Abstract
Numerous microbial pathogens exploit complement regulatory proteins such as factor H (FH) and factor H-like protein 1 (FHL-1) for immune evasion. Fba is an FHL-1 and FH binding protein expressed on the surface of the human pathogenic bacterium, Streptococcus pyogenes, a common agent of pharyngeal, skin, and soft-tissue infections. In the present study, we demonstrate that Fba and FHL-1 work in concert to promote invasion of epithelial cells by S. pyogenes. Fba fragments were expressed as recombinant proteins and assayed for binding of FHL-1 and FH by Western blotting, enzyme-linked immunosorbent assay, and surface plasmon resonance. A binding site for FHL-1 and FH was localized to the N-terminal half of Fba, a region predicted to contain a coiled-coil domain. Deletion of this coiled-coil domain greatly reduced FHL-1 and FH binding. PepSpot analyses identified a 16-amino-acid segment of Fba which overlaps the coiled-coil domain that binds both FHL-1 and FH. To localize the Fba binding site in FHL-1 and FH, surface plasmon resonance was used to assess the interactions between the streptococcal protein and a series of recombinant FH deletion constructs. The Fba binding site was localized to short consensus repeat 7 (SCR 7), a domain common to FHL-1 and FH. SCR 7 contains a heparin binding site, and heparin was found to inhibit FHL-1 binding to Fba. FHL-1 promoted entry of Fba+ group A streptococci into epithelial cells in a dose-dependent manner but did not affect invasion by an isogenic fba mutant. To our knowledge, this is the first report of a bacterial pathogen exploiting a soluble complement regulatory protein for entry into host cells.
Activation of the complement system in response to infectious agents is a key component of the innate immune response, resulting in opsonization and/or lysis of invading pathogens. Uncontrolled complement activation, however, damages host tissues and results in complement depletion (38). Therefore, complement activity is tightly regulated by nearly a dozen plasma and cell membrane regulatory proteins. Two fluid-phase regulators of complement activation (RCAs) are factor H (FH) and factor H-like protein 1 (FHL-1). The two proteins are encoded by the same gene and their transcripts are derived by alternative splicing (11, 15, 57). FH is a 150-kDa protein composed of 20 repeat elements known as short consensus repeats (SCRs). Each SCR constitutes an independently folded domain of approximately 60 amino acid residues and contains two invariant disulfide bonds that form the framework of the domain. FHL-1 is a 42-kDa protein composed of seven SCRs that are identical, with the exception of four amino acids at the C terminus, to SCRs 1 to 7 of FH (57).
A number of bacterial pathogens, including Streptococcus pyogenes (21), Streptococcus pneumoniae (9, 23, 49), Streptococcus agalactiae (1), Neisseria gonorrhoeae (48), and Borrelia burgdorferi (30), exploit FHL-1 and FH for host colonization. In the absence of an RCA, C3b deposited on the surface of a pathogen serves as a nucleus for formation of the alternative complement pathway C3 convertase C3bBb. Once formed, C3 convertase cleaves additional C3, thereby promoting further C3b deposition and C3bBb formation. Recruitment of FH or FHL-1 by microbes facilitates factor I-mediated cleavage of cell surface-bound C3b to iC3b. Because iC3b does not participate in C3bBb formation, C3b cleavage results in decreased deposition of C3 fragments on bacterial cells. Because C3 fragments can be bound by receptors on phagocytes, limiting the amount of C3 deposited on cells can lessen the probability that a pathogen will be phagocytized (21, 57).
The group A streptococcus (GAS) S. pyogenes is an important human pathogen that exploits FHL-1 and FH to inhibit opsonization by C3, a mechanism that contributes to GAS resistance to phagocytosis (21). GAS also utilize the cell membrane-bound RCA CD46 for adherence to human keratinocytes (41, 44). Numerous laboratories have reported that RCA binding by GAS is mediated by M proteins, a family of cell surface antiphagocytic proteins (13). We recently identified a novel GAS cell surface protein, Fba, that mediates binding of both FHL-1 and FH. Fba is the first non-M-like protein of GAS shown to bind human RCAs (43). The same protein was identified by Terao et al. (55) as a fibronectin binding protein involved in invasion of Hep-2 cells. Although Fba was initially identified in M1 serotype isolates, fba is present in at least 18 GAS serotypes (46, 55). The conservation of fba suggests that the gene product contributes to survival of GAS during infection. We previously demonstrated that Fba contributes to GAS resistance to phagocytosis and that M1+ Fba+ streptococci bind more FHL-1 than FH when exposed to human plasma or serum (43). The latter finding was surprising, as the concentrations of FH and FHL-1 in human plasma are 400 and 30 μg/ml, respectively (20). Selective binding of FHL-1 seems of little benefit for resistance to phagocytosis, as FHL-1 and FH seem equivalent in their capacities to promote C3b cleavage (57). A possible benefit of selective FHL-1 binding is in the interaction of GAS with human cells.
It was recently reported that FHL-1, and to a lesser extent FH, can serve as an adhesion molecule for mammalian cells (7, 20, 57). Human polymorphonuclear leukocytes bind to immobilized FH via integrin CD11b/CD18 (7). The capacity of FH to function as an adhesive ligand may be limited to polymorphonuclear leukocytes. In contrast, fibroblasts, epithelial cells, and melanoma cells can all bind to immobilized FHL-1, but not to FH (20). It has been proposed that FHL-1 binding by these cells is integrin mediated, as SCR 4 contains the amino acid sequence RGD, a motif that is recognized by members of the integrin family of cellular receptors (20, 22). In fact, cell binding to FHL-1 can be inhibited by soluble RGD peptides. The more limited capacity of FH to function as an adhesion molecule may be due to intramolecular interactions between the C- and N-terminal SCRs, resulting in masking of the cell binding site in FH (20, 57).
Bacterial pathogens, including GAS, can utilize cell adhesive proteins, such as fibronectin, vitronectin, and laminin, for gaining entry into the cytoplasm of mammalian cells (3, 12, 24). We and others have previously shown that GAS are capable of intracellular invasion, i.e., entry into and persistence within mammalian cells (4, 33, 36, 42, 55). Despite much recent progress in understanding the mechanisms underlying GAS entry into host cells, the contribution of invasion to pathogenesis is not entirely clear (45). There is strong evidence that intracellular invasion contributes to the frequency and severity of GAS disease by affording partial protection of GAS from immune factors and antibiotics (3, 40, 53) and contributing to GAS dissemination in an infected host (35). Intracellular invasion can also permit translocation of pathogens through epithelial and endothelial cell barriers (12).
The capacity of FHL-1 to function as a cell adhesion molecule prompted us to investigate whether the protein could affect the interaction of GAS with human cells. We report here that binding of FHL-1, via Fba, promotes entry of streptococci into human epithelial cells. This finding demonstrates a novel function of FHL-1 in streptococcal pathogenesis.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
Intracellular invasion experiments were performed with an Fba+ S. pyogenes strain, 90-226 emm1::km (4), and an isogenic Fba− strain, DC276 (43), hereafter referred to as Fba+ and Fba− GAS, respectively. Streptococci were grown in Todd-Hewitt broth containing 1% yeast extract (Difco Laboratories, Detroit, Mich.) at 37°C, without shaking, in 5% CO2-95% air. Solid media for streptococci were Todd-Hewitt or sheep blood agar. DC276 was grown in medium containing 100 μg of spectinomycin per ml. Escherichia coli XL1-Blue (Stratagene, La Jolla, Calif.) was used for cloning of fba fragments and maintenance of plasmids. The protease-deficient E. coli strain BL21 (Stratagene) was used for expression of recombinant Fba proteins. The E. coli strains were cultured in Luria-Bertani (LB) broth or on LB agar (52). Ampicillin was added to E. coli media at a concentration of 100 μg/ml, as appropriate.
Cloning and expression of recombinant Fba proteins.
Plasmids encoding truncated or internally deleted Fba proteins were constructed as follows. Regions of fba (accession number AB040536) were amplified by PCR using pSport-fba as the template (43). The oligonucleotide primers used are listed in Table 1. Forward primers for cloning of Fba37-324, Fba37-164, and Fba135-324 all contained BamHI restriction sites. The reverse primers all contained EcoRI restriction sites. PCR products were digested with BamHI and EcoRI, gel purified, and ligated with BamHI- and EcoRI-restricted pGEX-2T (Amersham Biosciences, Piscataway, N.J.). For construction of FbaΔ68-104, the oligonucleotide primers Fba forward 3 and Fba reverse 5 were used to amplify the coding region for Fba37-67. An AseI restriction site was added to the reverse primer. The PCR product was digested with BamHI and AseI. The Fba104-324 coding region of fba was amplified with the Fba forward 7 and Fba reverse 3 oligonucleotide primers. The PCR product was digested with AseI and EcoRI. The two restricted, gel-purified DNA fragments were ligated with BamHI- and EcoRI-digested pGex-2T to construct the FbaΔ68-104-encoding plasmid. All constructs were verified by DNA sequencing performed by the Kansas University Medical Center Biotechnology Support Facility.
TABLE 1.
Oligonucleotides used for this study
| Oligonucleotide | Sequencea | Nucleotidesb |
|---|---|---|
| fba forward 3 | GTTGGATCCGCCGTAGATGGCATCCCTCCAAT | 250-272 |
| fba forward 6 | GTTGGATCCCCAGTTGAGGCACCCGTGCAG | 135-324 |
| fba forward 7 | CATGATTAATCCAGACAAAGATAAGCTATTATTCACG | 329-356 |
| fba reverse 3 | AGTGAATTCTCACTGTTGAAGGCAAGTACTTACCTG | 1111-1091 |
| fba reverse 4 | AGTGAATTCCAGAATCAGTCGTAACTGATGTTGAATCACC | 154-163 |
| fba reverse 5 | GCATGATTAATAAGGCCATCCTTATCAATATGATGCCA | 188-218 |
Restriction endonuclease cleavage sites are underlined.
Based on the fba sequence (GenBank accession number AB040536).
The recombinant proteins were expressed as fusions to glutathione S-transferase (GST) and purified by chromatography on glutathione-Sepharose (Amersham Biosciences) according to the manufacturer's instructions. The GST tag was removed by thrombin cleavage, thrombin was removed by chromatography on benzamidine-Sepharose (Amersham Biosciences), and the proteins were dialyzed against phosphate-buffered saline (PBS). Protein concentrations were determined by the bicinchoninic acid method (Pierce Chemical Co., Rockford, Ill.), using bovine serum albumin as the standard.
FHL-1, FH, and FH deletion constructs.
FH was purchased from Quidel Corp., Santa Clara, Calif. Recombinant FHL-1 and the FH deletion mutants were expressed in Sf9 cells and purified by Ni2+ chelation chromatography as previously described (20, 32).
Antibodies and antisera.
Goat anti-human FH antibody was purchased from Quidel Corp. Donkey anti-goat immunoglobulin G (IgG), labeled with alkaline phosphatase, was from Chemicon International, Temecula, Calif. Normal donkey serum, donkey anti-rabbit IgG labeled with fluorescein isothiocyanate (FITC), and donkey anti-rabbit IgG conjugated with rhodamine red were obtained from Jackson Immunoresearch Laboratories, West Grove, Pa. Rabbit anti-goat IgG conjugated with FITC was obtained from Sigma Chemical Co. Rabbit anti-Fba serum was produced by Cocalico Biologicals Inc., Reamstown, Pa., using Fba37-324 as antigen. Antisera against FHL-1 (SCRs 1 to 4) and FH (SCRs 19 to 20) were raised in rabbits using the indicated purified recombinant fragments (31, 47).
SDS-PAGE, Western blotting, and enzyme-linked immunosorbent assay.
Recombinant Fba proteins were electrophoresed on sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) gels, using standard procedures. Protein molecular weight standards were purchased from Bio-Rad Laboratories, Hercules, Calif. To assay FHL-1 and FH binding by recombinant Fba proteins, 1 μg of each recombinant protein was subjected to SDS-PAGE and transferred to nitrocellulose. Membranes were blocked with TBST (20 mM Tris [pH 7.5], 0.5 M NaCl, 0.05% Tween 20) containing 0.5% gelatin and then incubated with TBST containing either 10 μg of FHL-1 per ml or 10 μg of FH per ml. Bound ligands were detected by using FH antiserum and a labeled secondary antibody.
Enzyme-linked immunosorbent assays to detect FH binding were performed as described previously (43). Wells of microtiter plates were coated with 0.1 ml of a 1-μg/ml solution of a recombinant Fba protein in carbonate buffer.
Surface plasmon resonance.
The binding of FHL-1, FH, and FH deletion mutants to Fba was assayed by the plasmon resonance technique, using a Biacore 3000 instrument as described previously (19). Briefly, Fba was immobilized via a standard amine-coupling procedure to flow cells of a sensor chip (carboxylated dextran chip CM5; Biacore AB, Uppsala, Sweden). Flow cells were activated and Fba diluted in coupling buffer (10 mM acetate buffer, pH 5.0) was injected into one of the flow cells until an appropriate level of coupling for the binding experiments (>4,000 resonance units) was reached. A flow cell that had no protein added was used as a control. Unreacted groups in both cells were inactivated by an injection of ethanolamine-HCl (35 μl). After the coupling procedure, flow cells were washed thoroughly by sequential injection of regeneration buffer (3 M NaCl in 10 mM acetate buffer, pH 4.6) and running buffer (1/3× Veronal-buffered saline, pH 7.4). Binding of FHL-1, FH, and FH deletion mutants to Fba was then tested. Each reaction mixture was injected separately into the flow cell with precoupled Fba or into a blank control at a flow rate of 5 μl/min at 22°C. Binding was assayed at least in triplicate.
PepSpot analyses.
The entire sequence of mature Fba protein was synthesized as subsequent peptides, with lengths of 13 amino acids and an overlap of 10 amino acids, and coupled to a Whatman 50 cellulose support (Jerini Peptide Technologies, Berlin, Germany). This membrane was blocked with 5% albumin (Sigma Chemical Co.) in buffer I (50 mM Tris, 1.37 mM NaCl, 2.7 mM KCl, pH 8.0) overnight, and the filter was washed three times with buffer II (50 mM Tris, 1.37 mM NaCl, 2.7 mM KCl [pH 8.0], 0.05% Tween). Following incubation with purified FHL-1 or FH for 3 h at 4°C, the filter was washed three times in buffer II and incubated with FHL-1-specific (anti-SCRs 1 to 4) or FH-specific (anti-SCRs 19 and 20) antiserum for 3 h (28). The membranes were washed thoroughly, incubated with a horseradish peroxidase-labeled anti-goat antibody, and washed again, and the reaction was developed with ECL (Amersham Biosciences) reagents and exposed to X-ray film.
Immunofluorescence microscopy.
To detect Fba and FH on the surfaces of streptococci, bacteria were harvested from log-phase cultures, washed twice with PBS, and diluted to approximately 108 CFU/ml. The suspensions were applied to empty wells of Lab-Tek II chamber slides (Nalge Nunc International, Naperville, Ill.), and unbound bacteria were removed by washing with PBS. Slides were blocked with 1% gelatin and 10% normal donkey serum diluted in PBS. To detect Fba, slides were successively incubated with Fba antiserum and donkey anti-rabbit IgG labeled with FITC. For detection of FH binding, slides were incubated with 10 μg of FH per ml, anti-FH serum, and FITC-labeled rabbit anti-goat IgG.
Immunofluorescence microscopy of infected A549 cells was performed as follows. A549 cells were cultured on 4-well chamber slides and then infected with Fba+ GAS in the presence of FHL-1. After incubation, monolayers were washed with PBS and fixed with 3.7% formaldehyde in PBS. Slides were blocked with 10% normal donkey serum and incubated with rabbit anti-Fba serum, followed by incubation with donkey anti-rabbit IgG labeled with rhodamine red. In order to detect intracellular GAS, unbound antibodies were removed by washing and cells were permeabilized with Triton X-100. Slides were then incubated successively with anti-Fba serum and FITC-labeled goat anti-rabbit IgG. Specimens were examined by fluorescence microscopy for intracellular (FITC-labeled) and extracellular (FITC- and rhodamine red-labeled) GAS.
Cell culture and epithelial cell invasion assay.
A549 human lung epithelial cells (ATCC CCL 185) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (Life Technologies, Rockville, Md.), 5 μg of penicillin per ml, and 100 μg of streptomycin (Sigma Chemical Co.) per ml.
Assays of streptococcal invasion were performed as described previously (4). A549 cells were cultured in standard 48-well tissue culture plates (105 cells/well). The monolayers were infected with 1 × 105 to 2 × 105 bacterial CFU and suspended in either RPMI medium or RPMI medium containing either FH, FHL-1, or FH8-20. Plates were then incubated for 2 h at 37°C in 5% CO2-95% air. Infected monolayers were then washed three times with 0.5 ml of Hanks balanced salt solution before addition of RPMI medium containing 10% fetal bovine serum, gentamicin (100 μg/ml), and penicillin (5 μg/ml). Following 2 h of incubation at 37°C, the monolayers were washed with Hanks balanced salt solution, dispersed by the addition of 0.25% trypsin-1 mM EDTA (Life Technologies), and then lysed by dilution into sterile distilled H2O. The numbers of bacterial CFU released from the lysed epithelial cells were determined by plating of lysates on Todd-Hewitt agar. The protein preparations used for invasion experiments contained no fibronectin, as confirmed by Western blots. For evaluation of inhibition of intracellular invasion by recombinant Fba proteins or Fba antiserum, standard invasion experiments were performed in the presence of 15 μg of FHL-1 per ml. Inhibition was determined relative to control wells lacking a potential inhibitor.
RESULTS
Localization of the FHL-1 and FH binding site in Fba protein.
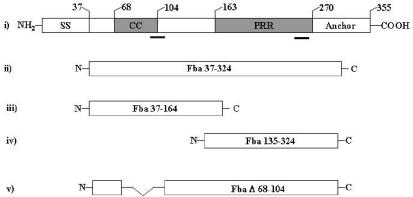
Secondary structure analysis of Fba predicted two distinct domains within the extracellular portion of the protein (Fig. 1). One is a 36-amino-acid segment in the N-terminal half of the protein (amino acid residues 69 to 104), predicted to form an α-helical coiled coil (34). The second domain is a 120-amino-acid region that lacks any predicted secondary structure and contains proline-rich repeats. In order to localize the FH binding region of Fba, fragments of fba were cloned into pGex-2T for expression of defined regions of the protein as fusions with GST. The recombinant proteins were expressed in E. coli and purified as described in Materials and Methods. Schematic representations of the recombinant Fba fragments are shown in Fig. 1. The Fba37-324 protein is analogous to mature Fba. Fba37-164 and Fba135-324 contain the coiled-coil domain and proline-rich repeats of Fba, respectively. The recombinant proteins all lack the N-terminal signal sequence and the C-terminal cell wall and cell membrane anchoring regions. The purified recombinant proteins separated by SDS-PAGE are shown in Fig. 2A.
FIG. 1.
Recombinant Fba constructs used for this study. Schematic representations of Fba and Fba derivatives are shown. (i) Structural and functional domains of Fba are as follows: SS, signal sequence; CC, coiled-coil region; PRR, proline-rich repeats; anchor, cell wall and membrane anchoring region. Numbers above the figure indicate the terminal amino acid residue of each domain. The bars below the figure indicate amino acid segments shown by PepSpot analyses to bind FHL-1 and FH. (ii to iv) Fba derivatives expressed as recombinant proteins. The numbers within the figures indicate Fba amino acid residues present in each derivative, where residue 1 is the N-terminal formylmethionine (accession number AB040536). (v) FbaΔ68-104 has an internal deletion of amino acid residues 68 to 104.
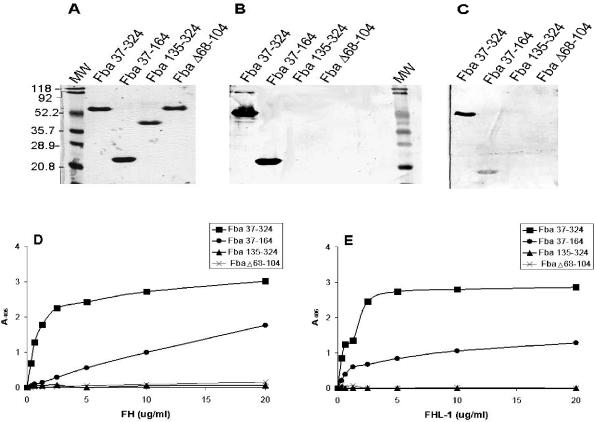
FIG. 2.
Localization of the FHL-1 and FH binding sites in Fba. (A) Coomassie blue-stained SDS-PAGE gel of the recombinant Fba proteins used for this study. The name of each Fba derivative is listed above the gel. MW, molecular mass standards. Numbers to the left of the figure indicate the sizes of each protein standard, in kilodaltons. (B) FH binding by Fba derivatives. The same proteins shown in panel A were transferred to nitrocellulose, blocked, and incubated successively with 10 μg of FH per ml, goat anti-human FH antibody, and an alkaline phosphatase-labeled secondary antibody. No bands were detectable on a duplicate blot incubated with only the primary and secondary antibodies (not shown). At least two independent experiments were performed with each Fba derivative. (C) FHL-1 binding by Fba derivatives. Binding was performed as described for panel B except that the membrane was incubated with FHL-1. (D) FH binding to immobilized Fba. The recombinant Fba derivatives were adsorbed onto microtiter plates and incubated with various concentrations of FH. Binding was detected with FH antiserum and a labeled secondary antibody. Data represent the means from a representative experiment, wherein each assay was performed in triplicate. Each recombinant protein was tested in at least two independent experiments. (E) FHL-1 binding by Fba derivatives. Binding assays were performed as described for panel D except that FHL-1 was substituted for FH. A405, absorbance at 405 nm.
To determine if the recombinant proteins could bind FHL-1 or FH, we subjected the proteins to SDS-PAGE and transferred them to nitrocellulose. Ligand binding was assessed by incubation of the membranes with FH (Fig. 2B) or FHL-1 (Fig. 2C), followed by detection of bound ligand with antibodies. FHL-1 and FH both bound to Fba37-324 and Fba37-164, but not to Fba135-324 or FbaΔ68-104. No signal was produced in control blots in which incubation with FH was omitted (data not shown). These results suggest that the coiled-coil domain represents the major binding site for FHL-1 and FH. Similar results were obtained in assays to measure FH (Fig. 2D) and FHL-1 (Fig. 2E) binding to the recombinant proteins immobilized in wells of microtiter plates. In this assay, FHL-1 and FH binding to Fba37-324 appeared stronger than that to Fba37-164, suggesting that the C-terminal portion of Fba contributes to ligand binding.
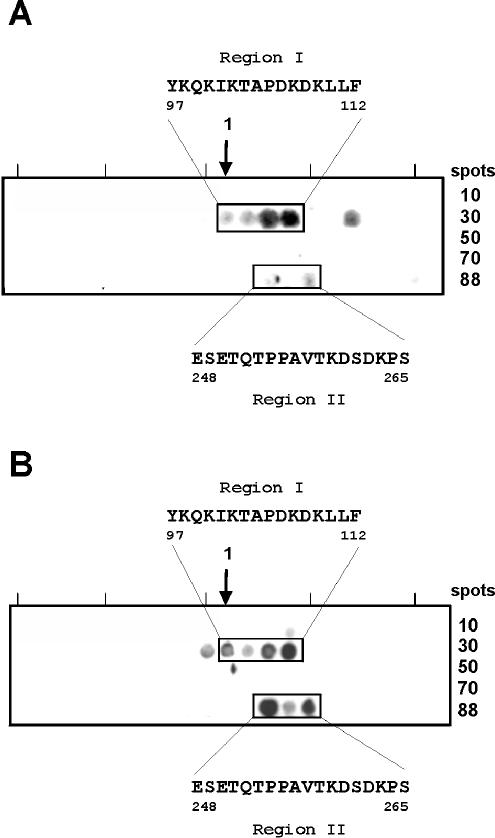
To localize the binding domain more precisely, a library of 88 peptides, representing the entire mature Fba protein, were synthesized and immobilized on a membrane. Each peptide was 13 amino acids in length and had an overlap of 10 amino acids. Membranes were incubated with FHL-1 (Fig. 3A) or FH (Fig. 3B) and ligand binding was detected with specific antibodies. The major binding site is represented by a linear 16-amino-acid region (positions 97 to 112) that overlaps the coiled-coil domain. The same experiments indicated the presence of a secondary FHL-1 and FH binding site in the C-terminal region of Fba. The secondary binding site is represented by an 18-amino-acid region encompassing Fba residues 248 to 265.
FIG. 3.
Mapping of the FHL-1 and FH binding sites in Fba by PepSpot analysis. A total of 88 peptides, spanning the entire Fba protein (excluding the signal sequence and cell anchoring domains), were synthesized and immobilized. Each peptide was 13 amino acid residues in length and had an overlap of 10 amino acids. The membranes were blocked and incubated with FHL-1 (A) or FH (B) and ligand binding was detected with specific antisera. The boxed regions indicate peptides that bound ligand. The amino acid sequences that mediate binding are listed. Numbers refer to amino acid residues of Fba (accession number AB040536), where residue 1 is the N-terminal formylmethionine.
Localization of the Fba binding site within FHL-1 and FH.
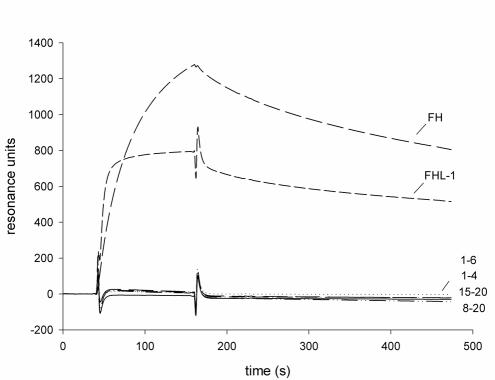
For localization of the Fba binding sites within FHL-1 and FH, recombinant proteins representing FHL-1 (SCRs 1 to 7), SCRs 1 to 6, SCRs 1 to 5, and SCRs 1 to 4, as well as fragments specific for FH (i.e., SCRs 8 to 20 and SCRs 15 to 20), were tested for Fba binding using the surface plasmon resonance technique (Fig. 4). FHL-1 bound to Fba, but fragments which lacked SCR 7, i.e., SCRs 1 to 6, SCRs 1 to 5, and SCRs 1 to 4, did not bind (Fig. 4 and data not shown), indicating that the binding site within FHL-1 is located at the most C-terminal SCR, i.e., SCR 7. Intact FH bound Fba, but fragments that represented SCRs 8 to 20 or SCRs 15 to 20 did not. These results indicate that Fba is bound by SCR 7, a site common to FHL-1 and FH.
FIG. 4.
Localization of the Fba binding site in FHL-1 and FH by the surface plasmon resonance technique. Fba37-324 was coupled to the surfaces of sensor chips, and either FHL-1, FH, or an FH deletion construct, as indicated, was injected into the flow cell. A flow cell that had no protein added was used as a control. Binding was detected as an increase in resonance units over time. Numbers, e.g., 1-6, 1-4, indicate the SCRs represented in each FH deletion construct. A representative sensorgram is shown. Binding experiments were performed three times, with similar results.
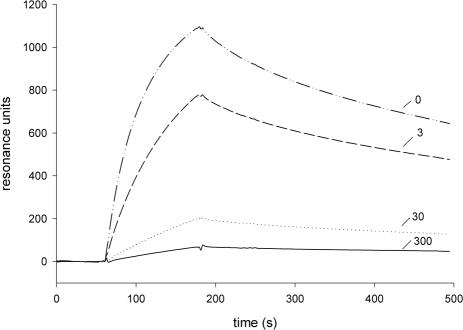
SCR 7 contains a heparin binding site; therefore, we tested whether heparin influences the interaction of FHL-1 and Fba (Fig. 5). Interaction of the proteins was inhibited by heparin in a dose-dependent manner. These results support the conclusion that Fba binds to SCR 7 and suggest that the heparin and Fba binding sites in FHL-1 and FH may overlap.
FIG. 5.
Inhibition of FHL-1 binding to Fba by heparin. Binding assays were performed as described for Fig. 4, except that reactions contained the indicated concentration (3, 30, or 300 μg per ml) of heparin.
Cell surface expression of Fba.
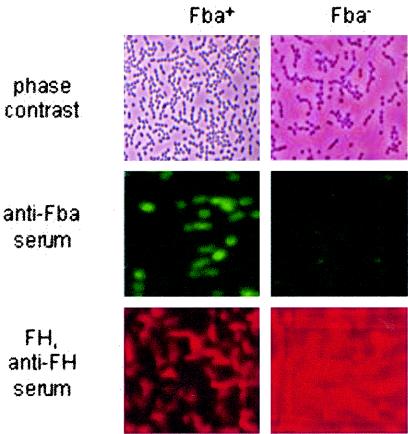
We hypothesized that binding of FHL-1 and FH by streptococci could affect their interaction with host cells. Therefore, immunofluorescence microscopy was performed to verify cell surface expression of Fba. As anticipated, Fba antiserum reacted with Fba+, but not Fba−, bacteria (Fig. 6, middle panels). Similar experiments were performed to verify binding of FH to the surface of the Fba+ strain (Fig. 6, lower panels). FH binding was detected only for Fba+ GAS.
FIG. 6.
Surface labeling of GAS with Fba antibodies and FH. Fba+ (90-226 emm1::km) and Fba− GAS (DC276) were incubated with Fba antiserum (middle) followed by FITC-labeled goat anti-rabbit IgG. Bacteria for the lower panels were incubated with FH, goat anti-human FH serum, and donkey anti-rabbit IgG conjugated to rhodamine red.
Involvement of Fba and FHL-1 in intracellular invasion.
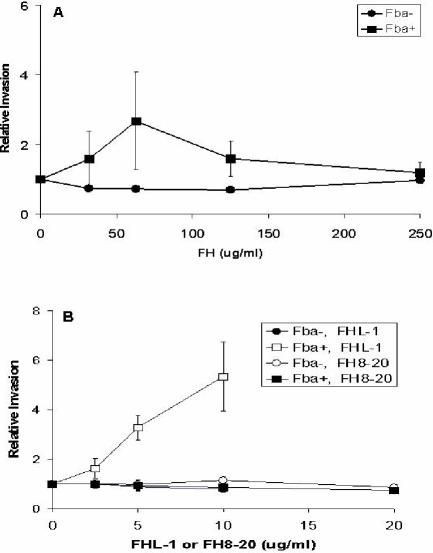
FH was tested for the capacity to promote ingestion of GAS by A549 (human lung epithelial) cells. A549 cells were infected with Fba+ or Fba− GAS in the presence of various concentrations of FH, and extracellular bacteria were eliminated by the addition of antibiotics. FH did not promote efficient internalization of either Fba+ or Fba− streptococci, even at concentrationsas high as 250 μg/ml (Fig. 7A). Conversely, recombinant FHL-1 stimulated ingestion of Fba+ GAS in a dose-dependent manner (Fig. 7B). Additionally, FHL-1 at a concentration of 10 μg/ml stimulated internalization of the wild-type GAS strain, 90-226 (4), by a factor of 3.6 (data not shown). FHL-1 had no effect on ingestion of the Fba− strain (Fig. 7B), consistent with the inability of the mutant to bind FHL-1 (43). The recombinant FH protein encompassing SCRs 8 to 20 was included as a negative control, as Fba does not bind to this region of FH (Fig. 4). As anticipated, recombinant SCRs 8 to 20 did not affect invasion by either the Fba+ or Fba− strain (Fig. 7B). These results demonstrate that Fba and FHL-1 function in concert to promote intracellular invasion.
FIG. 7.
Fba- and FHL-1-dependent invasion of A549 cells. (A) Fba+ (90-226 emm1::km) (squares) and Fba− (DC276) (circles) GAS were suspended in RPMI medium or in medium containing various levels of FH and inoculated onto monolayers of A549 cells. The numbers of internalized GAS were determined by a standard antibiotic protection assay, as described in Materials and Methods. For each experiment, the number of CFU recovered from monolayers infected with GAS in the absence of FH was assigned a value of 1.0, and other data are expressed relative to that value. Data are from three independent experiments in which each assay was performed in duplicate. Bars represent standard deviations. (B) Invasion in the presence of various concentrations of FHL-1 (open symbols) or an FH deletion construct that represented SCRs 8 to 20 (filled symbols). Data are presented as described for panel A. The maximum concentrations of FHL-1 and FH8-20 used (10 and 21 μg/ml, respectively) represent equimolar concentrations of the proteins. The overall efficiencies of internalization, in the absence of FHL-1 or FH, were 0.17% ± 0.03% for the Fba+ strain and 0.26% ± 0.12% for the fba mutant strain.
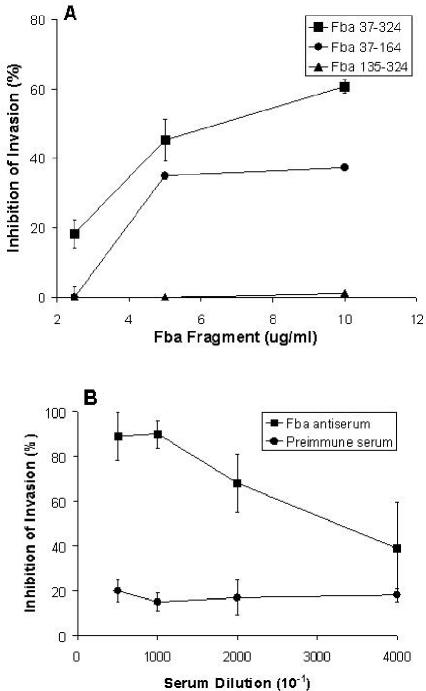
The essential role of Fba-FHL-1 interaction was confirmed by the inhibitory effects of Fba fragments on internalization of Fba+ GAS. Fba constructs capable of binding FHL-1 (Fba37-324 and Fba37-164) were effective inhibitors of invasion, whereas Fba135-324, a construct with weak binding activity, was ineffective as an inhibitor (Fig. 8A). Moreover, Fba37-324 was a more effective inhibitor than Fba37-164, reflecting the relative affinities of the proteins for FHL-1 and FH. Additionally, Fba antiserum, but not preimmune serum, could inhibit uptake of streptococci (Fig. 8B). Fluorescence microscopy of infected A549 cells was performed to verify the presence of intracellular GAS (Fig. 9).
FIG. 8.
Inhibition of intracellular invasion by recombinant Fba proteins and Fba antiserum. A549 cells were infected with GAS strain 90-226 emm1::km suspended in RPMI medium containing 15 μg of FHL-1 per ml and various concentrations of either a recombinant Fba derivative (A), Fba antiserum (B), or preimmune serum (B). The numbers of internalized GAS were determined by a standard antibiotic protection assay, as described in Materials and Methods. The percentages of invasion inhibition were calculated relative to those of control wells lacking either Fba or serum. Data are from at least two independent experiments in which each assay was performed in duplicate. Bars represent standard deviations.
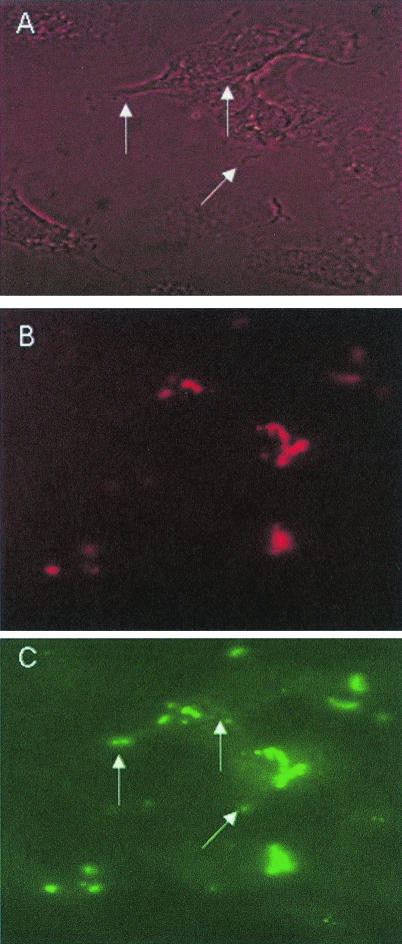
FIG. 9.
Immunofluorescence microscopy of FHL-1-Fba-mediated invasion. A549 cells were infected with GAS strain 90-226 emm1::km in the presence of FHL-1. After incubation, cells were fixed, blocked, and incubated with rabbit anti-Fba serum, followed by incubation with donkey anti-rabbit IgG labeled with rhodamine red. After removal of unbound antibodies, cells were permeabilized with Triton X-100 and incubated successively with anti-Fba serum and goat anti-rabbit IgG labeled with FITC. Specimens were examined by fluorescence microscopy for intracellular (FITC-labeled) and extracellular (FITC- and rhodamine red-labeled) GAS. (A) Phase-contrast microscopy of infected A549 cells. (B) The same field as that shown in the other panels, irradiated for visualization of extracellular GAS. (C) The same field viewed for the presence of intracellular and extracellular GAS. Arrows in panels A and C indicate intracellular streptococci.
DISCUSSION
The gram-positive bacterium S. pyogenes, or GAS, is an important and highly versatile human pathogen. A major virulence trait of GAS is the capacity of the organism to evade ingestion and/or killing by host phagocytes (6). Streptococcal resistance to phagocytosis is due, in part, to the binding of human complement regulatory proteins, such as FH, FHL-1, and C4b binding protein (21, 25, 29). Fba is a recently identified GAS cell surface protein capable of binding FHL-1, FH, and fibronectin (43, 55). The gene encoding Fba is present in at least 18 serotypes of streptococci, suggesting that the protein contributes to survival of GAS in an infected host. In fact, Fba has been shown to contribute to GAS virulence in mice (55). Previously, we demonstrated that Fba contributes to GAS resistance to phagocytosis (43). In the present study, we report that Fba, via binding to FHL-1, can promote the entry of streptococci into the cytoplasm of human epithelial cells. These data argue that Fba is an important, multifunctional virulence factor.
To localize the binding site for FHL-1 and FH, we expressed deletion derivatives of Fba as recombinant proteins and tested them for FH binding. Binding experiments demonstrated that an Fba construct encompassing the coiled-coil domain (Fba37-164) binds both ligands. Deletion of the coiled-coil region drastically decreased binding of FHL-1 and FH. PepSpot analyses confirmed that a linear domain that overlaps the coiled-coil region mediates binding to FHL-1 and FH. These results argue that the sequence YKQKIKTAPDKDKLLF (amino acids 97 to 112) represents the major FHL-1 and FH binding site in Fba. A similar positively charged FH binding domain is found at the C terminus of the B. burgdorferi CRASp-3 protein (30). This FH binding motif is conserved among several homologues of CRASp-3. In this context, it is important to note that not all microbial proteins that bind FH or FHL-1 possess ligand binding sites enriched in basic amino acid residues. For example, one of the FH binding sites of the GAS M6 protein has a net negative charge and hydrophobic interactions are involved in FH binding by the PspC protein of S. pneumoniae (9, 14).
Binding experiments indicated the presence of a secondary FHL-1 and FH binding site in the proline-rich repeat domain (amino acids 163 to 269) of Fba. PepSpot analyses identified an 18-amino-acid segment (amino acids 248 to 265) that shows weak binding of both ligands. Moreover, an Fba construct lacking the proline-rich repeats, Fba37-164, binds FHL-1 and FH with a lower affinity than the intact recombinant protein (Fba37-324). Fba37-164 is also a less effective inhibitor of intracellular invasion than Fba37-324. These results argue that the sequence ESETQTPPAVTKDSDKPS constitutes a secondary, low-affinity binding site for FHL-1 and FH.
The Fba binding site in FHL-1 and FH was localized to SCR 7, a domain common to both proteins. At least two types of M proteins, M5 and M6, also bind to SCR 7 (2, 29, 54). M6 protein binds to a cluster of positively charged amino acids on the surface of FH at the interface of SCRs 6 and 7 (17, 18). Molecular modeling suggests that similar binding sites are present in CD46 and C4b binding protein, both of which are M protein ligands (26, 41). FH contains three heparin binding sites, including one site in SCR 7 (56). Accordingly, we found that heparin inhibits FHL-1 binding to Fba in a dose-dependent manner, suggesting that the heparin and Fba binding sites in FHL-1 may overlap.
FHL-1 is a well-characterized complement regulator that also functions in cell adhesion (7). Bacterial pathogens, including streptococci, often exploit host adhesive proteins to facilitate adherence to and invasion of host cells (3, 4, 24). Binding of adhesive proteins can be regarded as a mechanism whereby microbial pathogens can extend their receptor repertoires. For example, invasion of human epithelial cells by GAS can be mediated by the fibronectin receptor integrin α5β1. Bacterial engagement of the integrin, however, depends on binding of fibronectin to the surfaces of bacterial cells. Evidence suggests that the role of fibronectin is that of forming a molecular bridge between a bacterial fibronectin binding protein and the integrin (5, 42).
We report here that FHL-1 also functions as an invasion agonist. Moreover, we demonstrated that the capacity of FHL-1 to facilitate invasion depends on cell surface expression of Fba by streptococci. Note that, even though FHL-1 promotes invasion of the wild-type (i.e., M1+ Fba+) GAS strain 90-226, our intracellular invasion experiments were performed with M1 mutant strains. There are two reasons for this. The first is that M1 protein does promote adherence to and invasion of A549 cells (4); thus, using M1-deficient strains simplified the performance and interpretation of our experiments. The second, and more important, reason is that the role of M1 protein in FH and FHL-1 binding is not completely clear. We reported previously that M1 protein does not significantly contribute to FH or FHL-1 binding by GAS strain 90-226 (43), but it has been shown that purified M1 protein binds FH (28) and there is at least one report that M1 contributes to FH binding by streptococci (27). Previous studies have established, however, that expression of M1 protein is not necessary for, nor does it inhibit, FHL-1 binding by streptococci (27, 43). We chose to avoid the issue of M1 interaction with FH and FHL-1, for the present, by using M1 mutants in our experiments.
FH binding by Fba+ GAS does not result in efficient invasion, a result in accordance with our previous report that FHL-1, but not FH, can serve as an attachment matrix for a variety of mammalian cells. Available evidence suggests that FHL-1 binding by epithelial cells and fibroblasts is mediated by integrins (20), but the specific receptor involved in binding has not been identified. It is tempting to speculate that Fba- and FHL-1-dependent invasion of A549 cells is integrin mediated, but this has not been substantiated. Whatever the nature of the Fba-FHL-1 receptor, recruitment of FHL-1 to the surfaces of GAS clearly affects the interaction between streptococci and host cells. GAS are versatile and genetically diverse pathogens. These properties are reflected in the organism's capacity to enter host cells by several distinct mechanisms. In addition to fibronectin and FHL-1, the adhesive protein laminin can facilitate GAS invasion and some GAS isolates express surface proteins that mediate bacterial internalization independent of a bridging molecule (4, 10, 37). The ability to utilize multiple invasion pathways may contribute to the bacterium's capacity to disseminate and colonize different tissues during infection. That is, different invasion mechanisms may be used for entry into different cell types. The mechanism of host cell entry can influence the fate of intracellular bacteria (8, 51). For example, Rohde et al. (50) have proposed that GAS internalized by caveola-dependent endocytosis avoid fusion with lysosomes, thereby increasing the likelihood of intracellular survival. The fate of GAS internalized by the FHL-1-dependent pathway has not been investigated. FHL-1-mediated invasion is likely to be most efficient for cells that are exposed to soluble FHL-1 and express an abundance of FHL-1 receptors. Streptococci are certain to be exposed to FHL-1 and FH in blood, but these regulatory factors are also present in inflammatory effusions (16, 39) and we have detected both proteins in oral secretions (E. Gregory and D. Cue, unpublished data). It seems likely that an inflammatory response induced by GAS colonization of the oral mucosa or skin leads to an influx of complement components, including FH and FHL-1, at the site of infection. As demonstrated here and in previous work (42), acquisition of these RCAs has the potential to impact subsequent interactions of GAS with both nonphagocytic and phagocytic cells.
In summary, we have described a novel mechanism for GAS invasion of epithelial cells, mediated by Fba protein and FHL-1. To our knowledge, this is the first description of a bacterial pathogen exploiting a soluble complement regulatory protein to gain access to an intracellular niche. Binding of FHL-1 and FH by bacteria, viruses, fungi, and protozoa is a common mechanism whereby pathogens attempt to evade the innate immune response of a host organism (38). Our data demonstrate, for the first time, that FHL-1 binding can affect other pathogen-host interactions. Utilization of FHL-1 to facilitate tissue adherence or invasion may represent a common theme in bacterial pathogenesis.
Acknowledgments
This work was supported by National Institutes of Health grant P20 RR16443-03 from the COBRE Program of the National Center for Research Resources, the Deutsche Forschungsgemeinschaft, and the Thüringer Ministerium für Wissenschaft Forschung and Technologie (TMWFK).
Editor: J. T. Barbieri
REFERENCES
- 1.Areschoug, T., M. Stalhammar-Carlemalm, I. Karlsson, and G. Lindahl. 2002. Streptococcal beta protein has separate binding sites for human factor H and IgA-Fc. J. Biol. Chem. 277:12642-12648. [DOI] [PubMed] [Google Scholar]
- 2.Blackmore, T. K., V. A. Fischetti, T. A. Sadlon, H. M. Ward, and D. L. Gordon. 1998. M protein of the group A streptococcus binds to the seventh short consensus repeat of human complement factor H. Infect. Immun. 66:1427-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cue, D., P. E. Dombek, and P. P. Cleary. 2000. Intracellular invasion by Streptococcus pyogenes: invasins, host receptors and relevance to human disease, p. 27-33. In V. A. Fischetti et al. (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C. [DOI] [PMC free article] [PubMed]
- 4.Cue, D., P. E. Dombek, H. Lam, and P. P. Cleary. 1998. Streptococcus pyogenes serotype M1 encodes multiple pathways for entry into human epithelial cells. Infect. Immun. 66:4593-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cue, D., S. O. Southern, P. J. Southern, J. Prabhakar, W. Lorelli, J. M. Smallheer, S. A. Mousa, and P. P. Cleary. 2000. A nonpeptide integrin antagonist can inhibit epithelial cell ingestion of Streptococcus pyogenes by blocking formation of integrin α5β1-fibronectin-M1 protein complexes. Proc. Natl. Acad. Sci. USA 97:2858-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiScipio, R. G., P. J. Daffern, I. U. Schraufstatter, and P. Sriramarao. 1998. Human polymorphonuclear leukocytes adhere to complement factor H through an interaction that involves αMβ2 (CD11b/CD18). J. Immunol. 160:4057-4066. [PubMed] [Google Scholar]
- 8.Duncan, M. J., J. S. Shin, and S. N. Abraham. 2002. Microbial entry through caveolae: variations on a theme. Cell. Microbiol. 4:783-791. [DOI] [PubMed] [Google Scholar]
- 9.Duthy, T. G., R. J. Ormsby, E. Giannakis, A. D. Ogunniyi, U. H. Stroeher, J. C. Paton, and D. L. Gordon. 2002. The human complement regulator factor H binds pneumococcal surface protein PspC via short consensus repeats 13 to 15. Infect. Immun. 70:5604-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elsner, A., B. Kreikemeyer, A. Braun-Kiewnick, B. Spellerberg, B. A. Buttaro, and A. Podbielski. 2002. Involvement of Lsp, a member of the LraI-lipoprotein family in Streptococcus pyogenes, in eukaryotic cell adhesion and internalization. Infect. Immun. 70:4859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estaller, C., W. Schwaeble, M. Dierich, and E. H. Weiss. 1991. Human complement factor H: two factor H proteins are derived from alternatively spliced transcripts. Eur. J. Immunol. 21:799-802. [DOI] [PubMed] [Google Scholar]
- 12.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischetti, V. A. 2000. Surface proteins on gram-positive bacteria, p. 11-24. In V. A. Fischetti et al. (ed.), Gram-positive pathogens. American Society for Microbiology, Washington D.C.
- 14.Fischetti, V. A., R. D. Horstmann, and V. Pancholi. 1995. Location of the complement factor H binding site on streptococcal M6 protein. Infect. Immun. 63:149-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friese, M. A., J. Hellwage, T. S. Jokiranta, S. Meri, H. H. Peter, H. Eibel, and P. F. Zipfel. 1999. FHL-1/reconectin and factor H: two human complement regulators which are encoded by the same gene are differently expressed and regulated. Mol. Immunol. 36:809-818. [DOI] [PubMed] [Google Scholar]
- 16.Friese, M. A., T. Manuelian, S. Junnikkala, J. Hellwage, S. Meri, H. H. Peter, D. L. Gordon, H. Eibel, and P. F. Zipfel. 2003. Release of endogenous anti-inflammatory complement regulators FHL-1 and factor H protects synovial fibroblasts during rheumatoid arthritis. Clin. Exp. Immunol. 132:485-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giannakis, E., T. S. Jokiranta, D. A. Male, S. Ranganathan, R. J. Ormsby, V. A. Fischetti, C. Mold, and D. L. Gordon. 2003. A common site within factor H SCR 7 responsible for binding heparin, C-reactive protein and streptococcal M protein. Eur. J. Immunol. 33:962-969. [DOI] [PubMed] [Google Scholar]
- 18.Giannakis, E., T. S. Jokiranta, R. J. Ormsby, T. G. Duthy, D. A. Male, D. Christiansen, V. A. Fischetti, C. Bagley, B. E. Loveland, and D. L. Gordon. 2002. Identification of the streptococcal M protein binding site on membrane cofactor protein (CD46). J. Immunol. 168:4585-4592. [DOI] [PubMed] [Google Scholar]
- 19.Hellwage, J., T. S. Jokiranta, M. A. Friese, T. U. Wolk, E. Kampen, P. F. Zipfel, and S. Meri. 2002. Complement C3b/C3d and cell surface polyanions are recognized by overlapping binding sites on the most carboxyl-terminal domain of complement factor H. J. Immunol. 169:6935-6944. [DOI] [PubMed] [Google Scholar]
- 20.Hellwage, J., S. Kuhn, and P. F. Zipfel. 1997. The human complement regulatory factor-H-like protein 1, which represents a truncated form of factor H, displays cell-attachment activity. Biochem. J. 326:321-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horstmann, R. D., H. J. Sievertsen, J. Knobloch, and V. A. Fischetti. 1988. Antiphagocytic activity of streptococcal M protein: selective binding of complement control protein factor H. Proc. Natl. Acad. Sci. USA 85:1657-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hynes, R. O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell 110:673-687. [DOI] [PubMed] [Google Scholar]
- 23.Janulczyk, R., F. Iannelli, A. G. Sjoholm, G. Pozzi, and L. Bjorck. 2000. Hic, a novel surface protein of Streptococcus pneumoniae that interferes with complement function. J. Biol. Chem. 275:37257-37263. [DOI] [PubMed] [Google Scholar]
- 24.Joh, D., E. R. Wann, B. Kreikemeyer, P. Speziale, and M. Hook. 1999. Role of fibronectin-binding MSCRAMMs in bacterial adherence and entry into mammalian cells. Matrix Biol. 18:211-223. [DOI] [PubMed] [Google Scholar]
- 25.Johnsson, E., A. Thern, B. Dahlback, L. O. Heden, M. Wikstrom, and G. Lindahl. 1996. A highly variable region in members of the streptococcal M protein family binds the human complement regulator C4BP. J. Immunol. 157:3021-3029. [PubMed] [Google Scholar]
- 26.Johnsson, E., A. Thern, B. Dahlback, L. O. Heden, M. Wikstrom, and G. Lindahl. 1997. Human C4BP binds to the hypervariable N-terminal region of many members in the streptococcal M protein family. Adv. Exp. Med. Biol. 418:505-510. [DOI] [PubMed] [Google Scholar]
- 27.Kihlberg, B. M., M. Collin, A. Olsen, and L. Bjorck. 1999. Protein H, an antiphagocytic surface protein in Streptococcus pyogenes. Infect. Immun. 67:1708-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kihlberg, B. M., J. Cooney, M. G. Caparon, A. Olsen, and L. Bjorck. 1995. Biological properties of a Streptococcus pyogenes mutant generated by Tn916 insertion in mga. Microb. Pathog. 19:299-315. [DOI] [PubMed] [Google Scholar]
- 29.Kotarsky, H., J. Hellwage, E. Johnsson, C. Skerka, H. G. Svensson, G. Lindahl, U. Sjobring, and P. F. Zipfel. 1998. Identification of a domain in human factor H and factor H-like protein-1 required for the interaction with streptococcal M proteins. J. Immunol. 160:3349-3354. [PubMed] [Google Scholar]
- 30.Kraiczy, P., J. Hellwage, C. Skerka, M. Kirschfink, V. Brade, P. F. Zipfel, and R. Wallich. 2003. Immune evasion of Borrelia burgdorferi: mapping of a complement-inhibitor factor H-binding site of BbCRASP-3, a novel member of the Erp protein family. Eur. J. Immunol. 33:697-707. [DOI] [PubMed] [Google Scholar]
- 31.Kuhn, S., C. Skerka, and P. F. Zipfel. 1995. Mapping of the complement regulatory domains in the human factor H-like protein 1 and in factor H1. J. Immunol. 155:5663-5670. [PubMed] [Google Scholar]
- 32.Kuhn, S., and P. F. Zipfel. 1995. The baculovirus expression vector pBSV-8His directs secretion of histidine-tagged proteins. Gene 162:225-229. [DOI] [PubMed] [Google Scholar]
- 33.LaPenta, D., C. Rubens, E. Chi, and P. P. Cleary. 1994. Group A streptococci efficiently invade human respiratory epithelial cells. Proc. Natl. Acad. Sci. USA 91:12115-12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lupas, A. 1996. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21:375-382. [PubMed] [Google Scholar]
- 35.Medina, E., O. Goldmann, A. W. Toppel, and G. S. Chhatwal. 2003. Survival of Streptococcus pyogenes within host phagocytic cells: a pathogenic mechanism for persistence and systemic invasion. J. Infect. Dis. 187:597-603. [DOI] [PubMed] [Google Scholar]
- 36.Molinari, G., and G. S. Chhatwal. 1999. Role played by the fibronectin-binding protein SfbI (protein F1) of Streptococcus pyogenes in bacterial internalization by epithelial cells. J. Infect. Dis. 179:1049-1050. [DOI] [PubMed] [Google Scholar]
- 37.Molinari, G., M. Rohde, C. A. Guzman, and G. S. Chhatwal. 2000. Two distinct pathways for the invasion of Streptococcus pyogenes in non-phagocytic cells. Cell. Microbiol. 2:145-154. [DOI] [PubMed] [Google Scholar]
- 38.Morgan, P. B., and C. L. Harris. 1999. Complement regulatory proteins. Academic Press, San Diego, Calif.
- 39.Narkio-Makela, M., J. Hellwage, O. Tahkokallio, and S. Meri. 2001. Complement-regulator factor H and related proteins in otitis media with effusion. Clin. Immunol. 100:118-126. [DOI] [PubMed] [Google Scholar]
- 40.Neeman, R., N. Keller, A. Barzilai, Z. Korenman, and S. Sela. 1998. Prevalence of internalisation-associated gene, prtF1, among persisting group-A streptococcus strains isolated from asymptomatic carriers. Lancet 352:1974-1977. [DOI] [PubMed] [Google Scholar]
- 41.Okada, N., M. K. Liszewski, J. P. Atkinson, and M. Caparon. 1995. Membrane cofactor protein (CD46) is a keratinocyte receptor for the M protein of the group A streptococcus. Proc. Natl. Acad. Sci. USA 92:2489-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozeri, V., I. Rosenshine, A. Ben-Ze'Ev, G. M. Bokoch, T. S. Jou, and E. Hanski. 2001. De novo formation of focal complex-like structures in host cells by invading streptococci. Mol. Microbiol. 41:561-573. [DOI] [PubMed] [Google Scholar]
- 43.Pandiripally, V., E. Gregory, and D. Cue. 2002. Acquisition of regulators of complement activation by Streptococcus pyogenes serotype M1. Infect. Immun. 70:6206-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez-Casal, J., N. Okada, M. G. Caparon, and J. R. Scott. 1995. Role of the conserved C-repeat region of the M protein of Streptococcus pyogenes. Mol. Microbiol. 15:907-916. [DOI] [PubMed] [Google Scholar]
- 45.Podbielski, A., and B. Kreikemeyer. 2001. Persistence of group A streptococci in eukaryotic cells—a safe place? Lancet 358:3-4. [DOI] [PubMed] [Google Scholar]
- 46.Podbielski, A., M. Woischnik, B. Pohl, and K. H. Schmidt. 1996. What is the size of the group A streptococcal vir regulon? The Mga regulator affects expression of secreted and surface virulence factors. Med. Microbiol. Immunol. (Berlin) 185:171-181. [DOI] [PubMed] [Google Scholar]
- 47.Prodinger, W. M., J. Hellwage, M. Spruth, M. P. Dierich, and P. F. Zipfel. 1998. The C-terminus of factor H: monoclonal antibodies inhibit heparin binding and identify epitopes common to factor H and factor H-related proteins. Biochem. J. 331:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ram, S., D. P. McQuillen, S. Gulati, C. Elkins, M. K. Pangburn, and P. A. Rice. 1998. Binding of complement factor H to loop 5 of porin protein 1A: a molecular mechanism of serum resistance of nonsialylated Neisseria gonorrhoeae. J. Exp. Med. 188:671-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren, B., A. J. Szalai, O. Thomas, S. K. Hollingshead, and D. E. Briles. 2003. Both family 1 and family 2 PspA proteins can inhibit complement deposition and confer virulence to a capsular serotype 3 strain of Streptococcus pneumoniae. Infect. Immun. 71:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rohde, M., E. Muller, G. S. Chhatwal, and S. R. Talay. 2003. Host cell caveolae act as an entry-port for group A streptococci. Cell. Microbiol. 5:323-342. [DOI] [PubMed] [Google Scholar]
- 51.Russel, D. G. 2000. Where to stay inside the cell: a homesteader's guide to intracellular parasitism, p. 131-152. In P. Cossart et al. (ed.), Cellular microbiology. American Society for Microbiology, Washington, D.C.
- 52.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 53.Sela, S., R. Neeman, N. Keller, and A. Barzilai. 2000. Relationship between asymptomatic carriage of Streptococcus pyogenes and the ability of the strains to adhere to and be internalised by cultured epithelial cells. J. Med. Microbiol. 49:499-502. [DOI] [PubMed] [Google Scholar]
- 54.Sharma, A. K., and M. K. Pangburn. 1997. Localization by site-directed mutagenesis of the site in human complement factor H that binds to Streptococcus pyogenes M protein. Infect. Immun. 65:484-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terao, Y., S. Kawabata, E. Kunitomo, J. Murakami, I. Nakagawa, and S. Hamada. 2001. Fba, a novel fibronectin-binding protein from Streptococcus pyogenes, promotes bacterial entry into epithelial cells, and the fba gene is positively transcribed under the Mga regulator. Mol. Microbiol. 42:75-86. [DOI] [PubMed] [Google Scholar]
- 56.Zipfel, P. F. 2001. Complement factor H: physiology and pathophysiology. Semin. Thromb. Hemost. 27:191-199. [DOI] [PubMed] [Google Scholar]
- 57.Zipfel, P. F., and C. Skerka. 1999. FHL-1/reconectin: a human complement and immune regulator with cell-adhesive activity. Immunol. Today 20:135-140. [DOI] [PubMed] [Google Scholar]