+) that has the multiple cloning site in the reverse orientation. Our experiments showed that in general the pMCH88 clone had slightly lower expression of NspA than pGMS 1.0. Mutagenesis of 8047 and MCH88 NspA in E. coli was performed using the QuikChange site-directed mutagenesis kit (Stratagene) following the protocol described by the manufacturer. The primers and DNA templates used are provided as supplementary material (http://www.chori.org/investigators/granoff_supplementary/primers.html). All mutants were confirmed by sequencing using T3 and T7 promoter primers that flank the inserted NspA gene.
MV preparation and immunoblotting.
Microvesicles (MV) were prepared from transformed E. coli strain XL2-Blue ultracompetent cells (Stratagene). The cells were grown in 25 ml of sterile Luria-Bertani broth containing 100 μg of ampicillin/ml overnight at 37°C with shaking. These cultures were then used to inoculate 500 ml of sterile Luria-Bertani broth containing 100 μg of ampicillin/ml and grown with vigorous shaking at 37°C until the OD620 reached 0.9 to 1.0 (4 to 5 h). The cells were then pelleted by centrifugation (11,000 × g) of the cultures for 30 min at 4°C. Blebbed MV were harvested from the cell-free culture supernatants by adding solid ammonium sulfate (390 g/liter, final concentration) slowly with stirring. After the ammonium sulfate was added and completely dissolved, the mixture was left at 4°C overnight. The precipitate containing the MV was collected by centrifugation at 11,000 × g for 30 min at 4°C. The pellet was resuspended in 5 ml of PBS and centrifuged again at 16,000 × g for 15 min at 4°C. The low-speed pellet was discarded, and the MV, which remained in the supernatant, were collected by centrifugation at 100,000 × g for 2 h at 4°C. The final MV-containing pellet was resuspended in 200 μl of PBS and sterile filtered using a Millex-HV 0.45-μm-pore-size filter (Millipore, Molsheim, France). The NspA concentration in the MV was estimated by densitometry of the NspA band on Western blots of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. NspA was detected using a polyclonal antiserum prepared in mice immunized with HisTag-NspA protein denatured in 8 M urea (see below). Approximately equivalent amounts of total NspA in MV were blotted onto nitrocellulose membranes (Bio-Rad) using a Scie-Plas DHM-96 hybridization manifold (Topac, Hingham, Mass.). Membranes were soaked in water for 10 min at ambient temperature prior to blotting. After blotting, membranes were blocked in blocking buffer (see ELISA methods, above). MAbs AL12 and 14C7 and goat anti-E. coli polyclonal antibody (RDI, Flanders, N.J.) were diluted in blocking buffer. Bound murine antibody was detected with rabbit anti-mouse IgG-, IgA-, and IgM-horseradish peroxidase-conjugated polyclonal antibody, and the goat antibody was detected with rabbit anti-goat IgG-, IgA-, and IgM-horseradish peroxidase-conjugated polyclonal antibody (both reagents from Zymed) using the Western Lightning Chemiluminescence Reagent Plus substrate (Perkin-Elmer Life Sciences, Boston, Mass.).
Reconstitution of HisTag-NspA in liposomes.
The NspA gene from strain 8047 was cloned into the BamHI and EcoRI sites of the HisTag expression vector pTrcHis A (Invitrogen, Carlsbad, Calif.). The resulting plasmid (pTrc8047) was used to express HisTag-NspA in E. coli strain BL21 (Stratagene). The HisTag-NspA protein was purified by nickel-nitrilotriacetic acid-Sepharose chromatography of cell lysates solubilized in 8 M urea using the materials and methods provided in the QiaExpress kit (Qiagen) as previously described (13). Liposomes containing HisTag-NspA were prepared by the ethanol injection method of Batzri and Korn (2). Soybean phosphatidyl choline (9 mg; Avanti Polarlipids, Alabaster, Ala.) in ethanol was rapidly injected through a bent-tip needle into a solution of HisTag-NspA (0.5 mg/ml) in solubilization buffer (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-HCl [pH 4.5], containing 0.6% [wt/vol] Zwittergent 3-14 [Calbiochem, La Jolla, Calif.] and 2% SDS). After stirring for 2 h, the mixtures were applied to a size-exclusion column (Toyopearl HW-75; Supleco, Bellefonte, Pa.) equilibrated with PBS. The mixture was pumped through the column at a flow rate of 1 ml/min using a BioCad workstation (Applied Biosystems, Foster City, Calif.). Fractions (5 ml) with absorbance at 280 nm were collected and concentrated by ultrafiltration using an XM300 membrane (Amicon; Millipore, Bedford, Mass.) with stirring under nitrogen. The vesicles were characterized with respect to size by gel filtration chromatography (2) and protein concentration by SDS-PAGE and bicinchoninic acid (BCA) assay (Pierce, Rockford, Ill.).
Reconstitution of HisTag-NspA in micelles.
HisTag-NspA in solubilization buffer was dialyzed stepwise against 0.6% (wt/vol) Zwittergent 3-14 in 0.1 M NaH2PO4, 0.01 M Tris-HCl buffer (pH 7.5) containing 150 mM NaCl and 6, 4, 2 M and, finally, no urea. Protein concentrations of the purified HisTag-NspA and the HisTag-NspA reconstituted in liposomes or micelles were determined by a BCA protein assay (Pierce).
RESULTS
Anti-NspA MAb binding to overlapping peptides.
The epitopes recognized by MAbs reactive with Opc, a 10-stranded β-barrel (17), and Opa, postulated to have an 8-stranded β-barrel topology similar to that of NspA, have been mapped using overlapping synthetic peptides (10, 12). With the idea of performing a similar mapping study of anti-NspA MAbs, overlapping synthetic peptides corresponding to the entire mature sequence of NspA from strain 8047 were synthesized on solid supports and tested for binding by using MAbs AL12 and 14C7. Preparation and testing of synthetic 10-mers (n = 146) on amino-polyethylene glycol-cellulose membranes (ABIMED,Langerfeld, Germany) were performed at the University of Siena, Siena, Italy, as previously described (7). In addition, synthetic 12-mers (n = 144) and cyclic 12-mers (n = 141) were prepared and tested for MAb binding by PepScan Systems (Lelystad, The Netherlands). Neither anti-NspA MAb showed significant binding above background to any of the peptides (data not shown). The results suggest that either the three-dimensional structure of the epitope recognized by the MAbs could not be reproduced by the shorter synthetic peptides, that the MAbs recognize a larger discontinuous epitope that was not contained within the synthetic peptides, or a combination of both possibilities. Further, neither MAb binds to denatured NspA, for example, on a Western blot of rNspA resolved on SDS-PAGE.
MAb binding to naturally occurring NspA variants.
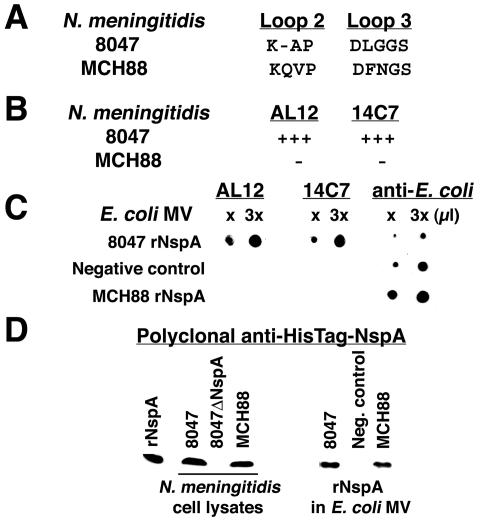
As an alternative to synthetic peptide mapping, we looked for naturally occurring variants of NspA that are not recognized by the MAbs. Cadieux et al. reported that a bactericidal anti-NspA MAb, Me-7, lacked bactericidal activity against N. meningitidis group A strain MCH88 (A:4:P1.10) and suggested that the lack of activity may have resulted from an NspA polymorphism (3) (Fig. 1A). Our anti-NspA MAbs AL12 and 14C7 also lacked bactericidal activity against strain MCH88 (50% bactericidal concentration, >500 μg/ml) and showed no binding to MCH88 in a whole-cell ELISA (Fig. 1B). Both MAbs bound strongly in the ELISA to cells from strain 8047 (Fig. 1B). The absence of binding and bactericidal activity against strain MCH88 were not a result of a failure to express NspA. Figure 1D (left panel) shows a portion (∼15- to 20-kDa range) of a Western blot of whole-cell lysate proteins from group B strains 8047 and MCH88 resolved by SDS-PAGE. The presence of NspA in both strains was shown by the reactivity of protein bands in the mass region expected for mature NspA as detected by mouse polyclonal antisera prepared to denatured HisTag-NspA.
FIG. 1.
(A) NspA amino acid polymorphisms found in loops 2 and 3 in N. meningitidis strains 8047 and MCH88. (B) Binding of anti-NspA MAbs AL12 and 14C7 to N. meningitidis bacterial cells from strains 8047 and MCH88 as measured by ELISA. The MAbs were tested at 5 μg/ml. +++, binding defined as an OD405 of >0.8; −, OD405 < 0.01. (C) Immunoblot using MAbs AL12 and 14C7 and polyclonal anti-E. coli antibody. Binding is shown to E. coli MV expressing rNspA encoded by the gene from N. meningitidis group B strain 8047 or group A strain MCH88. The negative control is E. coli MV prepared from cells containing the vector [pKS(+)] alone. (D) Western blot analysis of N. meningitidis whole-cell lysates (left) or E. coli MV (right) using polyclonal anti-HisTag NspA sera. The positive control is rNspA (lanes 1 and 5).
Amino acid sequence differences between NspA proteins expressed by strains 8047 and MCH88.
The respective NspA amino acid sequences inferred from genes of strains 8047 and MCH88 have been reported (11, 13). There are a total of five amino acid differences, two each in loops 2 and 3 (Fig. 1A) and one (not shown in the figure) located in a transmembrane domain at the carboxyl terminus (Met in 8047 versus Val in MCH88). In the segment of loop 2, MCH88 contains the sequence KQVP, compared to KAP in NspA from strain 8047. In the loop 3 segment, MCH88 contains the sequence DFNGS, compared to DLGGS in 8047. To determine which of the sequence differences contributes to differences in MAb binding, we constructed two E. coli expression vectors, one containing the NspA gene from MCH88 and the other with the NspA gene from 8047. The respective DNA sequences of the NspA genes cloned were identical to those previously reported.
When E. coli strain XL2-Blue is transformed with the expression vectors, the cells release outer membrane vesicle blebs into the culture medium that contain the respective rNspA (13). We isolated the blebbed vesicles and tested them for MAb binding to rNspA by Western blotting of the vesicles spotted onto nitrocellulose filters. It was necessary to use Western blotting to measure binding since this method allowed for spotting equivalent amounts of each NspA variant irrespective of how much was expressed in the blebbed vesicles. Since the rNspA contained in the vesicles has been processed by signal peptidase and inserted into the outer membrane, it is likely that the protein folds normally. Therefore, amino acid substitutions that result in changes in antibody binding are the result of discrete contacts between the residue and the MAb or changes in local conformation resulting from the substitutions.
As shown in Fig. 1C, we observed strong binding of MAbs AL12 and 14C7 to rNspA expressed from the 8047 NspA gene but not with MV prepared from control E. coli transformed with the same plasmid but lacking the NspA gene. Neither MAb reacted with rNspA expressed in E. coli MV from the MCH88 NspA gene. Both the 8047 and MCH88 NspA vesicle preparations contained approximately equivalent amounts of rNspA that were reactive with the anti-HisTag-NspA polyclonal antisera (Fig. 1D, right panel). These results confirm that the failure of the MAbs to bind to MCH88 NspA results from differences in the NspA protein and not from modifications occurring in the neisserial strains or from blocking by other neisserial surface antigens. Further, rNspA encoded by the NspA gene from group B strain 8047, which is released in blebbed outer membrane vesicles from E. coli cells, retains epitopes recognized by an anti-NspA MAb elicited by immunizing with native neisserial vesicles (14C7) and, therefore, is antigenically similar to native NspA.
Anti-NspA MAb binding to rNspA site-specific mutants.
To investigate the specific role of amino acid sequence differences in binding by MAbs AL12 and 14C7 between NspA from strain 8047 and that of strain MCH88, we constructed site-specific mutants in the rNspA 8047 and MCH88 expression vectors. We prepared E. coli MV from these mutants, spotted them onto nitrocellulose, and measured binding of MAb AL12 and 14C7 by immunoblotting. Total rNspA in each vesicle preparation was measured by densitometry of SDS-PAGE Western blots using polyclonal anti-HisTag-NspA antibody to detect rNspA to ensure that equal amounts of NspA were spotted onto the filter. The polyclonal antiserum was made to unfolded NspA and reacted with both native and unfolded NspA on immunoblots. The data from these experiments are summarized in Table 1.
TABLE 1.
Summary of immunoblotting results for anti-NspA MAbs AL12 and 14C7
| Construct no. | NspA gene | NspA segment amino acid sequence
|
Bindinga
|
||
|---|---|---|---|---|---|
| Loop 2 | Loop 3 | AL12 | 14C7 | ||
| 1 | MCH88 (wt)b | KQVP | DFNGS | − | − |
| 2 | MCH88 | KQVP | DLGGS | +++ | +++ |
| 3 | MCH88 | K-VP | DLGGS | +++ | +++ |
| 4 | MCH88 | K-AP | DLGGS | +++ | +++ |
| 5 | 8047 (wt) | K-AP | DLGGS | +++ | +++ |
| 6 | 8047 | K-VP | DLGGS | +++ | +++ |
| 7 | 8047 | K-AP | DFNGS | − | + |
| 8 | 8047 | K-AP | DAAGS | +++ | ++ |
| 9 | 8047 | K-AP | DLGAS | +++ | + |
| 10 | 8047 | K-AP | DAAAS | + | − |
| 11 | 8047 | KQAP | DLGGS | + | − |
| 12 | 8047 | KQAP | DAAGS | + | − |
Binding of MAb was measured by densitometry of the immuoblot with the following scoring system. Negative control E. coli MV averaged 4 densitometry units (DU), and values above 8 DU were considered positive. +, 8 to 15 DU; ++, 16 to 31 DU; +++, ≥32 DU.
wt, wild type.
Since MCH88 and 8047 NspA differ in both loops 2 and 3, the first question is whether the mutations in one or both loops account for the differences in binding by anti-NspA MAbs. As expected, both AL12 and 14C7 bound to a mutant of MCH88 in which both sequences of loops 2 and 3 were changed to the corresponding sequences in the 8047 strain (i.e., QV to -A in loop 2 and FN to LG in loop 3; construct 4). The respective binding to this mutant was similar to that of the wild-type 8047 construct (construct 5). When only the loop 3 sequence of MCH88 was changed to the loop 3 sequence of 8047 (FN to LG; construct 2), binding by both AL12 and 14C7 was restored. This result suggests that the insertion of Gln and the change of Ala to Val in loop 2 have little or no effect in the decreased binding of the MAbs to MCH88. In contrast, the LG-to-FN mutation in loop 3 of the 8047 construct resulted in complete loss of binding by AL12 and greatly decreased binding by 14C7 (construct 7). Together, the binding data from these two loop 2 and 3 mutants showed that the lack of binding of the MAbs to MCH88 was primarily the result of mutations in loop 3. However, some mutations in loop 2 can affect MAb binding. For example, insertion of Gln in loop 2 of NspA from 8047, which is adjacent to Ala instead of Val in MCH88, resulted in decreased binding of both MAbs (construct 11). Similar results were obtained with a second construct (construct 12) in which insertion of Gln in loop 2 (KQAP) resulted in decreased binding of both MAbs compared to the binding of the AAG loop 3 mutant having a wild-type 8047 loop 2 sequence (construct 8).
To investigate the role of individual residues in the apex segment of loop 3, we made Ala substitutions in the LGG segment of 8047 NspA. Binding by AL12 was unaffected by AAG or LGA substitutions (constructs 8 and 9, respectively), but AL12 showed only marginal binding to the AAA mutant (construct 10). Binding by 14C7 was decreased for AAG and LGA mutants and did not bind to the AAA mutant. Therefore, binding by the MAb 14C7, which was made to native NspA, is more sensitive to mutations in the LGG segment than MAb AL12, which was made to rNspA, since every Ala substitution in the LGG segment resulted in a substantial decrease in binding of 14C7. These results show that even though the GG segment in loop 3 has no side chains and is conformationally flexible, it constitutes a critical contact region for both MAbs. It is not clear whether the different epitope specificity of MAb 14C7 suggested by these results accounts for its superior functional activity compared to that of AL12, since the two MAbs also have different isotypes (IgG3 versus IgG2a, respectively).
Reconstitution of rNspA in liposomes.
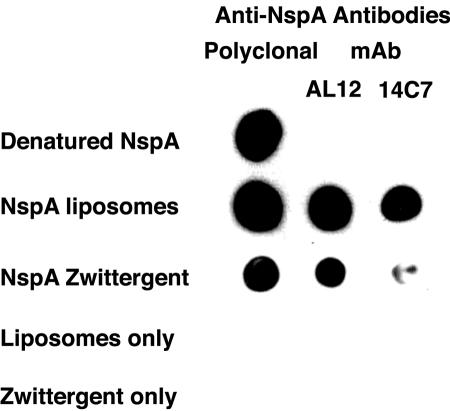
To determine whether the NspA epitopes recognized by the MAbs could be reconstituted from denatured NspA by the addition of detergent and/or lipids, we refolded HisTag-NspA in the presence of the detergent Zwittergent 3-14 and/or soy bean phospholipids. In Fig. 2, approximately equal amounts of HisTag-NspA as measured by BCA protein assay were spotted onto a nitrocellulose filter. Polyclonal antisera prepared to HisTag-NspA bound to both denatured and renatured HisTag-NspA (Fig. 2, left column). However, neither MAb showed detectable binding to denatured HisTag-NspA (Fig. 2, first row). In contrast, when HisTag-NspA was reconstituted in liposomes (Fig. 2, second row) or with the detergent Zwittergent 3-14, AL12 showed strong binding. The MAb made to native NspA, 14C7, also showed binding to NspA reconstituted in liposomes but bound poorly to HisTag-NspA reconstituted in detergent micelles (Fig. 2, third row). These results show that bactericidal anti-NspA MAbs bind to conformational epitopes that depend on the protein adopting a β-barrel tertiary structure (13, 20). Evidently, some epitopes present in native NspA are not reconstituted in Zwittergent 3-14 alone but also require a lipid bilayer.
FIG. 2.
Binding of anti-NspA antibodies to denatured HisTag-NspA reconstituted in liposomes or in detergent micelles. The polyclonal antibody was prepared in mice to HisTag-NspA. MAb AL12 was prepared against rNspA (without HisTag) expressed in E. coli MV (15). MAb 14C7 was prepared against native NspA expressed in N. meningitidis vesicles (14).
DISCUSSION
Using site-specific mutagenesis, we have shown that the epitopes recognized by two anti-NspA MAbs are located in loops 2 and 3. The most important contacts between the MAbs and NspA appear to be located within the LGG segment of loop 3. Since the residue side chains in this segment offer limited possibilities for binding interactions, it is likely that the backbone conformation in this segment has an important role in antibody recognition. Also, the conformation of loop 2 appears to be an important determinant, as particular combinations of residues in this segment can result in complete loss of antibody binding. Overall, our studies suggest that the two anti-NspA MAbs recognize discontinuous conformational features that result from the close proximity of loops 2 and 3 in the three-dimensional structure of NspA. As described further below, the fine antigenic specificities of the two MAbs also appear to be different from each other.
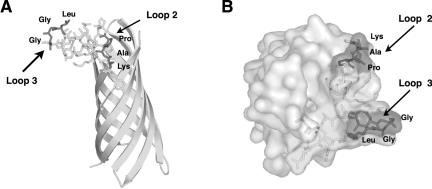
Based on the amino acid sequence homology between the opacity protein Opa and NspA, we proposed a structural model of NspA that contains eight transmembrane β-strands and four surface-exposed connecting loops (13). Recent crystallographic studies by Vandeputte-Rutten et al. of rNspA from N. meningitidis group B strain H44/76 showed that NspA adopts an eight-stranded β-barrel structure when reconstituted in detergent (Zwittergent 3-12) micelles (20). The structure determined is similar to that of our topological model. The amino acid sequences of the respective loop regions of NspA from strain H44/76 are identical to those of strain 8047. Our MAb binding results suggest that loop 2 and loop 3 of NspA are in close proximity to each other and that loop 3 is the dominant structural feature on the surface of the molecule. The crystal structure presented elsewhere (20) confirms the close proximity of loops 2 and 3, with loop 3 forming a prominent “knob” above and at the edge of the β-barrel.
A side view and a top view of the NspA structure determined by Vandeputte-Rutten et al. are shown in Fig. 3A and B, respectively. As shown in Fig. 3, loops 2 and 3 are a prominent feature on the outward facing surface of the molecule. The segments mutated are at or near the top of each loop. However, the residues mutated in loop 2 were not close to the residues mutated in loop 3. This arrangement is consistent with the observations described above that replacement of FN alone in loop 3 of MCH88 NspA with LG restored binding of the MAbs. Therefore, based on the crystal structure, the mutations in loop 2 (i.e., KAP to KQAP) that affect binding of the MAbs are likely to have resulted from an indirect effect on loop 3 caused by conformational changes in loop 2, rather than through altering direct contacts with the MAb. However, it remains a possibility that in comparison to the positions in the crystal, the loops are more flexible in solution and that the mutated regions are in close proximity upon MAb binding.
FIG. 3.
Structure of NspA (20). (A) Side view with respect to membrane orientation. Stick views of NspA loops 2 and 3 are shown. The shaded and labeled residues correspond to segments mutated in this study. (B) Top view with respect to the membrane orientation. Shown is a surface rendering of the molecule, with segments mutated in this study indicated by shading and labels. The figures were produced using the program Pymol (DeLano Scientific, San Carlos, Calif.).
Based on GenBank and neisserial genome database searches and sequences we have obtained from our strain collection, the NspA protein is highly conserved. Based on CLUSTAL W analysis (5), there is an average of 98% amino acid identity in N. meningitidis group B strains (n = 11) and an average of 97% identity in strains with other capsular groups (five group A, one group C, and one group W135 isolate). These strains are likely to be genetically diverse, having been collected for over 30 years from patients hospitalized in the United Kingdom, The Netherlands, Norway, the United States, and China. The group B strains include representatives of ET-37, ET-5, lineage A4, and three other distinct sequence or electrophoretic types. The loop 2 amino acid polymorphisms (Gln-Val) identified in group A strain MCH88, which decrease binding of bactericidal MAbs AL12 and 14C7, are found in some strains of N. meningitidis (for example, group A strain Z1073) and in other neisserial species, including Neisseria gonorrhea and Neisseria lactamica (unpublished data). The MCH88 loop 3 polymorphism (Phe-Asn), which appears to be in the portion of the molecule that is the most important contact between the MAbs and NspA, has been identified to date in only one strain, MCH88. Thus, a vaccine prepared from NspA from a single carefully chosen strain has the potential to prevent the majority of N. meningitidis disease.
Our findings, however, on the binding of anti-NspA MAbs AL12 and 14C7 imply that successful NspA-based vaccines will require NspA to be in a native conformation. This point also is illustrated by our group's previously published data with polyclonal anti-NspA antibody prepared in mice immunized with HisTag-NspA (13). By Western blotting, the polyclonal antibody recognized denatured NspA. However, the antiserum lacked complement-mediated bactericidal activity and did not bind by flow cytometry to the surface of live N. meningitidis cells. Finally, antisera prepared in mice to neisserial membrane vesicles treated with deoxycholate, a procedure commonly used to prepare vesicle vaccines for use in humans, had low anti-NspA activity compared to that of antisera raised in mice immunized with native vesicles (14). Taken together, the data suggest that vaccines using rNspA will require optimal refolding of the protein, for example in liposomes (for example, Fig. 2). Alternatively, optimal anti-NspA antibody responses may be obtained by immunizing with native vesicles prepared from N. meningitidis cells that have been either genetically detoxified (21; T. Kijet, M. Fisseha, B. Brandt, E. E. Moran, P. Chen, and W. Zollinger, 13th Int. Pathog. Neisseria Conf., p. 267, 2002) or detoxified by detergent treatments that do not disrupt expression of critical NspA epitopes.
Acknowledgments
This work was supported by grants RO1 AI45642 and AI46464 from the National Institute of Allergy and Infectious Disease of the National Institutes of Health.
We are grateful to Paolo Neri and Luisa Lozzi of the University of Siena, Siena, Italy, for performing the binding studies of overlapping 10-mer peptides. Alexander H. Lucas, Children's Hospital Oakland Research Institute, Oakland, Calif., provided helpful comments on the manuscript.
Editor: F. C. Fang
REFERENCES
- 1.Abdillahi, H., and J. T. Poolman. 1988. Neisseria meningitidis group B serosubtyping using monoclonal antibodies in whole-cell ELISA. Microb. Pathog. 4:27-32. [DOI] [PubMed] [Google Scholar]
- 2.Batzri, S., and E. D. Korn. 1973. Single bilayer liposomes prepared without sonication. Biochim. Biophys. Acta 298:1015-1019. [DOI] [PubMed] [Google Scholar]
- 3.Cadieux, N., M. Plante, C. R. Rioux, J. Hamel, B. R. Brodeur, and D. Martin. 1999. Bactericidal and cross-protective activities of a monoclonal antibody directed against Neisseria meningitidis NspA outer membrane protein. Infect. Immun. 67:4955-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cartwright, K., N. Noah, and H. Peltola. 2001. Meningococcal disease in Europe: epidemiology, mortality, and prevention with conjugate vaccines. Report of a European advisory board meeting Vienna, Austria, 6-8 October, 2000. Vaccine 19:4347-4356. [DOI] [PubMed] [Google Scholar]
- 5.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the CLUSTAL series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finne, J., M. Leinonen, and P. H. Makela. 1983. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. Lancet ii:355-357. [DOI] [PubMed] [Google Scholar]
- 7.Granoff, D. M., G. R. Moe, M. M. Giuliani, J. Adu-Bobie, L. Santini, B. Brunelli, F. Piccinetti, P. Zuno-Mitchell, S. S. Lee, P. Neri, L. Bracci, L. Lozzi, and R. Rappuoli. 2001. A novel mimetic antigen eliciting protective antibody to Neisseria meningitidis. J. Immunol. 167:3487-3496. [DOI] [PubMed] [Google Scholar]
- 8.Hayrinen, J., H. Jennings, H. V. Raff, G. Rougon, N. Hanai, R. Gerardy-Schahn, and J. Finne. 1995. Antibodies to polysialic acid and its N-propyl derivative: binding properties and interaction with human embryonal brain glycopeptides. J. Infect. Dis. 171:1481-1490. [DOI] [PubMed] [Google Scholar]
- 9.Jodar, L., I. Feavers, D. Salisbury, and D. M. Granoff. 2002. Development of vaccines against meningococcal disease. Lancet 359:1499-1508. [DOI] [PubMed] [Google Scholar]
- 10.Malorny, B., G. Morelli, B. Kusecek, J. Kolberg, and M. Achtman. 1998. Sequence diversity, predicted two-dimensional protein structure, and epitope mapping of neisserial Opa proteins. J. Bacteriol. 180:1323-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin, D., N. Cadieux, J. Hamel, and B. R. Brodeur. 1997. Highly conserved Neisseria meningitidis surface protein confers protection against experimental infection. J. Exp. Med. 185:1173-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merker, P., J. Tommassen, B. Kusecek, M. Virji, D. Sesardic, and M. Achtman. 1997. Two-dimensional structure of the Opc invasin from Neisseria meningitidis. Mol. Microbiol. 23:281-293. [DOI] [PubMed] [Google Scholar]
- 13.Moe, G. R., S. Tan, and D. M. Granoff. 1999. Differences in surface expression of NspA among Neisseria meningitidis group B strains. Infect. Immun. 67:5664-5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moe, G. R., P. Zuno-Mitchell, S. N. Hammond, and D. M. Granoff. 2002. Sequential immunization with vesicles prepared from heterologous Neisseria meningitidis strains elicits broadly protective serum antibodies to group B strains. Infect. Immun. 70:6021-6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moe, G. R., P. Zuno-Mitchell, S. S. Lee, A. H. Lucas, and D. M. Granoff. 2001. Functional activity of anti-neisserial surface protein A monoclonal antibodies against strains of Neisseria meningitidis serogroup B. Infect. Immun. 69:3762-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morley, S. L., and A. J. Pollard. 2001. Vaccine prevention of meningococcal disease, coming soon? Vaccine 20:666-687. [DOI] [PubMed] [Google Scholar]
- 17.Prince, S. M., M. Achtman, and J. P. Derrick. 2002. Crystal structure of the OpcA integral membrane adhesin from Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 99:3417-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenstein, N. E., B. A. Perkins, D. S. Stephens, L. Lefkowitz, M. L. Cartter, R. Danila, P. Cieslak, K. A. Shutt, T. Popovic, A. Schuchat, L. H. Harrison, and A. L. Reingold. 1999. The changing epidemiology of meningococcal disease in the United States, 1992-1996. J. Infect. Dis. 180:1894-1901. [DOI] [PubMed] [Google Scholar]
- 19.Rosenstein, N. E., B. A. Perkins, D. S. Stephens, T. Popovic, and J. M. Hughes. 2001. Meningococcal disease. N. Engl. J. Med. 344:1378-1388. [DOI] [PubMed] [Google Scholar]
- 20.Vandeputte-Rutten, L., M. P. Bos, J. Tommassen, and P. Gros. 2003. Crystal structure of neisserial surface protein A (NspA), a conserved outer membrane protein with vaccine potential. J. Biol. Chem. 278:24825-24830. [DOI] [PubMed] [Google Scholar]
- 21.van der Ley, P., L. Steeghs, H. J. Hamstra, J. ten Hove, B. Zomer, and L. van Alphen. 2001. Modification of lipid A biosynthesis in Neisseria meningitidis lpxL mutants: influence on lipopolysaccharide structure, toxicity, and adjuvant activity. Infect. Immun. 69:5981-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]