Abstract
A number of bacterial small RNAs (sRNAs) act as global regulators of stress responses by controlling expression of multiple genes. The sRNA SgrS is expressed in response to glucose–phosphate stress, a condition associated with disruption of glycolytic flux and accumulation of sugar–phosphates. SgrS has been shown to stimulate degradation of the ptsG mRNA, encoding the major glucose transporter. This study demonstrates that SgrS regulates the genes encoding the mannose and secondary glucose transporter, manXYZ. Analysis of manXYZ mRNA stability and translation in the presence and absence of SgrS indicate that manXYZ is regulated by SgrS under stress conditions and when SgrS is ectopically expressed. In vitro footprinting and in vivo mutational analyses showed that SgrS base pairs with manXYZ within the manX coding sequence to prevent manX translation. Regulation of manX did not require the RNase E degradosome complex, suggesting that the primary mechanism of regulation is translational. An Escherichia coli ptsG mutant strain that is manXYZ+ experiences stress when exposed to the glucose analogs α-methyl glucoside or 2-deoxyglucose. A ptsG manXYZ double mutant is resistant to the stress, indicating that PTS transporters encoded by both SgrS targets are involved in taking up substrates that cause stress.
INTRODUCTION
Trans-acting bacterial small RNAs (sRNAs) are encoded at chromosomal sites distal to those of their target mRNAs. These sRNAs act by base pairing with mRNAs and either positively or negatively affect translation or mRNA stability (1). For enterobacterial sRNAs, pairing requires the RNA chaperone, Hfq, which is thought to stimulate base pairing interactions by remodeling RNA structures and by increasing local concentrations of the sRNA and mRNA (2). The mechanism of negative regulation most commonly described involves base pairing between an sRNA and the 5′-untranslated region (UTR) of a target mRNA to block ribosome binding and inhibit translation (1). In numerous cases, the endoribonuclease RNase E and its associated proteins (collectively known as the degradosome) act on the sRNA–mRNA complex subsequent to translational repression. This results in coupled degradation of the sRNA and mRNA, rendering the regulation irreversible (1). There are other examples of sRNA-mediated negative regulation that do not adhere to the aforementioned model. The GcvB sRNA binds upstream of the ribosome binding sites of some target mRNAs and inhibits translation by blocking translational enhancer sites (3). The MicC sRNA binds within the coding region of one of its targets, ompD mRNA (4). The pairing occurs too far downstream to inhibit ompD translation initiation; instead, RNase E-dependent degradation is required for MicC-mediated repression of ompD (4). sRNA-mediated regulation of polycistronic transcripts can result in discoordinate regulation of genes in an operon. galK in the galETKM operon and iscS in the iscRSUA operon are negatively regulated by the sRNAs Spot 42 and RyhB, respectively (5,6). Both sRNAs bind to the ribosome binding sites of their target mRNAs to inhibit translation. Spot 42 inhibition of galK allows for translation of the upstream genes, galET (6) while RyhB inhibition of iscS causes degradation of the iscSUA portion of the transcript but generates a stable iscR transcript available for translation (5). Recently, coordinate regulation of the genes in an operon by the sRNA RsaE in Staphylococcus aureus was suggested by the identification of RsaE interactions with two different cistrons on a polycistronic transcript (7); no such example has been found for Hfq-dependent sRNAs. Since novel mechanisms of bacterial sRNA-mediated regulation are still being found, it is important to continue characterizing interactions of individual sRNA–mRNA pairs.
The transcription of many bacterial sRNAs is activated under specific stress conditions and their activities aid cells in recovery from those stresses (1). Transcription of the sRNA SgrS is activated under glucose–phosphate stress conditions in Escherichia coli (8,9). These conditions arise if glycolysis is blocked, e.g. by a mutation in the pgi gene (so that glucose-6-phosphate accumulates) or if cells are exposed to a non-metabolizable glucose analog α-methylglucoside (αMG) (so that αMG-6-phosphate accumulates) (9,10). Growth of wild-type E. coli cells is transiently inhibited by this stress, while sgrS mutant cells are unable to continue growing under stress conditions (9). SgrS seems to promote growth recovery by two independent mechanisms. The first is a base pairing-dependent mechanism whereby SgrS pairs in an Hfq-dependent manner with sequences overlapping the ribosome binding site of ptsG mRNA. ptsG encodes the EIICBGlc component of the Phosphoenolpyruvate Phosphotransferase System (PTS) glucose transporter in E. coli. Pairing between SgrS and ptsG mRNA prevents ptsG translation, thereby stopping synthesis of new glucose transporters (9,11). As part of this regulation, the SgrS–ptsG mRNA complex is degraded by the RNase E degradosome complex (12,13). In addition to the base pairing activity, SgrS encodes the small protein, SgrT. When SgrT is produced, it prevents glucose (or αMG) uptake by a mechanism independent of the base pairing activity (14). Likewise, the base pairing function of SgrS does not depend on production of the SgrT protein (14,15). The combination of these two SgrS functions was proposed to allow cells to stop sugar–phosphate accumulation and overcome growth inhibition. However, we recently determined that in stressed E. coli K12 cells, little SgrT is produced and the base pairing function appears to be primarily responsible for stress recovery (15).
Since several other characterized bacterial sRNAs target multiple mRNAs and the base pairing function of E. coli SgrS is required for rescue from glucose–phosphate stress, a preliminary microarray experiment was conducted to identify other putative SgrS targets. For this experiment, plasmid-borne SgrS was induced from a heterologous promoter for 5 min in an sgrS mutant host growing in rich medium. The transcriptomes of vector control and SgrS-expressing cells were compared and genes that were up- or downregulated were identified (our unpublished data). As expected, ptsG mRNA levels were decreased upon SgrS induction, as were levels of another mRNA encoding a PTS transporter, manXYZ. ManXYZ is a broad-substrate range sugar transporter that has been shown to transport mannose, glucose, 2-deoxyglucose, αMG, fructose, glucosamine, N-acetylglucosamine and trehelose (16–18). The manX gene encodes the EIIABMan cytoplasmic component of the transporter while manY and manZ encode the membrane components EIICMan and EIIDMan, respectively (18). The three genes are operonic and transcriptionally controlled in a manner similar to ptsG (18–20).
In this study, we show that the manXYZ polycistronic mRNA is negatively regulated by SgrS post-transcriptionally through a base pairing mechanism involving sequences downstream of the manX ribosome binding site. Translation of manX is inhibited and the entire manXYZ mRNA is degraded in an SgrS-dependent manner under stress conditions. However, SgrS-mediated repression of manX translation does not require RNase E, which suggests a ribosome occlusion mechanism of translational inhibition. The physiological relevance of SgrS-mediated repression of manXYZ was demonstrated by the finding that EIIMan transports sugar analogs that induce the SgrS stress response.
MATERIALS AND METHODS
Strain and plasmid construction
The strains and plasmids used in this study are listed in Supplementary Table S1 and oligonucleotides are listed in Supplementary Table S2. Alleles were moved between strains by P1 transduction or inserted via lambda Red recombination (21). Translational LacZ fusions were constructed as described previously (22). Strain XL10 Gold (Stratagene) was used for the QuikChange Mutagenesis procedure. LacIq (harbored in several strains, including JH111, Supplementary Table S1) was used to control expression from the Plac promoter.
The manX′–′lacZ translational fusion was created as described previously (22) using primers O-JH129/O-JH130 to fuse lacZ to the 34th codon of manX. The fusion was transduced into various backgrounds (see Supplementary Table S1) to create JH114, JH115, JH116, JH149, JH236 and JH255.
kanR-Cp19-manXYZ was created by PCR-amplifying the kanR-linked Cp19 promoter from JNB024 (N. Majdalani, J. Benhammou and S. Gottesman, unpublished results) chromosomal DNA using oligos containing manX homology (O-JH104/O-JH137). The resulting PCR product was recombined into NM200. kanR-Cp19–manXYZ from this strain was transduced into wild-type (DJ480) and ΔsgrS (CS104) backgrounds to yield strains JH124 and JH125.
The kanR-Cp19-manX′–′lacZ fusion was created as follows: kanR-Cp19–manXYZ was transduced into NM200 to create JH131. pCP20 was used to flip out the kanR cassette, resulting in strain JH136. Then the manX′–′lacZ fusion was created in the JH136 strain as described above.
Strains containing manX 5′ UTR mutations were created as follows: the manX′–′lacZ fusion was transduced into NM200 to create JH149. A tetR-Cp19 PCR product generated using oligonucleotides O-JH161/O-JH175 was used as a PCR template to generate tetR-Cp19–manX (O-JH173/O-JH163) and tetR-Cp19–manX30 (O-JH174/O-JH163) products. These PCR products were recombined into JH149 to create JH175 and JH181, respectively.
A ptsG′–′lacZ translational fusion (in strains JH169, JH172 and JH238) was created so that lacZ was fused in frame to ptsG codon 141 using methods described previously (22). Oligonucleotides O-JH153/O-JH154 were used to obtain the PCR product. The PtsG portion of the PtsG-LacZ fusion protein contains four transmembrane domains; LacZ remains in the cytoplasm. The tetR-Cp19–ptsG′–′lacZ fusion was created by recombining a tetR-Cp19 cassette (amplified using oligonucleotides O-JH170/O-JH154) upstream of the ptsG 5′ UTR.
Chromosomal mutations in manX were made using the following method: a tetR cassette (generated with oligonucleotides O-JH194/O-JH195) was inserted into manX so that nucleotides to be targeted for mutation were deleted. The resulting strain was named JH197. A PCR product (obtained using oligonucleotides O-JH199/O-JH130 or O-JH211/O-JH130 and strain JH113 as a template) containing the C143T or CAC to GUG manX mutations and a kanR cassette at the desired lacZ fusion junction was recombined into JH197 creating JH208 and JH227, respectively. The manX′–′lacZ translational fusion was then constructed as described previously by flipping out the kanR cassette and replacing with ′lacZ (22) to create JH212 and JH228. Another tetR cassette was inserted into manX at a different site using a PCR product generated by primers O-JH220/O-JH195; the resulting strain was JH239. To make the strain JH240 containing the manX GTG to ATG mutation, a PCR product containing the mutation linked to a kanR cassette (amplified using O-JH221/O-JH130) was recombined into JH239 and colonies were screened for tet sensitivity. The manX′–′lacZ translational fusion was constructed as described previously by flipping out the kanR cassette and replacing with ′lacZ (22) to create JH241.
manXYp′–′lacZ and manXEcar′–′lacZ fusions were created by recombining a PCR product containing upstream and downstream homology to E. coli manX flanking the Yersinia pestis or Erwinia carotovora manX 5′ UTR and a kanR marker into JH197. The kanR marker was replaced by lacZ as described previously (22) to create JH232 (manXYp′–′lacZ) and JH264 (manXEcar′–′lacZ). The PCR products used to create these strains were derived by stitching two PCR products: (i) a product with homology upstream of E. coli manX and the Y. pestis or E. carotovora manX sequence and (ii) a product with homology to the Y. pestis or E. carotovora manX sequence linked to a kanR marker. For Y. pestis manX, O-JH212/O-JH213 were used to create product 1. O-JH214/O-JH130 were used to create product 2. The same procedure was used to create the E. carotovora manX′–′lacZ fusion. The primers used were O-JH215/O-JH216, O-JH217/O-JH130 and O-JH215/O-JH130.
PsgrS–lacZ was constructed by B. Hussain in our laboratory using the previously described ‘Materials and Methods’ section (9). PsgrS was amplified using primers O-BAH102 and O-CV142 containing EcoRI and BamHI sites. This product was cloned into pRS1553. The resulting fusion was recombined into the λatt site and the resulting strain was named BAH100. The original strain carrying ΔptsG::cat was kindly provided by H. Aiba. ΔptsG::cat was transduced into PsgrS–lacZ (BAH100) to yield strain CS177. The original strain carrying ΔmanXYZ::kanR was obtained from Amos Oppenheimer. ΔmanXYZ::kanR was transduced into PsgrS–lacZ (BAH100) and ΔptsG::cat (CS177) backgrounds to yield strains CS178 and CS179.
Plasmid pBRJH19, containing the GUG to CAC mutation in SgrS, was created using the QuikChange Mutagenesis kit (Stratagene) with primers O-JH209/O-JH210 and Plac-sgrS (pLCV1) plasmid as template. Plasmid pBRJH23 was created using the QuikChange Mutagenesis kit with primer O-JH252 and Plac-sgrS10 (pBRJH19) as the template. Plasmid pBRJH26, containing the G168A mutation, was created using the QuikChange Mutagenesis kit with primer O-JH260 and the pLCV1 plasmid as a template.
Media and reagents
Bacteria were cultured in LB medium or on LB agar plates at 37°C unless otherwise noted. TB medium (Bacto Tryptone, BD) was used for β-galactosidase assays. Media were supplemented with 100 μg/ml ampicillin where indicated. IPTG was used at a concentration of 0.1 mM for induction of Plac-sgrS. MOPS (morpholinepropanesulfonic acid) rich defined medium (Teknova) with 0.2% mannose as a carbon source was used for culturing cells for RNA extraction for 5′ RACE. MacConkey indicator plates supplemented with 1% lactose were used to measure PsgrS–lacZ activity.
β-galactosidase assays
Strains containing translational fusions were grown overnight in TB medium and subcultured 1:200 to fresh medium. Cultures were grown to an OD600 ∼0.1 at which time 0.5% αMG or 0.1 mM IPTG was added to cells. Samples were taken at times indicated and assayed for β-galactosidase activity as described previously (23). Activities (in Miller Units) were normalized to ΔsgrS or empty vector control to give the percentage relative activity for experimental samples.
RNA extraction and northern blot analysis
Strains were grown in LB medium to OD600 ∼0.5 and exposed to 0.5% αMG. RNA was extracted at times indicated by the hot phenol method as described previously (24). The RNA concentration was determined spectrophotometrically. Fifteen microgram of total RNA was used for electrophoresis and run on a 1.2% agarose gel alongside the Millenium Marker (Ambion) at 90 V for ∼1.5 h. RNA was transferred to a membrane as described previously (25). Prehybridization was performed in ULTRAhyb (Ambion) solution at 42°C for at least 45 min. Blots were subsequently probed overnight with a 5′-biotinylated probe, O-JH108 for manY or SsrA-bio for ssrA. Detection was performed according to BrightStar BioDetect kit (Ambion) specifications.
5′ RACE
5′ RACE was performed as described previously (26) using the manY-specific primer O-JH139 with RNA isolated from DJ480 cells grown to an OD600 ∼0.5 in minimal MOPS + 0.2% Mannose.
SgrS random mutagenesis
The sgrS region of Plac-sgrS (pLCV1) was mutated using the GeneMorph EZclone II kit (Stratagene) and the resulting mutated plasmid library transformed into a manX′–′lacZ, ΔsgrS, lacIq+ strain (JH116). Mutant plasmids that were unable to regulate manX′–′lacZ activity were isolated and transformed into a ptsG′–′lacZ, ΔsgrS, lacIq+ strain (JH171). Plasmids that were able to negatively regulate ptsG′–′lacZ activity were sequenced.
In vitro RNA footprinting
In vitro transcription templates were generated by PCR using gene-specific oligonucleotides containing the T7 promoter sequence. Oligos O-JH218/O-JH169 were used to generate a manX template (from −115 to +102 relative to the translational start site), oligos O-JH236/O-JH241 were used to generate a ptsG template (from −103 to +104 relative to the translational start site) and oligos O-JH219/O-JH119 were used to generate an sgrS template. In vitro transcription was performed with the PCR templates using the MEGAscript T7 Kit (Ambion). The resulting RNA was 5′-end labeled using the KinaseMax Kit (Ambion). Footprinting reactions were performed as described previously (5) with the following changes. Ten picomol of unlabeled SgrS, manX mRNA or ptsG mRNA was incubated with 0.1 pmol of 5′-end labeled manX mRNA, ptsG mRNA or SgrS at 37°C for 15 min in 1× Structure Buffer (Ambion) in the presence or absence of 300 nM Hfq.
RNA Structure Analysis
RNA structure was analyzed using the RNA folding program Mfold (27). Structures with the lowest ΔG that were most consistent with experimental data from footprinting experiments (if available) are shown.
RESULTS
manXYZ is post-transcriptionally regulated by SgrS
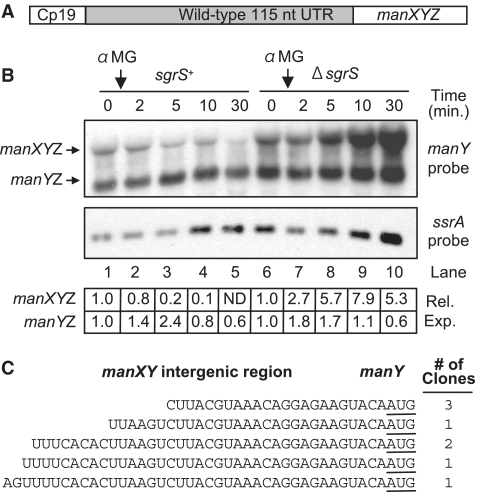
Preliminary microarray results suggested that manXYZ mRNA is negatively regulated by SgrS (our unpublished data). In order to investigate whether regulation of the polycistronic manXYZ mRNA by SgrS was direct, a translational manX′–′lacZ fusion was created at the manX locus. A synthetic constitutive promoter, Cp19 (28), was used to drive transcription of the manX′–′lacZ fusion in order eliminate indirect effects due to transcriptional regulation (Figure 1A). β-Galactosidase activities of wild-type and ΔsgrS mutant cells harboring the fusions were monitored after stress was induced by addition of αMG to cultures. Wild-type cells had ∼65% lower manX′–′lacZ fusion activity than ΔsgrS cells (Figure 1B). These data indicate that manX is post-transcriptionally regulated in an SgrS-dependent manner under glucose–phosphate stress conditions.
Figure 1.
manX translation is repressed in an SgrS-dependent manner. (A) Chromosomal lacZ translational fusions were constructed at the native loci. The native promoters of ptsG and manX were replaced with the constitutive Cp19 promoter (28). (B) Cp19-manX′–′lacZ strains in ΔsgrS (JH143) or sgrS+ (wild-type, JH138) backgrounds were grown to early log phase and 0.5% αMG was added. Samples were harvested 60 min after αMG addition and assayed for β-galactosidase activity. Specific activities in all samples were normalized to the levels in the ΔsgrS strain to yield percentage relative activity (reported in the graph). Specific activity values in Miller units are reported below the graph. (C) Strains with Cp19-manX′–′lacZ (JH193) or Cp19-ptsG′–′lacZ (JH184) carrying an empty vector or Plac-sgrS were grown to early log phase and 0.1 mM IPTG was added. Samples were harvested 60 min after IPTG addition and assayed for β-galactosidase activity. Specific activities were normalized and data reported as in part (B).
To determine whether SgrS could regulate manX in the absence of other stress-inducible factors, a Plac-sgrS plasmid and an empty vector control were transformed into an sgrS mutant host containing the same reporter constructs shown in Figure 1A. Regulation of manX translation in cells containing Plac-sgrS was similar to that in cells where chromosomal sgrS expression was induced by αMG. Expression of the fusion was reduced upon sgrS induction by 75% (Figure 1C). These results suggest that SgrS is sufficient to regulate manX translation in the absence of other stress-inducible factors. In order to compare SgrS regulation of manX and ptsG, a Cp19-ptsG′–′lacZ fusion was also analyzed for regulation by Plac-sgrS. The ptsG′–′lacZ fusion activity responded similarly to the manX′–′lacZ fusion; upon sgrS induction, it was downregulated by ∼75% (Figure 1C). Since ptsG mRNA is an established target of SgrS (9,11), this finding lends further weight to the hypothesis that SgrS directly regulates manXYZ. Because manX′–′lacZ is regulated by SgrS in the absence of the downstream manY and manZ sequences, we hypothesized that base pairing between SgrS and the manXYZ transcript involves the manX translation initiation region.
A manX′–′lacZ fusion under control of the native manX promoter was also tested for regulation by SgrS induced under stress conditions or from a plasmid. In all cases, the results were analogous to those for the Cp19-controlled fusions (data not shown).
SgrS stimulates degradation of manXYZ mRNA
To assess whether SgrS-dependent regulation of manX translation affected manXYZ mRNA stability, northern blots were performed on RNA samples from wild-type and ΔsgrS strains carrying Cp19-manXYZ constructs (Figure 2A). A manX-specific probe hybridized to a ∼2.8 kb band corresponding to the full-length manXYZ mRNA (data not shown). A manY-specific probe hybridized to the ∼2.8 kb band as well as an additional ∼1.8 kb band (Figure 2B). When RNA from a ΔmanXYZ strain was probed with the manY-specific probe, neither of these bands was present (data not shown). The 1.8 kb band is the size predicted for a manYZ transcript. 5′ RACE analysis (26) was utilized to distinguish whether manYZ was a newly initiated transcript or a processed product. This analysis revealed that the manYZ mRNA contained a monophosphate end, indicative of a processed transcript (Supplementary Figure S1). Furthermore, several different 5′-ends of the manYZ species mapped to a region between manX and manY that was AU-rich (Figure 2C). These sequences resemble RNase E recognition and cleavage sites (29–31). A transcriptional manY′–lacZ fusion confirmed that there is no promoter activity associated with the manX–manY intergenic region (data not shown). All together, these data suggested that manYZ is processed from manXYZ.
Figure 2.
manXYZ mRNA is destabilized in an SgrS-dependent manner and manYZ mRNA is processed in an SgrS-independent manner. (A) The Cp19 constitutive promoter replaced the manXYZ promoter at the native locus. (B) Cp19-manXYZ in wild-type sgrS+ (JH124) or ΔsgrS (JH125) backgrounds were grown to mid-log phase and 0.5% αMG was added to the cultures. RNA was extracted at times indicated and analyzed by northern Blot using manY- or ssrA-specific probes. SsrA served as the loading control. ImageJ was used to quantify band intensities and correct for uneven loading. Corrected intensities for each band were then normalized to time 0 (set at 1.0). Expression relative to time zero is reported below the blot (‘Rel. Exp.’). (C) 5′ RACE analysis of manY was performed on RNA extracted from wild-type (DJ480) cells grown to mid-log phase. The sequences of manY 5′-ends and the frequency of their occurrence in sequenced clones (out of eight total) are shown. The manY start codon is underlined.
We proceeded by examining SgrS regulation of both manXYZ and manYZ mRNA levels. In wild-type cells, a decrease in the levels of the manXYZ transcript was apparent following αMG addition, while levels of the manYZ mRNA remained relatively steady (Figure 2B, lanes 1–5). In contrast, cells lacking sgrS displayed greatly increased levels of manXYZ transcript and relatively constant levels of manYZ transcript after αMG addition (Figure 2B, lanes 6–10). Because the transcription of manXYZ in this experiment is driven by the constitutive Cp19 promoter (28), we hypothesize that changes in steady-state levels of manXYZ mRNA reflect changes in stability rather than rates of synthesis. Therefore, these results suggest that SgrS is responsible for decreasing the stability of the full-length manXYZ transcript but has no significant effect on the stability of the manYZ transcript.
To test whether the apparent accumulation of manXYZ mRNA in sgrS mutant cells (Figure 2B, lanes 6–10) reflected a manXYZ-specific post-transcriptional mechanism of regulation, we examined levels of the ptsG mRNA expressed from the Cp19 promoter in sgrS+ and sgrS mutant cells exposed to αMG. This experiment showed that ptsG mRNA does not accumulate in the sgrS mutant strain after αMG addition (data not shown), in contrast to the manXYZ mRNA. We also examined manXYZ and manYZ mRNAs (expressed from the Cp19 promoter) in ΔsgrS cells in the absence of αMG and found no accumulation over time in non-stressed cells (data not shown). Together, these data suggest that the accumulation of manXYZ mRNA in sgrS mutant cells reflects a post-transcriptional mechanism of regulation that is stress dependent.
Biochemical demonstration of SgrS base pairing with target mRNAs
SgrS regulates the ptsG mRNA by a base pairing mechanism involving the region surrounding the ptsG ribosome binding site. In order to narrow down the region of manX required for regulation by SgrS, the 115 nt 5′ UTR of the manX′–′lacZ fusion was truncated by moving the Cp19 promoter closer to the manX translational start site. A fusion containing 30 nt upstream of the manX start codon was constructed (denoted as manX30′–′lacZ) (Supplementary Figure S2A). Activity of the manX30′–′lacZ fusion was monitored in the presence or absence of ectopically expressed sgrS. Cells showed a pattern of regulation by SgrS that was similar to the regulation of the full-length Cp19-manX′–′lacZ fusion, i.e. a 65% decrease in β-galactosidase activity when SgrS was present (Supplementary Figure S2B). Truncating the 5′-UTR further (to 20 nt) resulted in an unstable manXYZ transcript regardless of the presence or absence of SgrS (data not shown). Therefore it was concluded that at most, 30 nt of the manX 5′-UTR and 102 nt of manX coding sequence are needed for regulation by SgrS.
Given that the region contained in the manX30′–′lacZ fusion is sufficient for regulation by SgrS, potential base pairing interactions between SgrS and the manX sequences contained in this region were examined. A region of complementarity between the 3′ region of SgrS and the coding region of manX was identified (Figure 3A). To test the base pairing prediction, footprinting analyses were performed with in vitro-transcribed SgrS and manX RNAs. Experiments using labeled manX mRNA revealed SgrS-dependent protection of manX bases from C139 to G147 (Figure 3C). This region corresponds to the longest contiguous stretch of predicted base pairing located from 23 to 33 nt downstream of the manX translational start (Figure 3A). The reverse analysis was done with labeled SgrS to determine the region that interacts with manX mRNA. The region of SgrS predicted to base pair with manX (from C159 to A180, Figure 3A) was mostly single stranded in the absence of target mRNA (Figure 3C), while a footprint spanning SgrS bases from A163 to G172 was observed in the presence of manX mRNA (Figure 3C). This result was consistent with interactions in the longest contiguous stretch of base pairing indicated in Figure 3A.
Figure 3.
SgrS base pairing with mRNA targets. (A) Predicted base pairing between manX mRNA and SgrS. Pairing confirmed by footprinting and genetic analyses is indicated by vertical lines and pairing predicted but not experimentally supported is indicated by asterisks. The manX RBS is denoted by a line above the sequence; the manX start codon is underlined; half of the inverted repeat of the SgrS terminator is denoted by the long arrow below the sequence. The boxed bases were mutated to test base pairing. Arrows and allele names (SgrS1, SgrS26 and SgrS10) indicate positions of mutations in different SgrS mutants (also see Supplementary Table S1). (B) Base pairing between SgrS and ptsG mRNA. See part (A) for further description. (C) In vitro transcribed manX mRNA containing the entire manX sequence present in the manX′–′lacZ fusion was end-labeled with 32P (manX*) and incubated with unlabeled SgrS where noted and treated with: T1, RNase T1; OH, alkaline hydrolysis; PbAc, lead acetate. The positions of G residues are shown. In vitro transcribed SgrS was labeled with 32P (SgrS*) and incubated with unlabeled manX where noted. (D) Interactions between SgrS and ptsG mRNA were mapped as described in part (C). Abbreviations are as described in part (C). (E) manX′–′lacZ (JH116), manXC143T′–′lacZ (JH216) and ptsG′–′lacZ (JH171) strains carrying an empty vector, Plac-sgrS, Plac-sgrS1 or Plac-sgrS26 were analyzed for β-galactosidase activity as described in Figure 1C.
The sequences in SgrS that base pair with manX in footprinting experiments partially overlap with sequences predicted to pair with ptsG mRNA (Figure 3A and B). Several studies (11,15) have genetically demonstrated the importance of interactions between SgrS and ptsG mRNA that encompass residues from C174 to U181 of SgrS and from A82 to G89 of ptsG mRNA (Figure 3B). In particular, mutations in residues G176 and G178 of SgrS effectively destroy regulation of ptsG mRNA, while compensatory mutations in ptsG restore regulation (11). Additional complementarity on either side of this ‘core’ SgrS-ptsG mRNA interaction is predicted, but interactions have not been verified either genetically or biochemically. Therefore, footprinting experiments were carried out to map SgrS binding sites on ptsG mRNA and vice versa. These experiments showed a clear SgrS footprint on ptsG mRNA spanning residues from A82 to G89 (Figure 3D), which is consistent with the core interaction region demonstrated genetically. There was also weak protection of ptsG mRNA residues from G76 to A78 (upstream of the core region) and from C92 to C95 (downstream of the core region). A ptsG mRNA footprint on SgrS showed protection of SgrS bases from G168 to U181 (Figure 3D), consistent with interactions in the core region and downstream (with respect to the ptsG mRNA). Together, the footprinting experiments demonstrated two important things: (i) different residues of SgrS are involved in specific base pairing interactions with different targets and (ii) base pairing involves different regions of different targets, i.e. overlapping the ribosome binding site of ptsG and within the coding sequence of manX.
Genetic demonstration of SGRS Base pairing with manX mRNA
As a second confirmation of SgrS–manX mRNA interactions, a genetic approach utilizing random mutagenesis of SgrS was performed. A screen was conducted to identify SgrS mutants that failed to regulate manX′–′lacZ but retained regulation of ptsG′–′lacZ. These criteria were used in order to avoid isolating mutations that had non-specific deleterious effects on SgrS structure and function. Numerous mutations were found that impaired regulation of both fusions, but mutations in only two residues, T169 and G170, had a specific phenotype for regulation of manX′–′lacZ (data not shown); these residues are encompassed within the region protected in footprinting experiments (Figure 3).
To further genetically test the base pairing interactions between SgrS and manX mRNA, wild-type and mutant alleles of SgrS were tested for regulation of wild-type and compensatory mutant manX′–′lacZ fusions as well as a wild-type ptsG′–′lacZ fusion (Figure 3E). Mutant SgrS alleles contained either G176C, G178C (sgrS1), which should interfere with regulation of ptsG (15) or G168A (sgrS26), which should interfere with regulation of manX. The predicted structures of both SgrS mutants were identical to that of wild-type SgrS [according to Mfold (27), data not shown]. Wild-type SgrS and SgrS1 (G176C, G178C) regulated manX′–′lacZ to the same extent (∼80% repression), while SgrS26 (G168A) was defective in regulating manX′–′lacZ (∼35% repression) (Figure 3E, left panel). The results for regulation of wild-type ptsG′–′lacZ by each of these mutants were the opposite; wild-type SgrS and SgrS26 regulated ptsG′–′lacZ to the same extent (∼80% repression), whereas SgrS1 completely lost the ability to repress (Figure 3E, right panel). These results strongly suggest that each of these mutants retains the appropriate structure and that loss of regulation reflects disruption of target-specific base pairing interactions. To further confirm direct base pairing between SgrS and manX mRNA, a compensatory mutation in manX (C143T), which should restore pairing with SgrS26 and disrupt pairing with wild-type SgrS was constructed in the context of the reporter fusion (designated manXC143T′–′lacZ). Consistent with this prediction, wild-type SgrS was deficient in regulating manXC143T′–′lacZ (∼60% repression) whereas SgrS26 gave full 80% repression (Figure 3E, center panel).
It is worth noting that the footprinting experiments accurately report relevant in vivo base pairing interactions. Protection of SgrS residues 174–186 and manX mRNA residues 125–137 was not observed (Figure 3C), despite the complementarity in this region (Figure 3A) and the apparently single-stranded conformation in vitro (based on susceptibility to cleavage by lead acetate, Figure 3C). Consistent with this, mutations in SgrS (in the sgrS1 allele) that would disrupt the base pairing of this region (complementarity denoted by asterisks, Figure 3A) had no effect on the regulation in vivo. Similarly, for SgrS:ptsG mRNA interactions, the most strongly protected residues in vitro (SgrS residues 174–181 and ptsG mRNA residues 82–89) were the most relevant for in vivo regulation (Figure 3B, D and E).
The structure of SgrS alone (Fig. 3C, D, left panels) corresponded very well to the Mfold (27) prediction (Figure 4B). The SgrS bases that participate in pairing with manX mRNA are mostly single stranded, with the exception of three residues that participate in formation of a short stem (residues 163–165, Figures 3C and 4B). Residues 159 and 160 are located in a small bubble in SgrS alone; in the presence of manX mRNA residues 159–160 and 163–172 are protected and residues 161 and 162 become hypersensitive (Figure 3C). In the course of testing certain SgrS and manX mutant pairs, we observed that mutation of some SgrS residues resulted in significant predicted structural changes in SgrS in the region that interacts with manX mRNA. For example, mutation of SgrS residues 168–170 from GUG to CAC (in allele sgrS10, Figure 3A, Supplementary Table S1) not only disrupted complementarity with wild-type manX, but also resulted in sequestration of the majority of the manX mRNA pairing region of SgrS in a stem (compare the wild-type SgrS structure, Figure 4B, to the mutant structure, Figure 4C). This sequestration could be partially alleviated by making an additional mutation in SgrS (A5U, allele sgrS23, Fig. 4D). To determine how these changes would affect regulation of manX, we assayed the activity of these mutants in wild-type manX′–′lacZ and mutant manXGUG′–′lacZ strains (the latter fusion contains mutations that would restore complementarity with the SgrS mutant alleles). As predicted, wild-type SgrS repressed wild-type manX′–′lacZ and both mutant alleles (SgrS10 and SgrS23) were strongly impaired for regulation (Figure 4A, left panel). In the manXGUG′–′lacZ background, wild-type SgrS lost the ability to regulate, again consistent with the involvement of these residues in SgrS:manX mRNA base pairing interactions (Figure 4B, right panel). SgrS10 regulated manXGUG′–′lacZ only slightly better than wild-type SgrS, despite having the compensatory changes that should have restored base pairing. This was consistent with the predicted sequestration of most of the base pairing region (Figure 4C). The additional mutation in SgrS23 gave a slight improvement in the regulation of manXGUG′–′lacZ compared to the wild-type and SgrS10 alleles, but full repression was not restored, likely because the structure of SgrS23 is still altered compared to wild-type SgrS (compare Figure 4B and D). Northern blots showed that SgrS10 and SgrS23 were produced at levels similar to wild-type SgrS and both of these mutants retained regulation of ptsG′–′lacZ (data not shown), which was not surprising since the residues in SgrS important for interaction with ptsG mRNA remained unpaired in the mutant structures (see residues from 174 to 181 in Figure 4C and D). Failure of compensatory mutations to fully restore wild-type levels of regulation has been observed for other sRNA:mRNA pairs (5,32,33) and has been interpreted to mean that both sequence and structure of the two RNAs are important for their interactions and that the structures of one or both molecules are altered by certain mutations. These results are certainly consistent with that explanation.
Figure 4.
Structural changes in SgrS mutants. (A) manX′–′lacZ (JH116) and manXGUG′–′lacZ (JH229) strains carrying an empty vector, Plac-sgrS, Plac-sgrS10 or Plac-sgrS23 were analyzed for β-galactosidase activity as described in Figure 1C. (B) Mfold prediction of wild-type SgrS structure, which is consistent with footprinting of SgrS alone (Figure 3C). (C) Mfold prediction of SgrS10 structure. Strong base pairing interactions are indicated by a red line. Mutated bases are circled in red. (D) Mfold prediction of SgrS23 structure. Strong base pairing interactions are indicated by a red line. Mutated bases are circled in red.
Degradation of manXYZ is not required for SgrS regulation of manX translation
Although SgrS promotes degradation of the ptsG message, this is not required for translational regulation (11). SgrS-mediated translational repression of ptsG is thought to occur through a ribosome occlusion mechanism, since residues of the ptsG Shine-Dalgarno sequence are involved in base pairing interactions with SgrS (9,11). Given that SgrS base pairs downstream of the manX ATG and we showed that the manXYZ mRNA is degraded when SgrS is expressed, we wanted to determine if SgrS-mediated translational repression of manX requires degradation of the manXYZ mRNA. For that reason, activities of manX and ptsG translational fusions were compared in wild-type and rne131 backgrounds in the presence or absence of sgrS. The E. coli rne131 allele encodes a C-terminally truncated RNase E that does not associate with Hfq or degradosome proteins. C-terminal RNase E mutants have been shown to be defective for degrading other sRNA–mRNA complexes, including SgrS-ptsG mRNA (34–36). In accordance with previous findings (36), the rne131 allele did not affect the ability of SgrS to repress translation of ptsG′–′lacZ (Figure 5A). Similar results were observed with manX′–′lacZ (Figure 5A) suggesting that like ptsG, manX regulation by SgrS does not require degradosome-mediated mRNA degradation.
Figure 5.
SgrS-dependent manX regulation does not require the degradosome. (A) ptsG′–′lacZ strains carrying an empty vector or Plac-sgrS were analyzed in a wild-type (JH258) or rne131 (JH248) background and compared to manX′–′lacZ strains carrying an empty vector or Plac-sgrS in wild-type (JH255) or rne131 (JH246) backgrounds. β-galactosidase assays were performed as described in Figure 1C. (B) manXYZ mRNA from sgrS+ (DJ480) or ΔsgrS (CV100) backgrounds were compared to manXYZ mRNA from rne131 strains containing wild-type sgrS (EM1377) or the sgrS deletion (JH273). RNA was harvested when cells reached mid-log phase (time 0) and at times indicated following addition of 0.5% alpha-MG. RNA was analyzed by Northern blotting as described in Figure 2B. ImageJ was used to quantify band intensities and correct for uneven loading. Corrected intensities for each band were then normalized to time zero (set at 1.0). Expression relative to time zero is reported below the blot.
To confirm that SgrS-mediated degradation of manXYZ mRNA was abolished in the rne131 background, Northern blot analysis was performed on sgrS+ and ΔsgrS cells in wild-type and rne131 strains. As expected, manXYZ levels decreased in sgrS+ cells upon αMG addition (Figure 5B, lanes 1–3) but accumulated in ΔsgrS cells (Figure 5B, lanes 4–6). In contrast, sgrS+ and ΔsgrS cells both had increased levels of manXYZ mRNA following αMG addition in the rne131 background (Figure 5B, lanes 7–12), confirming that degradation was abolished in these strains. Interestingly, the manYZ transcript was present at much lower levels in the rne131 strains (Figure 5B, lanes 7–12), suggesting that manYZ processing involves RNase E. Combined with the results from translational fusions in the rne131 background (Figure 5A), these data are consistent with the hypothesis that manX regulation by SgrS occurs primarily at the level of translational inhibition, rather than mRNA degradation.
The weak start codon of manX does not influence SgrS regulation
Work by Kawamoto et al. (36) suggested that co-translational membrane localization of ptsG mRNA is required for ptsG regulation by SgrS. The data support a model whereby ptsG mRNA membrane localization decreases translational efficiency of the ptsG message, thus allowing SgrS to compete effectively with ribosomes for binding to the translation initiation region of the ptsG mRNA. Since manX encodes the cytoplasmic component of the transporter, the manX′–′lacZ transcript would not be localized to the membrane, therefore this cannot be a contributing factor to regulation by SgrS. We did notice, however, that manX has a conserved GTG start codon (Figure 7B), which promotes decreased rates of translation initiation compared with ATG start codons (37). We therefore tested the hypothesis that the weaker start codon might be important for SgrS regulation of manX. A GTG to ATG mutation was made in the context of the manX′–′lacZ chromosomal fusion. Though the ATG mutation did increase basal levels of manX′–′lacZ translation as expected, it did not alter the fold-repression when SgrS was induced from the chromosome (Supplementary Figure S3A) or a plasmid (Supplementary Figure S3B), indicating that the weaker start codon is not required for regulation by SgrS.
Figure 7.
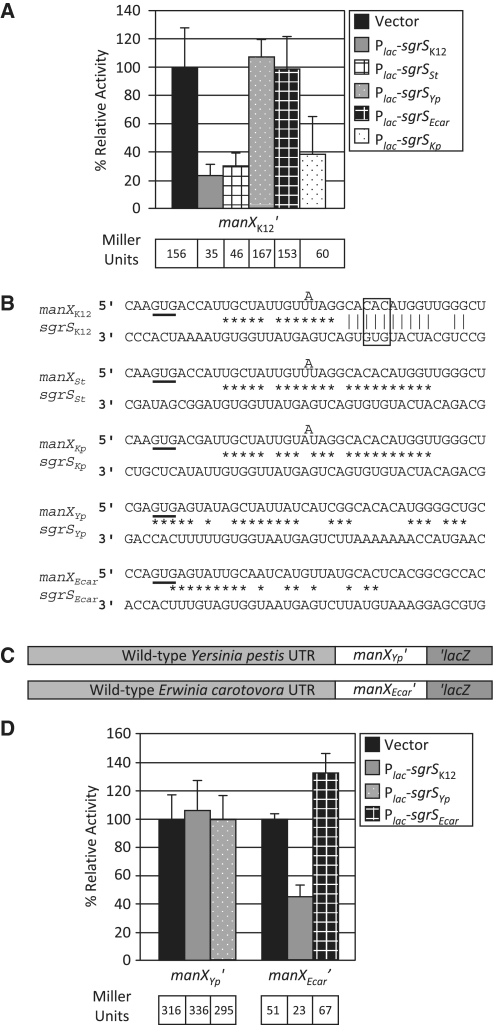
SgrS-dependent regulation of manX is not entirely conserved among enteric species. (A) The E. coli manX′–′lacZ strain (JH116) carrying different SgrS orthologs was analyzed for β-galactosidase activity as described in Figure 1C. Abbreviations: K12, Escherichia coli K12; St, Salmonella typhimurium; Yp, Yersinia pestis; Ecar, Erwinia carotovora; Kp, Klebsiella pneumoniae. (B) Alignments of SgrS orthologs and their cognate manX mRNAs highlights predicted differences in base pairing interactions. The boxed region represents bases of E. coli manX and SgrS analyzed by mutational analysis in Figure 3E. Start codons for manX are underlined. Experimentally verified base pairing interactions are indicated by vertical lines. Other potential base pairing interactions are indicated by asterisks. (C) manX sequences from Y. pestis or E. carotovora were swapped into the manXK12′–′lacZ locus as diagramed. (D) manXYp′–′lacZ (JH232) and manXEcar′–′lacZ (JH264) carrying an empty vector, Plac-sgrSK12 or the cognate Plac-sgrS ortholog were analyzed for β-galactosidase activity as described in Figure 1C.
ManXYZ proteins transport stress-inducing sugar analogs
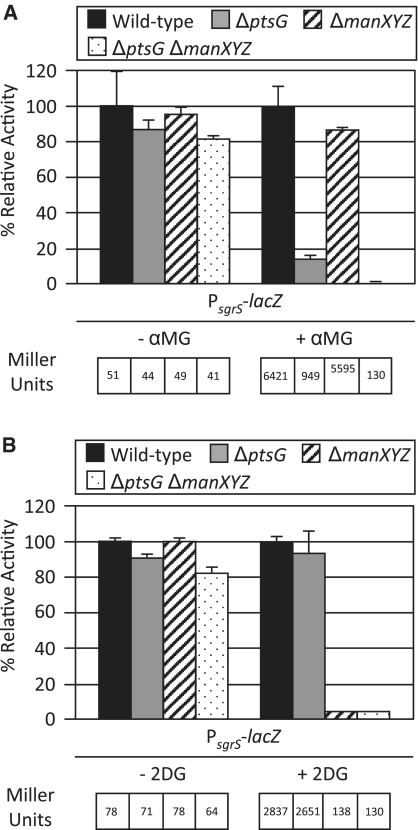
It is well established that EIICBGlc (PtsG) is involved in generating glucose–phosphate stress because it transports and phosphorylates sugars or sugar analogs like αMG that accumulate under certain conditions. SgrS relieves the stress by regulating ptsG mRNA, preventing synthesis of new EIICBGlc and therefore decreasing accumulation of sugar–phosphates (8–10,38). We hypothesized that manXYZ also contributes to glucose–phosphate stress since it transports glucose and αMG and is regulated by SgrS. Another glucose analog, 2-deoxyglucose (2DG) is a good substrate for the EIIMan system (ManXYZ) but only weakly transported by EIICBGlc (39–43); upon transport, 2DG becomes 2-deoxyglucose-6-phosphate (2DG6P). It was previously shown that high levels of 2DG6P cause growth inhibition in E. coli cells (44), however, we had not tested whether 2DG6P induces the SgrS-mediated stress response. To test the role of manXYZ in stress caused by these two glucose analogs, strains were constructed with mutations in ptsG and manXYZ individually and in combination. Induction of the stress response was monitored using a chromosomal PsgrS–lacZ reporter. The reporter fusion was not induced in the absence of sugar analogs (Figure 6A and B, left panels), as expected. When wild-type cells were exposed to 0.1% αMG, PsgrS–lacZ activity was induced by 126-fold over –αMG levels (see Miller Units, Figure 6A), indicating that the stress response was induced. The reporter was still induced slightly in the ΔptsG strain in the presence of αMG, albeit to a lower level (21.5-fold compared to uninduced), suggesting that cells take up sufficient αMG by an EIICBGlc-independent route to slightly induce the stress response. In the ΔptsG, ΔmanXYZ double mutant the reporter was not activated in the presence of αMG, indicating that in the absence of EIICBGlc, the EIIMan system is responsible for transporting the αMG that induces the stress response.
Figure 6.
Glucose–phosphate stress is induced by sugar analogs transported by EIICBGlc and EIIMan. Wild-type (BAH100), ΔptsG (CS177), ΔmanXYZ (CS178) or ΔptsG ΔmanXYZ (CS179) strains carrying the PsgrS–lacZ reporter were grown to early log phase at which time 0.1% αMG (A) or 0.1% 2DG (B) was added. Samples were harvested 120 min after αMG/2DG addition and assayed for β-galactosidase activity. Miller units from all samples were normalized to the wild-type levels to yield relative relative activity. Specific activity values (Miller units) are reported below each graph.
In the presence of 0.1% 2DG, the PsgrS–lacZ fusion was activated in wild-type cells by 36-fold compared to uninduced levels (see Miller Units, Figure 6B). This indicated that 2DG does induce the glucose–phosphate stress response. In the ΔptsG background, induction of the reporter was similar to the induction in the wild-type strain, implying that EIICBGlc does not contribute significantly to sgrS induction in response to 2DG. In contrast, in the ΔmanXYZ strain, the PsgrS–lacZ reporter was induced only 1.7-fold (Figure 6B). The reporter was also induced by 1.7-fold in the ΔptsG ΔmanXYZ double mutant. These results suggest that EIIMan is almost fully responsible for transporting 2DG, which induces the stress response. Cumulatively, these findings suggest that SgrS-mediated regulation of manXYZ is physiologically relevant for minimizing the stress caused by accumulation of non-metabolizable sugar–phosphates.
The SgrS regulon does not appear to be fully conserved among different organisms
Two recent studies from our laboratory (15,45) suggested that regulation of ptsG by SgrS is conserved among enterobacterial species that possess SgrS orthologs. We investigated whether this was also true for SgrS translational regulation of manX. SgrS orthologs from Salmonella typhimurium, Klebsiella pneumoniae, E. carotovora and Y. pestis regulate E. coli ptsG translation when expressed ectopically (15). Therefore, we tested these orthologs (expressed from plasmids under the control of the Plac promoter) for their ability to regulate E. coli manX′–′lacZ. Northern blots performed in a previous study (15) showed that at the concentration of inducer used here, similar levels of all SgrS orthologs are produced. E. carotovora and Y. pestis SgrS orthologs did not repress E. coli manX translation while S. typhimurium and K. pneumoniae orthologs did (Figure 7A). This result suggested that sequences in SgrS required for regulation of manX translation may not be as conserved as those involved in regulating ptsG.
Sequence comparisons showed that the while the complementarity in the relevant base pairing region of SgrS and manX is conserved for S. typhimurium and K. pneumoniae SgrS and manX mRNA orthologs, it is not conserved for E. carotovora or Y. pestis orthologs (Figure 7B). It remained possible, however, that E. carotovora and Y. pestis SgrS orthologs regulated their cognate manX transcripts through the remaining regions of complementarity (Figure 7B). To test this, E. carotovora and Y. pestis manX sequences were swapped into the E. coli chromosome in the context of the manX′–′lacZ fusion (Figure 7C). The resulting manXYp′–′lacZ and manXEcar′–′lacZ fusion activities were analyzed in the presence of empty vector, Plac-sgrSK12 (E. coli SgrS) and the cognate Plac-sgrS constructs (Figure 7D). The manXYp′–′lacZ fusion was not regulated by either E. coli SgrS or by Y. pestis SgrS. Loss of regulation of Y. pestis manX by Y. pestis SgrS can be explained by the lack of complementarity in the region corresponding to the important E. coli pairing region. However, it was initially surprising that E. coli SgrS failed to regulate Y. pestis manX since the E. coli and Y. pestis manX sequences are nearly identical in the relevant base pairing region (Figure 7B). We note that the accessibility of the SgrS CU residues at positions 159 and 160 (Figure 4B–D) seemed to correlate with the ability to regulate manX. We hypothesize that pairing of these residues helps open up the short stem downstream (sequestering residues 162–165) to allow better pairing in the longer stretch of complementarity (residues 163–175). For Yersinia manX and E. coli SgrS, the U residue of the CU pair is not complementary, so perhaps opening of the stem and accessibility of the downstream region for pairing is impaired. The manXEcar′–′lacZ was repressed by E. coli SgrS, suggesting that base pairing between this heterologous pair is sufficient to yield regulation. For Erwinia manX and E. coli SgrS, the pattern of predicted pairing is different. There is additional complementarity encompassing SgrS residues downstream of the stem, so perhaps the CU residues are not needed for regulation in this case. Regardless, manXEcar′–′lacZ was not regulated by E. carotovora SgrS, suggesting that regulation of manX does not occur in E. carotovora. Together with the previous studies from our laboratory (15,45), these data suggest that regulation of some targets, e.g. ptsG, is conserved, while regulation of other targets, e.g. manXYZ, occurs in only a subset of organisms with SgrS orthologs.
DISCUSSION
SgrS is one of the best-studied Hfq-dependent bacterial small RNAs. Studies from our laboratory (9,11,14) and the Aiba group (11) have shown that SgrS mediates the glucose–phosphate stress response in bacterial cells by two different mechanisms: (i) base pairing-dependent translational regulation of target mRNAs encoding sugar transporters and (ii) SgrT-dependent post-translational regulation of sugar transport proteins. Prior to this study, only the ptsG mRNA had been confirmed as a target of SgrS. Here we have demonstrated that SgrS also post-transcriptionally regulates expression of the manXYZ genes, which encode the mannose and alternative glucose PTS transporter. A manX translational fusion (Figure 1) and footprinting experiments (Figure 3C) indicate that sequences in the manX coding sequence, between codons 8 and 11, are involved in SgrS–manXYZ mRNA base pairing interactions that lead to translational repression of manX. Additional complementarity between SgrS and manX is predicted to encompass codons 4–7, though this region was not protected in footprints and mutations that would disrupt these interactions did not affect SgrS regulation of manX′–′lacZ (Figure 3E). Bouvier et al. (46) reported that base pairing within the first five codons of an mRNA coding sequence can inhibit translation through direct occlusion of ribosome binding. Although the experimentally confirmed pairing for SgrS and manX mRNA falls outside this window, manX′–′lacZ translational regulation by SgrS did not require degradation via the RNase E degradosome (Figure 5) suggesting that translational inhibition of manX is the primary mechanism of regulation. It is possible that other base pairing interactions not evident in footprints occur in vivo and contribute to a ribosome occlusion mechanism of translational inhibition. If this is the case, destabilization of the full-length manXYZ mRNA may be caused by polarity resulting from translational repression at manX. Microarrays that identified putative targets of the sRNA RyhB suggest that RyhB carries out a similar type of regulation on other operons (47).
In northern blots using a manY-specific probe, two bands appeared. The first was the size expected for the full-length manXYZ transcript, and the second was the size of manYZ mRNA. The second band did not appear in blots using a manX-specific probe, consistent with the idea that it was a manYZ transcript. Furthermore, 5′ RACE analysis identified a transcript with a processed 5′-end in the manX–manY intergenic region. Our 5′ RACE results and a manY′–lacZ transcriptional fusion (data not shown) were both consistent with the idea that manYZ is processed from the full-length manXYZ transcript. Northern blots show that the processing of manYZ is SgrS-independent since the manYZ transcript is observed in both wild-type and ΔsgrS mutants. Interestingly, analysis of the RNA extracted from rne131 mutants showed that degradosome assembly on RNase E is involved in the processing (Figure 5B). Degradosome assembly is also required for RNase E-mediated turnover of SgrS–ptsG mRNA and SgrS–manX mRNA turnover [Figure 5A and B; and (12)]. RNase E catalytic activity (but not degradosome assembly) is required for processing of RNAs like rRNAs and tRNAs (48–50). The apparent involvement of the degradosome in generating the manYZ species may be somewhat unique. It remains to be seen whether the manYZ transcript is translated and what physiological function the processing serves.
A previous study suggested that one parameter affecting SgrS regulation of ptsG mRNA is the translational efficiency of ptsG. The translation rate is apparently reduced by co-translational membrane localization of the ptsG mRNA via SRP-dependent membrane insertion of nascent PtsG protein. Increased translational efficiency of ptsG greatly reduced SgrS’s ability to downregulate ptsG translation (36). These observations prompted us to look at how translational efficiency of manX impacts SgrS regulation. We noticed that the GTG start codon of manX was highly conserved among different species (data not shown). GTG is a weaker start codon with decreased translational efficiency compared to ATG. Changing the GTG start codon of manX to an ATG increased activity of the manX′–′lacZ fusion but did not influence regulation by SgrS (Supplementary Figure S3), suggesting that at least this factor affecting the translational efficiency of manX does not play a role in its regulation by SgrS. Our experiments also demonstrated that membrane localization is not required for manX regulation, since our manX′–′lacZ fusions were regulated in the absence of the transmembrane segment coding sequences of manY and manZ (Figure 1). These data are consistent with the idea that although a small RNA can have multiple targets, the mechanisms by which it regulates each one may be different.
In addition to mannose, the ManX, ManY and ManZ proteins are capable of transporting numerous sugars including mannose, glucose, 2DG, αMG, fructose, glucosamine, N-acetylglucosamine and trehelose (17). We demonstrated that SgrS expression is induced in response to growth on two different glucose analogs: α-methlyglucoside (αMG) and 2-deoxyglucose (2DG) (Figure 6). Analysis of ptsG mutants, manXYZ mutants and double mutants demonstrated that SgrS expression is induced in the absence of ptsG by both glucose analogs. Both EIICBGlc (encoded by ptsG) and EIIMan (encoded by manXYZ) contribute to stress induction in the presence of αMG, though EIICBGlc clearly plays the dominant role. In contrast, a functional EIIMan is required for induction of the stress by 2DG (Figure 6). These results indicated regulation of manXYZ is important under specific growth conditions where sugar–phosphates accumulate and further suggests that there may be more conditions that induce glucose–phosphate stress that have yet to be determined.
Computational and experimental analyses of different SgrS orthologs revealed that regulation of ptsG by SgrS is likely to be universally conserved in enteric species possessing SgrS (15,45). In contrast, we noted that the region of SgrS important for regulating manX in E. coli is not very highly conserved (data not shown), and our experiments indicate that SgrS orthologs from E. carotovora and Y. pestis do not regulate their cognate manX orthologs (Figure 7). The fact that SgrS regulation of manX is not as conserved as regulation of ptsG implies that in at least some enterobacterial species, the major source of sugar–phosphates that cause stress is PtsG (EIICBGlc) and not ManXYZ (EIIMan). The true nature of glucose–phosphate stress including the cellular targets and pathways affected is still mysterious. However, this study provides additional evidence that PTS-mediated transport of sugars and their subsequent metabolism are at the center of the stress.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
American Cancer Society Research Scholar Grant (ACS2008-01868); University of Illinois Department of Microbiology James R. Beck Fellowship (to J.R.). Funding for open access charge: American Cancer Society Grant.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Nadim Majdalani, Jihane Benhammou, Susan Gottesman, Hiroji Aiba, Caryn Wadler, Basil Hussain and the late Amos Oppenheimer for strains and James Slauch and Susan Gottesman for critical reading of the manuscript and valuable suggestions and comments. We thank Eric Massé and members of his laboratory for strains and purified Hfq. We are extremely grateful for the advice of Hubert Salvail, Guillaume Desnoyers and Kai Pappenfort on RNA footprinting as well as Jörg Vogel for extending his facilities for footprinting training. We extend a special thank you to Paul Babitzke for fruitful discussions about RNA structure. We are grateful to members of the Vanderpool lab, including past member Basil C. Hussain, for moral support and stimulating discussions.
REFERENCES
- 1.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Sharma C, Darfeuille F, Plantinga T, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes & Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 2009;16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 5.Desnoyers G, Morissette A, Prevost K, Masse E. Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. EMBO J. 2009;28:1551–1561. doi: 10.1038/emboj.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Møller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 2002;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohn C, Rigoulay C, Chabelskaya S, Sharma CM, Marchais A, Skorski P, Borezee-Durant E, Barbet R, Jacquet E, Jacq A, et al. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 2010;38:6620–6636. doi: 10.1093/nar/gkq462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vanderpool CK. Physiological consequences of small RNA-mediated regulation of glucose-phosphate stress. Curr. Opin. Microbiol. 2007;10:146–151. doi: 10.1016/j.mib.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 9.Vanderpool CK, Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- 10.Kimata K, Tanaka Y, Inada T, Aiba H. Expression of the glucose transporter gene, ptsG, is regulated at the mRNA degradation step in response to glycolytic flux in Escherichia coli. EMBO J. 2001;20:3587–3595. doi: 10.1093/emboj/20.13.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- 12.Morita T, Mochizuki Y, Aiba H. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc. Natl Acad. Sci. USA. 2006;103:4858–4863. doi: 10.1073/pnas.0509638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl Acad. Sci. USA. 2007;104:20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wadler CS, Vanderpool CK. Characterization of homologs of the small RNA SgrS reveals diversity in function. Nucleic Acids Res. 2009;37:5477–5485. doi: 10.1093/nar/gkp591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott J, Arber W. E. coli K-12 pel mutants, which block phage lambda DNA injection, coincide with ptsM, which determines a component of a sugar transport system. Mol. Gen. Genet. 1978;161:1–8. doi: 10.1007/BF00266608. [DOI] [PubMed] [Google Scholar]
- 17.Meadow ND, Fox DK, Roseman S. The bacterial phosphoenolpyruvate: glycose phosphotransferase system. Annu. Rev. Biochem. 1990;59:497–542. doi: 10.1146/annurev.bi.59.070190.002433. [DOI] [PubMed] [Google Scholar]
- 18.Erni B, Zanolari B, Kocher HP. The mannose permease of Escherichia coli consists of three different proteins. Amino acid sequence and function in sugar transport, sugar phosphorylation, and penetration of phage lambda DNA. J. Biol. Chem. 1987;262:5238–5247. [PubMed] [Google Scholar]
- 19.Plumbridge J. Control of the expression of the manXYZ operon in Escherichia coli: Mlc is a negative regulator of the mannose PTS. Mol. Microbiol. 1998;27:369–380. doi: 10.1046/j.1365-2958.1998.00685.x. [DOI] [PubMed] [Google Scholar]
- 20.Plumbridge J. Regulation of PTS gene expression by the homologous transcriptional regulators, Mlc and NagC, in Escherichia coli (or how two similar repressors can behave differently) J. Mol. Microbiol. Biotechnol. 2001;3:371–380. [PubMed] [Google Scholar]
- 21.Yu DG, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellermeier CD, Janakiraman A, Slauch JM. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene. 2002;290:153–161. doi: 10.1016/s0378-1119(02)00551-6. [DOI] [PubMed] [Google Scholar]
- 23.Miller JH. Experiments in Bacterial Genetics. New York: Cold Spring Harbor Laboratory, Cold Spring Harbor; 1972. [Google Scholar]
- 24.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 25.Majdalani N, Chen S, Murrow J, St. John K, Gottesman S. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- 26.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 27.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen PR, Hammer K. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl. Environ. Microbiol. 1998;64:82–87. doi: 10.1128/aem.64.1.82-87.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDowall KJ, Lin-Chao S, Cohen SN. A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J. Biol. Chem. 1994;269:10790–10796. [PubMed] [Google Scholar]
- 30.Mackie GA. Specific endonucleolytic cleavage of the mRNA for ribosomal protein S20 of Escherichia coli requires the product of the ams gene in vivo and in vitro. J. Bacteriol. 1991;173:2488–2497. doi: 10.1128/jb.173.8.2488-2497.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackie GA. Secondary structure of the mRNA for ribosomal protein S20. Implications for cleavage by ribonuclease E. J. Biol. Chem. 1992;267:1054–1061. [PubMed] [Google Scholar]
- 32.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl Acad. Sci. USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guillier M, Gottesman S. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res. 2008;36:6781–6794. doi: 10.1093/nar/gkn742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morita T, Kawamoto H, Mizota T, Inada T, Aiba H. Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol. Microbiol. 2004;54:1063–1075. doi: 10.1111/j.1365-2958.2004.04329.x. [DOI] [PubMed] [Google Scholar]
- 36.Kawamoto H, Morita T, Shimizu A, Inada T, Aiba H. Implication of membrane localization of target mRNA in the action of a small RNA: mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes Dev. 2005;19:328–338. doi: 10.1101/gad.1270605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy P, Peterkofsky A, McKenney K. Translational efficiency of the Escherichia coli adenylate cyclase gene: mutating the UUG initiation codon to GUG or AUG results in increased gene expression. Proc. Natl Acad. Sci. USA. 1985;82:5656–5660. doi: 10.1073/pnas.82.17.5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morita T, El-Kazzaz W, Tanaka Y, Inada T, Aiba H. Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J. Biol. Chem. 2003;278:15608–15614. doi: 10.1074/jbc.M300177200. [DOI] [PubMed] [Google Scholar]
- 39.Amaral D, Kornberg HL. Regulation of fructose uptake by glucose in Escherichia coli. J. Gen. Microbiol. 1975;90:157–168. doi: 10.1099/00221287-90-1-157. [DOI] [PubMed] [Google Scholar]
- 40.Henderson PJ, Giddens RA, Jones-Mortimer MC. Transport of galactose, glucose and their molecular analogues by Escherichia coli K12. Biochem. J. 1977;162:309–320. doi: 10.1042/bj1620309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curtis SJ, Epstein W. Phosphorylation of D-glucose in Escherichia coli mutants defective in glucosephosphotransferase, mannosephosphotransferase, and glucokinase. J. Bacteriol. 1975;122:1189–1199. doi: 10.1128/jb.122.3.1189-1199.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stock JB, Waygood EB, Meadow ND, Postma PW, Roseman S. Sugar transport by the bacterial phosphotransferase system. The glucose receptors of the Salmonella typhimurium phosphotransferase system. J. Biol. Chem. 1982;257:14543–14552. [PubMed] [Google Scholar]
- 43.Rephaeli AW, Saier MH., Jr Substrate specificity and kinetic characterization of sugar uptake and phosphorylation, catalyzed by the mannose enzyme II of the phosphotransferase system in Salmonella typhimurium. J. Biol. Chem. 1980;255:8585–8591. [PubMed] [Google Scholar]
- 44.Kadner RJ, Murphy GP, Stephens CM. Two mechanisms for growth inhibition by elevated transport of sugar phosphates in Escherichia coli. J. Gen. Microbiol. 1992;138(Pt 10):2007–2014. doi: 10.1099/00221287-138-10-2007. [DOI] [PubMed] [Google Scholar]
- 45.Horler RS, Vanderpool CK. Homologs of the small RNA SgrS are broadly distributed in enteric bacteria but have diverged in size and sequence. Nucleic Acids Res. 2009;37:5465–5476. doi: 10.1093/nar/gkp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol. Cell. 2008;32:827–837. doi: 10.1016/j.molcel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 47.Massé E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Z, Deutscher MP. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA. 2002;8:97–109. doi: 10.1017/s1355838202014929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopez PJ, Marchand I, Joyce SA, Dreyfus M. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol. Microbiol. 1999;33:188–199. doi: 10.1046/j.1365-2958.1999.01465.x. [DOI] [PubMed] [Google Scholar]
- 50.Ow MC, Liu Q, Kushner SR. Analysis of mRNA decay and rRNA processing in Escherichia coli in the absence of RNAse E-based degradosome assembly. Mol. Microbiol. 2000;38:854–866. doi: 10.1046/j.1365-2958.2000.02186.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.