Abstract
Ikaros is associated with both gene transcriptional activation and repression in lymphocytes. Ikaros acts also as repressor of human γ-globin (huγ-) gene transcription in fetal and adult erythroid cells. Whether and eventually, how Ikaros can function as a transcriptional activator in erythroid cells remains poorly understood. Results presented herein demonstrate that Ikaros is a developmental-specific activator of huγ-gene expression in yolk sac erythroid cells. Molecular analysis in primary cells revealed that Ikaros interacts with Gata-1 and favors Brg1 recruitment to the human β-globin Locus Control Region and the huγ-promoters, supporting long-range chromatin interactions between these regions. Additionally, we demonstrate that Ikaros contributes to transcription initiation and elongation of the huγ-genes, since it is not only required for TBP and RNA Polymerase II (Pol II) assembly at the huγ-promoters but also for conversion of Pol II into the elongation-competent phosphorylated form. In agreement with the latter, we show that Ikaros interacts with Cyclin-dependent kinase 9 (Cdk9), which contributes to efficient transcription elongation by phosphorylating the C-terminal domain of the large subunit of Pol II on Serine 2, and favours Cdk9 recruitment to huγ-promoters. Our results show that Ikaros exerts dual functionality during gene activation, by promoting efficient transcription initiation and elongation.
INTRODUCTION
The transcription factor Ikaros is widely expressed in hematopoietic cells where it regulates various aspects of hematopoiesis (1,2). Ikaros has been associated with gene activation (3–5), potentiation (6), priming (7) and transcriptional repression (8,9). In proliferating T cells, a high fraction of Ikaros co-purifies with Mi-2, a core component of the NuRD complex (10,11). In addition, Ikaros and Mi-2 control transcriptional regulation of the Cd4 locus during T cell differentiation (12). Nevertheless, a significant fraction of Ikaros is associated with a Brg1-based SWI/SNF-like complex (10). The relevance of the latter interaction has been indicated by several observations: (i) Ikaros has been associated with gene activation mediated by SWI/SNF-like complexes in T cells (3); (ii) Ikaros co-fractionates and co-immunoprecipitates with Brg1 (10,13); and (iii) Ikaros and Brg1 are also components of a SWI/SNF-like complex in mouse erythroleukemia (MEL) cells (14).
The human β- (huβ-) globin locus has been widely used as a model to explore the effects of transcription factors and co-factors on tissue- and developmental-specific gene expression. The huβ-globin locus contains five developmentally regulated genes (ε-Gγ-Aγ-δ-β). The locus control region (βLCR), which is located upstream of the globin genes, provides high-level globin gene expression in erythroid cells (EryC). The βLCR is composed of five DNase I hypersensitive sites (HSs) which are particularly rich in transcription factor binding sites (15). In EryC the βLCR favors high-level transcription through close interaction with gene promoters, and is a major determinant of locus chromatin organization (16). The transcription factors EKLF (17), Ikaros (18), BCL11A (19), GATA-1 and its co-factor FOG-1 (Friend of GATA-1) (20), as well as the nuclear factor NLI/Ldb1 (21) and the chromatin remodeling co-regulator Brg1 (22) have all been shown to be required for efficient long-range chromatin interactions between the βLCR and β-like globin gene promoters.
Beside transcription initiation, modulation of transcription elongation is also likely implicated in globin gene regulation. Indeed, it has been shown that the βLCR enhances the transition from transcription initiation to elongation for the β major globin (βmaj) gene (23). Additionally, globin gene expression can be reversibly inhibited by the ATP analog 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) (24,25), an inhibitor of cyclin-dependent kinase 9 (Cdk9) (26). Cdk9 is the catalytic subunit of the positive transcription elongation factor (P-TEFb). Cdk9 contributes to efficient transcription elongation by phosphorylating the C-terminal domain (CTD) of the large subunit of RNA Polymerase II (Pol II) on Ser2 (Pol II phospho Ser2). Several gene-specific regulators interact with P-TEFb but only a very limited number of transcription activators have been shown to recruit it to gene promoters (27). Two Cdk9 isoforms are found in mammalian cells and classified according to apparent molecular weight, namely Cdk942 and Cdk955. In general Cdk9 is essential for definitive but not primitive erythropoiesis in zebrafish (28), and deregulation of the Cdk9 pathway is implicated in the establishment and/or maintenance of a transformed cell phenotype, as documented for a number of human tumors and lymphomas (26).
Primitive hematopoiesis is the transient production of large, nucleated erythroid and megakaryocyte progenitors, and takes place in the yolk sac (29). Mouse primitive EryC express the embryonic globin genes εy and βh1, whereas the human β-like globin genes expressed at the same stage of development in transgenic mice (carrying the complete huβ-globin locus) are the embryonic ε- (huε-) and the fetal γ- (huγ-) genes (30,31). Huγ-genes are transcriptionally silenced in adult definitive EryC by the contribution of several transcription regulators, such as Gata-1 (32,33), Ikaros (18,34) the TR2/TR4 heterodimer (35) that are present throughout primitive as well as definitive erythropoiesis or the recently characterized Bcl11a protein, which however appears to be expressed only in definitive but not primitive, yolk sac-derived EryC (30,36). We have recently shown that Ikaros and Gata-1 cooperate to silence the huγ-genes in EryC at the fetal stage of development when huγ- and huβ-genes are expressed (18). To investigate if Ikaros can also act as transcriptional activator in EryC and to define the molecular mechanisms of Ikaros-dependent transcriptional control, we studied the expression profile of huγ-genes in embryonic day 10.5 (e10.5) yolk sac EryC. We used line 2 (ln2) mice, which carry a 70 Kb DNA fragment containing the entire huβ-globin locus and express the human β-like globin genes in a developmental-specific manner (31), as well as Ikaros null (Iknull) mice that are null for Ikaros proteins due to a deletion in the last exon that leads to protein instability (37). Our results reveal that Ikaros is a transcriptional activator of huγ-gene expression in primitive EryC. We provide the evidence that Ikaros contributes to efficient recruitment of Brg1, Gata-1, Pol II and Cdk9 to the huγ-promoters, thus supporting active chromatin re-organization, transcription initiation and elongation of the huγ-genes.
MATERIALS AND METHODS
Transgenic mouse lines
All mutant embryos were compared with wild type littermates, to reduce difference in genetic background or developmental stage. Embryos were genotyped by PCR as described in (37). Homozygous ln2 (ln2+/+) (31): heterozygous Ikaros (Iknull+/−) (37) mice were bred with ln2−/−:Iknull+/− females in order to avoid contamination of embryos with adult blood of the mother, and ln2+/−:IkWt (ln2) or ln2+/−:Iknull−/− (ln2-Iknull) e10.5 yolk sacs were isolated. Animals were sacrificed by cervical dislocation. Yolk sacs were rinsed several times with phosphate-buffered saline (PBS) to remove maternal blood. Blood was collected by piercing and compressing the yolk sac with forceps. Animal experiments were conducted in accordance with the Canadian Council on Animal Care (CCAC) guidelines and approved by the Maisonneuve-Rosemont Hospital animal care committee.
Wright-Giemsa staining
This protocol was carried out as previously described (18).
Chromatin immunoprecipitation and quantitative real-time PCR analyses
Chromatin immunoprecipitation (ChIP) procedure and quantification were as previously described (18) starting with 5 × 105 e10.5 yolk sac EryC. Cells were fixed with 1% formaldehyde for 10 min at 37°C. Chromatin was reduced in size by sonication in order to obtain fragments of 400–500 bp. About 1/30th of immunoprecipitated and unbound (input) material was used as template for quantitative real-time PCR (qPCR) with SYBR Green (Invitrogen) on an iCycler iQTM (Bio-Rad) system; the kidney-specific Tamm-Horsfall gene promoter (Thp) was used as internal control. All data shown are the results of at least four independent ChIP experiments with qPCR reactions from each ChIP performed in triplicate; the data are plotted as the mean ± standard deviation (SD) of the measurements.
Protein analysis
Western blot (WB) and co-immunoprecipitation (co-IP) analyses were basically as previously described (18). For co-IP, 5 × 106 e10.5 yolk sac EryC, 1 × 106 COS-7 cells or 10 × 106 Jurkat cells were lysed in 1 ml/107 cells of ice-cold lysis buffer (20 mM Hepes pH 8.0, 25% glycerol, 0.2 mM EDTA, 150mM NaCl, 1.5 mM MgCl2, 0.5% NP-40) containing protease inhibitors (Protease Inhibitor Cocktail; Sigma). Where detailed, co-IP washing buffers contained higher salt (200 mM) or NP-40 (1%) concentration. Ikaros proteins were not detected by WB in yolk sac EryC co-IP input samples, which represents 2% volume of the total lysate, i.e. ∼105 yolk sac EryC. Therefore, more concentrated yolk sac protein lysates, i.e. 2.5 × 106 yolk sac EryC per lane, were loaded onto SDS–PAGE in order to identify Ikaros-specific proteins.
Cell transfection
Cells were transfected with murine Ikaros 2 cDNA, which is, together with Ikaros-1, the most abundantly expressed isoform in hematopoietic cells (4) and/or murine Gata-1. Stable Jurkat cell clones were obtained by retroviral transduction of wild type Jurkat cells with the pOZN-FH-Ik expression vector (18), which allows expression of Ikaros in double N-terminally epitope-tagged (Flag and Hemoagglutinin -FH-) fusion protein. Plasmid characteristic, cell transfection and selection were carried out as described in Nakatani et al. (38). COS-7 cells were transiently transfected with Flag/HA Ikaros (pCMV-FH-Ik) or with Gata-1 (pCMV-Gata) CMV-based expression vector using the FuGene system (FuGene 6, Roche), according to the manufacturer’s instructions. Cells were harvested 36 h post-transfection.
Immunofluorescence
For IF studies, transiently transfected COS-7 cells were seeded on glass slide 24 h post-transfection and allowed to adhere for additional 12 h. Cells were then fixed in 4% formaldehyde for 30 min at RT, washed once in PBS and subjected to heat-mediated antigen retrieval treatment. Slides were heated three times to 90°C for 2 min in the retrieval solution (10 mM citric acid pH 6.0, 0.05% Tween-20) and then allowed to cool down for 20 min at RT. Next, slides were treated with PBS/0.5% Triton for 20 min at RT, washed twice in PBS and blocked in PBS/0.5% NFDM. Primary (listed in Supplementary Figure S1) as well as secondary antibodies (all from Jackson Immunoresearch) were diluted in PBS containing 0.1% Bovine Serum Albumin and 0.01% Triton. Slides were dehydrated by treatment with 70, 90 and 100% ethanol and covered with Dapi-Dabco: Vectashield solution (1:1) in glycerol. Images were acquired with a LSM 510 META Zeiss confocal microscope with z-resolution of 200 µM equipped with Zeiss LSM Image Browser. Red–green–blue (RGB) profiles were generated by the WCIF ImageJ(®) program (NIH).
Quantitative reverse transcription–PCR
Total RNA was extracted with Trizol (Invitrogen) and treated with DNaseI–RNase free (Invitrogen). Reverse transcription reactions were performed with oligo(dT)15 or random primers and SuperScript III reverse transcriptase (Invitrogen). qPCR was carried out on an iCycler iQTM (Bio-Rad) system using specific primer sets; quantification was carried out as previously reported (18). Data shown are the results of at least five independent experiments with qPCR reactions from each cDNA performed in triplicate; the data are expressed as the mean ± SD of the measurements.
Chromosome conformation capture
The chromosome conformation capture (3C) protocol was basically as previously described (18) using 1–2 × 106 e10.5 yolk sac EryC; data shown are the results of five independent experiments with qPCR reactions from each 3C reaction performed in triplicate; the data are plotted as the mean ± SD of the measurements.
Statistical analysis
Unpaired ‘Student’s t-test’ was used to determine the level of statistical significance (P-value).
Primer sequences and antibodies
Primer sequences and antibodies are published as Supplementary Figure S1.
RESULTS
Ln2-Iknull e10.5 yolk sacs do not have gross developmental defects
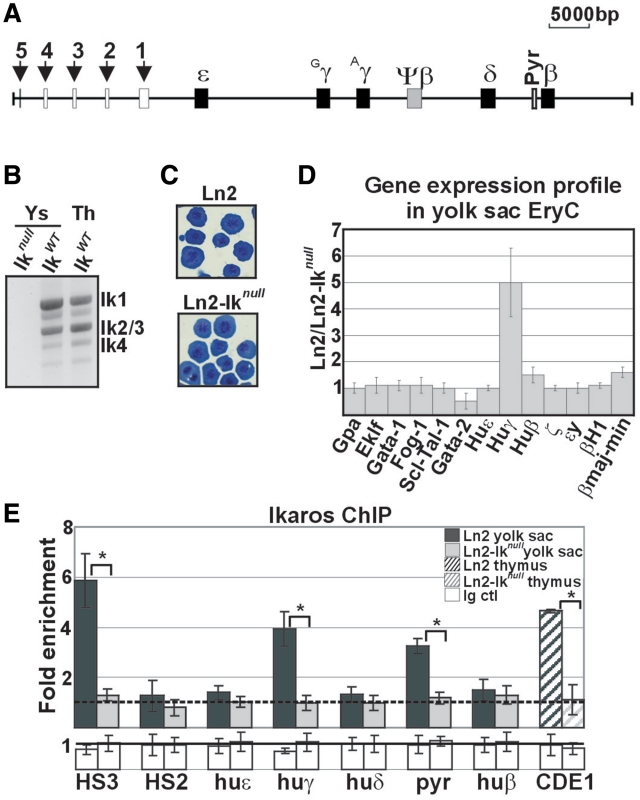
Primitive EryC are characterized by their large size and expression of embryonic β-like globin genes, i.e. εy and βh1 in mice and ε in humans. In transgenic mice carrying the huβ-globin locus (Figure 1A), such as ln2 mice, both huγ- and huε-genes are expressed in yolk sac EryC (30,31). Ikaros is expressed throughout development in EryC and can be detected in yolk sac and fetal liver at e11 (2). The DNA-binding isoforms Ikaros-1, -2/3 and -4 are the predominant isoforms expressed in hematopoietic cells (4). To assess the expression pattern of Ikaros isoforms in yolk sac EryC, total RNA was retro-transcribed and cDNA amplified using Ikaros exon 1- and 7-specific primers. As shown in Figure 1B, Ikaros-1, -2/3 and -4 isoforms are expressed in yolk sac EryC at a relative molecular ratio resembling that observed in adult thymus cells. We then investigated whether erythroid development and differentiation are affected in ln2-Iknull yolk sac EryC. First, we evaluated yolk sac cellularity. Keeping in mind the difficulties associated with quantitative analysis of e10.5 blood cells, no direct correlation between Ikaros genotype and total number of nucleated EryC collected per embryo could be established, suggesting that loss of Ikaros does not generate a quantitative defect of primitive erythropoiesis; also, ln2-Iknull embryos were not visibly anemic, had apparently normal morphology, and were indistinguishable from wild-type counterparts (data not shown). Second, we performed Wright-Giemsa staining of ln2 and ln2-Iknull yolk sac EryC. In both genetic backgrounds, embryonic blood was predominated by large basophilic erythroblasts (Figure 1C), indicating similar erythroid differentiation behaviour in ln2 versus ln2-Iknull yolk sac EryC. Third, we evaluated transcript levels of the erythrocyte cell-surface molecule Gpa (Glycophorin A) and the transcription factors Eklf, Gata-1, Fog-1 and Scl/Tal-1, which are co-regulators critical for erythroid differentiation and globin gene expression (39). Quantitative reverse transcription–PCR (qRT–PCR) analysis revealed that Gpa, Eklf, Gata-1, Fog-1 and Scl/Tal-1 expression levels do not significantly vary between ln2 and ln2-Iknull yolk sac EryC (n ≥ 4) whereas, as observed in e12.5 fetal liver EryC (18), loss of Ikaros is associated with increased Gata-2 levels (Figure 1D; P = 0.03). Overall, these results suggest that loss of Ikaros does not impair yolk sac EryC development or homeostasis.
Figure 1.
Morphological and molecular characterization of ln2 and ln2-Iknull yolk sac EryC. (A) Map of the human β-globin locus; black arrows indicate βLCR HSs; genes are indicated by black boxes; (B) RT–PCR performed on ln2-Iknull (Iknull), ln2 (IkWT) yolk sac EryC or ln2 (IkWT) thymus (Th) cells; (C) Wright-Giemsa staining of ln2 or ln2-Iknull yolk sac cells; (D) RT–qPCR of hematopoietic as well as globin gene transcripts performed on equal amounts of ln2 and ln2-Iknull yolk sac EryC; transcript quantification was calculated according to Pfaffl (73) using mouse Actin cDNA as internal control; ln2/ln2-Iknull ratios are plotted as the mean ± SD of the measurements; n ≥ 4; (E) ChIP on ln2 and ln2-Iknull yolk sac EryC or thymus cells carried out with Ikaros antibodies; immunoprecipitated and input chromatin samples were used as template in qPCR; quantification was carried out according to the 2−ΔΔCt method, using mouse kidney-specific Tamm-Horsfall protein (Thp) promoter as internal control; fold enrichments (y-axis) of globin regions relative to the control and the input samples are plotted as the mean ± SD of the measurements; a value of 1 (dashed line) indicates no enrichment; *P ≤ 0.05 by Student’s t-test; huε, huε-promoter; huγ, huγ-promoters; huδ, huδ-promoter; pyr, Pyr region; and huβ, huβ-promoter; dark gray bars: ln2 yolk sac EryC; light gray bars: ln2-Iknull yolk sac EryC; dark gray dashed bars: ln2 thymus cells; light gray dashed bars: ln2-Iknull thymus cells; white bars: isotype-matched Ig (Ig ctl).
Ikaros-dependent huγ-globin gene expression in yolk sac EryC
We have recently shown that Ikaros contributes to huγ-gene transcriptional silencing in e12.5 fetal liver EryC (18). To investigate the role of Ikaros in globin gene regulation in yolk sac EryC, levels of huε-, huγ- and huβ-gene expression were established by RT–qPCR, using oligo(dT) primers for cDNA synthesis, intron–exon junction sequence-specific primers for qPCR and Actin as internal control. Loss of Ikaros leads to reduced huγ-gene expression levels in yolk sac EryC (Figure 1D; P = 0.005). Moreover expression of the embryonic huε- and mouse ζ-, εy- and βH1-globin genes is unchanged in ln2-Iknull yolk sac EryC (n ≥ 4), and in these cells we detected only a slight but significant reduction of huβ- and mouse βmaj/βmin globin gene expression (n = 6; P ≤ 0.05). Similar results were obtained when Gpa was used as internal control. For instance, in ln2 versus ln2-Iknull cells, the relative level of huγ-gene expression was 5.9 (SD 1.2; n = 6; P = 0.05). Gpa is an erythroid-specific membrane marker gene used as control to ensure that variations in globin gene expression are not merely due to changes in the number of viable EryC. Altogether, the above data suggest that Ikaros specifically contributes to huγ-gene transcriptional activation in yolk sac EryC.
Ikaros promotes efficient recruitment of Brg1 to βLCR HS3 and huγ-promoters in ln2 yolk sac EryC
It has recently been demonstrated that Ikaros is recruited to βLCR HS3 (HS3), huγ-promoters and a pyrimidine stretch (Pyr region) located 1 kilobase upstream of the huδ-gene that contributes to huγ-gene silencing in adult-type erythroid cells; however, Ikaros does not bind to HS2 nor to huε- and huβ-promoters even though the Ikaros consensus sequence, TGGGAA, is found in the latter [data not shown and (1,14,18,40)]. To investigate occupancy by Ikaros of these regulatory regions in yolk sac EryC in vivo, we performed chromatin immunoprecipitation (ChIP) analysis using chromatin isolated from ln2 or ln2-Iknull yolk sac EryC and Ikaros antibodies (18). Ikaros is efficiently recruited to HS3, huγ-promoters and Pyr region in yolk sac EryC and to CD4 enhancer 1 region (CD4E1, used as control) in adult thymus cells (12), whereas no significant recruitment was detected at HS2, huε- or huβ-promoter (Figure 1E).
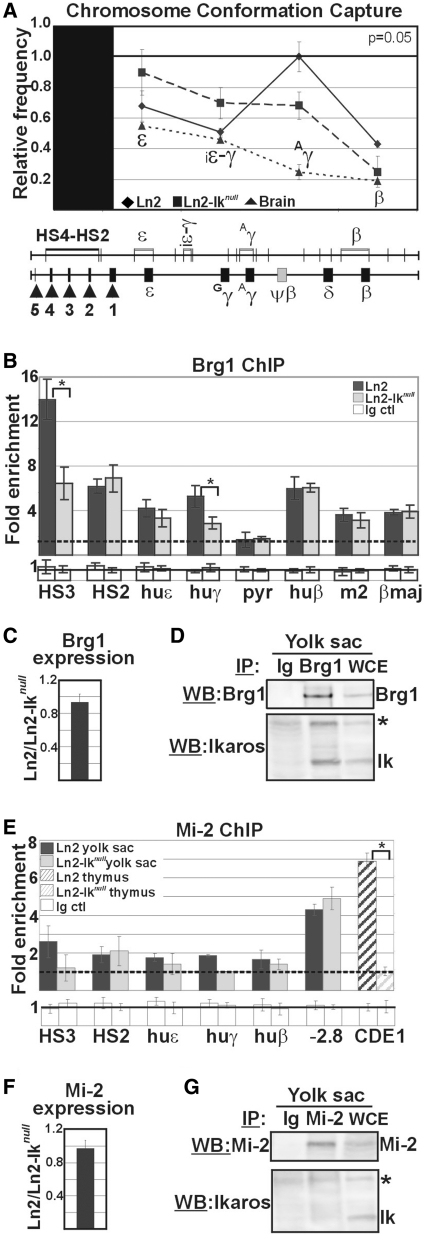
Ikaros co-fractionates and co-immunoprecipitates with Brg1 (10,13,14), and furthermore acts as a potentiator of gene expression during T-cell development possibly by recruiting Brg1-containing chromatin remodelling complexes to appropriate lineage-specific target genes (4,6). In addition, Brg1 is recruited to mouse HS3, is required for embryonic and adult β-globin gene transcription, erythroblast survival (41,42), and long-range chromatin interactions at the β-globin locus (22). Therefore, we studied the influence of Ikaros, when acting as transcription activator, on long-range chromatin interactions. As such we exploited the 3C assay to determine the physical proximity between chromosomal regions that are normally located many kilobases apart in vivo. Using the βLCR HS4-HS2 region as ‘fixed’ fragment (43), we observed that long-range chromatin interactions between the βLCR and the huγ-promoters are decreased in ln2-Iknull yolk sac EryC (Figure 2A; n = 5; P = 0.05). Next, we analyzed in vivo recruitment of Brg1 to several regions across the huβ-globin locus by ChIP. Brg1 recruitment to HS3 and huγ-promoters is reduced in ln2-Iknull EryC but is not affected at HS2, huε- or huβ-promoters or mouse HS2 and βmaj promoter (Figure 2B). To exclude the possibility that Ikaros might influence Brg1 transcription, we compared Brg1 expression levels in ln2 and ln2-Iknull yolk sac EryC by RT–qPCR and found Brg1 transcription to be essentially unchanged in the two backgrounds (Figure 2C). Next, we tested whether Ikaros co-immunoprecipitates with Brg1 in yolk sac EryC by protein co-IP assay. Brg1 is immunoprecipitated and recognized by Brg1 antibodies by WB; additionally, Ikaros is detected upon Brg1 IP, thus indicating that Ikaros and Brg1 proteins co-immunoprecipitate in these cells (Figure 2D).
Figure 2.
Brg1 and Mi-2 recruitment to the huβ-globin locus in ln2 and ln2-Iknull yolk sac EryC. (A) Ln2 and ln2-Iknull yolk sac EryC were subjected to 3C assay; nuclei were digested with EcoRI and genomic DNA was ligated and used as template for qPCR; βLCR HS4-HS2 EcoRI fragment was used as ‘fixed’ point and primer sets were designed in order to amplify the genomic regions corresponding to the ε, ε-gene; iε-γ, inter-ε-γ region; Aγ, Aγ-gene and β-gene region; relative crosslinking frequencies (y-axis) of the ‘fixed’ fragment with globin fragments were defined using naked DNA encompassing the whole huβ-globin as control and normalized to endogenous mouse Actin; a value of 1 was attributed to the highest crosslinking frequency; the data are plotted as the mean ± SD of the measurements of five independent experiments; x-axis: position across the locus; rhomboids: ln2 yolk sac EryC; squares: ln2-Iknull yolk sac EryC; triangles: brain cells; (B and E) ChIP on ln2 and ln2-Iknull yolk sac EryC or thymus cells carried out with Brg1 or Mi-2 antibodies; fold enrichments (y-axis) were calculated as described in Figure 1E and are plotted as the mean ± SD of the measurements; a value of 1 (dashed line) indicates no enrichment; *P ≤ 0.05 by Student’s t-test; huε, huε-promoter; huγ, huγ-promoters; pyr, Pyr region; huβ, huβ-promoter; m2, mouse HS2; βmaj, βmajor promoter; −2.8: −2.8 kb region upstream of the Gata-2 promoter and CD4E1: CD4 enhancer 1 region; dark gray bars: ln2 yolk sac EryC; light gray bars: ln2-Iknull yolk sac EryC; dark gray dashed bars: ln2 thymus cells; light gray dashed bars: ln2-Iknull thymus cells; white bars: isotype-matched Ig (Ig ctl); (C and F) RT–qPCR of Brg1 and Mi-2 gene transcripts performed on equal amounts of ln2 and ln2-Iknull yolk sac EryC; transcript quantification was calculated according to Pfaffl (73) using mouse Actin cDNA as internal control; ln2/ln2-Iknull ratios are plotted as the mean ± SD of the measurements; n = 6; (D and G) Protein co-IP of total cell lysates prepared from ln2 yolk sac EryC; cell lysates were immunoprecipitated with Mi-2 or Brg1 antibodies as well as isotype-matched Ig; samples were resolved on SDS–PAGE, immunoblotted and WB membranes were cut in half; the higher half was probed with either Mi-2 or Brg1 antibodies, whereas the lower half was probed with Ikaros antibodies; asterisks indicate non-specific bands; WCE: yolk sac EryC total cell lysate.
Ikaros is also known to interact with the nucleosome remodelling ATPase Mi-2 (a subunit of the NuRD chromatin remodeling complex) (10,11), and recruits this co-factor to specific chromatin regions (18). Even though Mi-2 is widely perceived as a transcriptional repressor, the Mi-2-NuRD complex has recently been linked to both gene activation and repression in the case of certain Gata-1/Fog-1 target genes (44). For this reason, we investigated in vivo recruitment of Mi-2 to the huβ-globin locus in ln2 as well as ln2-Iknull yolk sac EryC by ChIP. As depicted in Figure 2E, no significant recruitment of Mi-2 either to HS3, HS2 or to huε-, huγ- and huβ-promoters could be detected. Nonetheless under the same experimental conditions, in confirming the technical feasibility of Mi-2 ChIP, we could demonstrate efficient Mi-2 recruitment to the −2.8 kb region upstream of the Gata-2 promoter in yolk sac EryC (Figure 2E) (45), the CD4E1 region (12) in freshly-isolated thymic cells (Figure 2E), and several regions of the huβ-globin locus in EryC isolated from e12.5 fetal livers (18). Due to the possibility that absence of significant Mi-2 recruitment to critical regulatory regions of the huβ-globin locus might depend upon lower levels of Mi-2 expression in ln2-Iknull yolk sac EryC, we next examined Mi-2 gene expression by RT–qPCR. We observed that Mi-2 expression levels are the same in ln2 and ln2-Iknull yolk sac EryC (Figure 2F). However, under the same stringency conditions that allowed co-IP of Ikaros and Brg1 (150 mM NaCl, 0.5% NP-40), Ikaros and Mi-2 could not be co-immunoprecipitated in yolk sac EryC with specific antibodies (Figure 2G). These results argue against a direct role of Mi-2 for huγ-gene activation in yolk sac EryC, and rather suggest that Ikaros-dependent recruitment of Brg1 to huγ-promoters and HS3 is required for efficient huγ-gene expression in these cells.
Ikaros facilitates Gata-1 recruitment to huγ-regulatory regions in yolk sac EryC
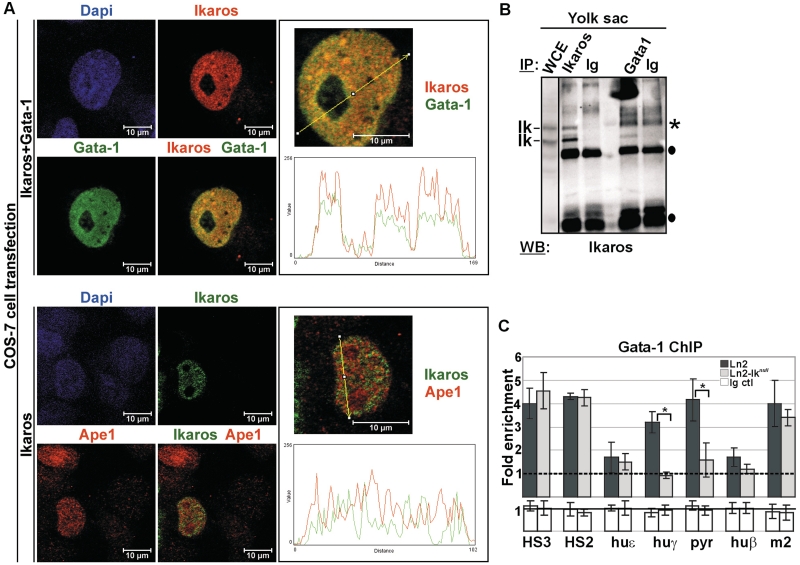
We previously reported that Ikaros physically interacts with Gata-1 and facilitates Gata-1 recruitment to HS3 and huγ-promoters in e12.5 fetal liver EryC (18). To better define Ikaros-Gata-1 protein interaction, we first analyzed the subcellular localization of Ikaros and Gata-1 at the single-cell level by immunofluorescence (IF) and confocal microscopy. COS-7 cells, which lack any Ikaros or Gata-1 protein, were transfected with Flag/HA-Ikaros (pCMV-FH-Ik) together with Gata-1 (pCMV-Gata) expression vectors, fixed with formaldehyde, permeabilized and immunolabeled with antibodies specific for HA (i.e. Ikaros detection) or Gata-1. Ikaros and Gata-1 were detected using secondary antibodies conjugated to Texas red (TR) or Fluorescein isothiocyanate (FITC) fluorochromes, respectively. The use of cross-absorbed and affinity-purified secondary antibodies minimized background and non-specific reactivity to undetectable fluorescent levels (data not shown). Confocal microscopy analysis revealed that both Ikaros (red signals) and Gata-1 (green signals) have a dominant nuclear localization (Figure 3A), whereas COS-7 cells transfected with empty vectors (mock control) showed undetectable levels of green or red stain (Supplementary Figure S2A). Interestingly, the merged image obtained in COS-7 cells that express both Ikaros and Gata-1, leads to the emergence of several yellow nuclear signals per focal plane (Figure 3A). Physical overlap between Ikaros and Gata-1 was further defined by the ImageJ(®)program, which revealed that the line profile plots and fluorescent peaks of Ikaros and Gata-1 often overlap whereas the fluorescence profiles of Ikaros and Ape1, a nuclear protein that does not interact with Ikaros used as negative control (46), are discordant.
Figure 3.
Ikaros-Gata-1 cooperative binding. (A) Confocal immunofluorescence of COS-7 cells expressing Flag/HA-Ikaros and Gata-1 proteins (Ikaros+Gata-1) or Flag/HA-Ikaros only (Ikaros); Ikaros+Gata-1 transfected cells were stained with mouse antibodies to the HA tag and with rat antibodies to Gata-1; the secondary staining was carried out with TR-conjugated anti-mouse as well as FITC-conjugated anti-rat antibodies; a single COS-7 cell is shown where Ikaros is detected as red signals, Gata-1 as green signals and Ikaros-Gata-1 co-localization as yellow signals in the magnified merged image; Ikaros-transfected cells were stained with rabbit antibodies to the HA tag and with mouse antibodies to Ape1; the secondary staining was carried out with FITC-conjugated anti-rabbit as well as TR-conjugated anti-mouse antibodies; a single COS-7 cell is shown where Ikaros is detected as green signals and Ape1 as red signals; line profile plots were obtained by the WCIF ImageJ(®) program (NIH); (B) Protein co-IP of total cell lysates prepared from ln2 yolk sac EryC; cell lysates were immunoprecipitated with Ikaros or Gata-1 antibodies as well as isotype-matched rabbit (for Ikaros co-IP) or rat (for Gata-1 co-IP) Ig and WB was carried out with Ikaros antibodies; filled circles represent Ig bands; asterisks indicate non-specific bands; WCE: yolk sac EryC total cell lysate; (C) ChIP on ln2 and ln2-Iknull yolk sac EryC carried out with Gata-1 antibodies; fold enrichments (y-axis) were calculated as described in Figure 1E and are plotted as the mean ± SD of the measurements; a value of 1 (dashed line) indicates no enrichment; *P ≤ 0.05 by Student’s t-test; huε, huε-promoter; huγ, huγ-promoters; pyr, Pyr region; huβ, huβ-promoter; m2, mouse HS2; dark gray bars: ln2 yolk sac EryC; light gray bars: ln2-Iknull yolk sac EryC; white bars: isotype-matched Ig (Ig ctl).
Next, Ikaros-Gata-1 protein interaction was investigated by protein co-IP. Protein lysates of ln2 yolk sac EryC were immunoprecipitated with Ikaros or Gata-1 antibodies as well as isotype-matched rabbit (for Ikaros co-IP) or rat (for Gata-1 co-IP) immunoglobulins (Ig) and immunoblots were probed with Ikaros antibodies. The results show that Ikaros can be immunoprecipitated by Gata-1 antibodies but not by control Ig (Figure 3B), suggesting that Ikaros and Gata-1 can be part of the same protein complex also in yolk sac EryC.
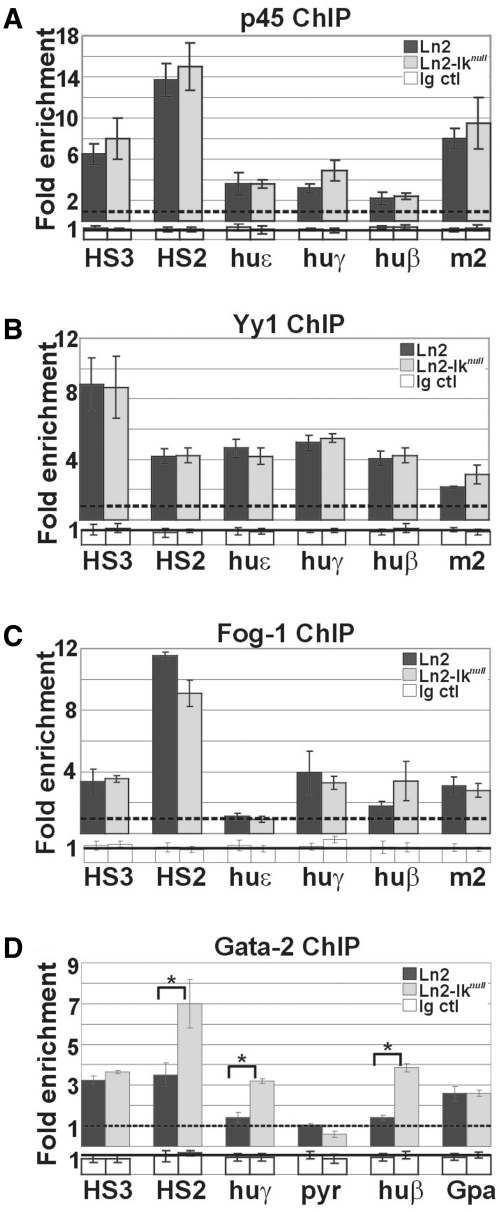
Ikaros-Gata-1 cooperative binding to the huβ-globin locus was then investigated by ChIP. In ln2-Iknull yolk sac EryC, Gata-1 binding is diminished at huγ-promoters and Pyr region whereas is normal at HS3 and HS2 (Figure 3C). In order to investigate whether loss of Ikaros specifically decreases Gata-1 binding or induces a more general alteration of transcription factor recruitment, we analyzed p45/Nf-e2 and Yy1occupancy since these proteins bind to huγ-promoters. No significant difference in p45/Nf-e2 (Figure 4A) or Yy1 (Figure 4B) recruitment was detected at tested chromatin regions in ln2 versus ln2-Iknull yolk sac EryC. We then examined whether the Ikaros-Gata-1 complex could modulate recruitment of the Gata-specific cofactor Fog-1 by ChIP. Fog-1 is recruited to HS3, HS2, huγ- and huβ-promoters in ln2 and, unexpectedly, ln2-Iknull yolk sac EryC (Figure 4C), even though Gata-1 binding to these promoters is close to background levels (Figure 3C). This outcome suggests that Fog-1 recruitment to huγ- and huβ-promoters in ln2-Iknull cells does not depend solely on Gata-1. Since Gata-1 and Gata-2 (i) bind to the same DNA motifs in vivo and in vitro (47–49); (ii) function redundantly to promote primitive erythroblast development (50); (iii) exert their respective functions in a dose-dependent fashion (39,51); and (iv) bind Fog-1 protein (52), we set out to investigate Gata-2 occupancy at globin promoters in ln2-Iknull EryC. Loss of Ikaros correlates with increased Gata-2 recruitment to HS2, huγ- and huβ-promoters, whereas Gata-2 recruitment does not significantly change at the Gpa promoter, which was used as a control for Gata-2 binding (Figure 4D) (53). These results suggest that Ikaros contributes to Gata-1 binding at huγ-promoters and Pyr region in ln2 yolk sac EryC. They also indicate that increased Gata-2/Gata-1 ratio, as observed in absence of Ikaros (Figure 1D), might enhance the interaction between Fog-1 and Gata-2, thus allowing Gata-2 to efficiently support Fog-1 recruitment to huγ-promoters in yolk sac EryC.
Figure 4.
Recruitment of transcriptional regulators to the huβ-globin locus in yolk sac EryC. (A–D) ChIP on ln2 and ln2-Iknull yolk sac EryC carried out with p45/Nf-e2 (p45), Yy1, Fog-1 and Gata-2 antibodies; fold enrichments (y-axis) were calculated as described in Figure 1E and are plotted as the mean ± SD of the measurements; a value of 1 (dashed line) indicates no enrichment; *P ≤ 0.05 by Student’s t-test; huε, huε-promoter; huγ, huγ-promoters; huβ, huβ-promoter; m2, mouse HS2; Gpa, Glycophorin A promoter; dark gray bars: ln2 yolk sac EryC; light gray bars: ln2-Iknull yolk sac EryC; white bars: isotype-matched Ig (Ig ctl).
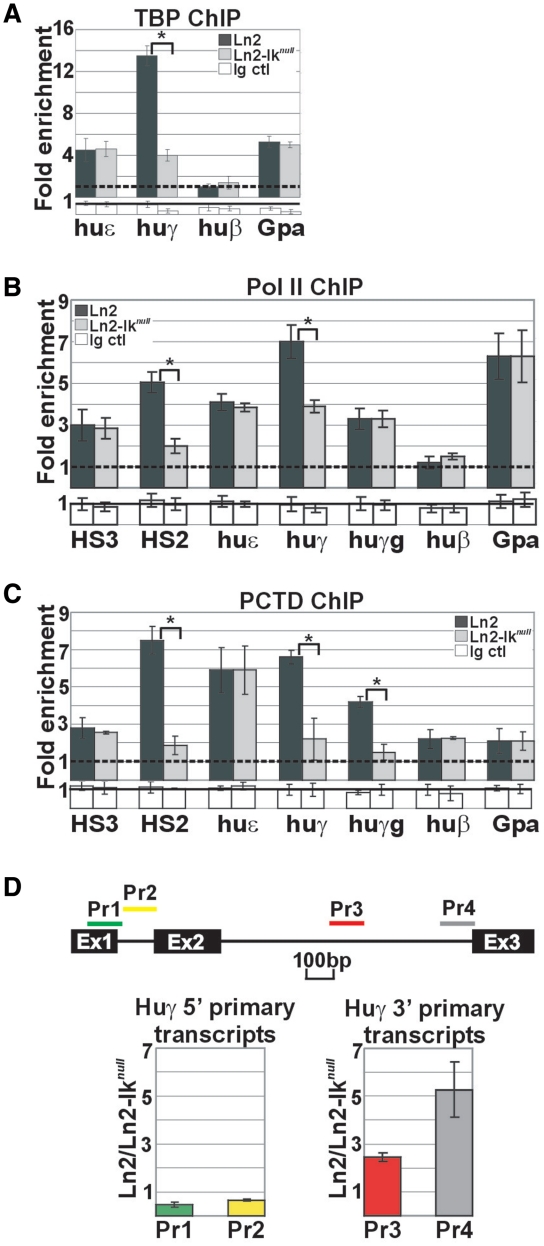
Ikaros contributes to efficient PIC assembly and transcription elongation of huγ-promoters in ln2 yolk sac EryC
A critical step during eukaryotic gene transcription is the assembly of the PIC consisting of Pol II and general transcription factors (27). Both chromatin conformation and gene-specific activators influence PIC assembly and hence, transcriptional efficiency. We proceeded to define Ikaros contribution to PIC assembly at the huγ-promoters by ChIP. As expected, TBP and Pol II were each efficiently detected at huε- and huγ-promoters in ln2 yolk sac EryC; in ln2-Iknull cells, TBP and Pol II recruitment is affected at huγ- but not huε-or Gpa-promoters and Pol II recruitment is also diminished at the HS2 region in ln2-Iknull yolk sac EryC (Figure 5A and B). Since Pol II recruitment to huγ-promoters is only moderately affected by loss of Ikaros, we examined whether Ikaros could also control the efficiency of transcription elongation by studying: (i) levels of Pol II phospho Ser2 (PCTD) recruitment to the huγ-genes, because it is known that the levels of this phosphorylated residue is required and increase during transcription elongation (27); (ii) the relative levels of huγ-globin 5′- and 3′-primary transcripts, because primary transcript levels accurately reflect the rate of transcription. ChIP assays revealed that PCTD recruitment to HS2 as well as huγ-genes and -promoters is significantly affected in absence of Ikaros (Figure 5C). Primary transcript expression was determined by RT–qPCR using random primers for cDNA synthesis and huγ-gene and mouse Gapdh intronic sequence-specific primers for qPCR. In line with reduced PCTD occupancy at huγ regions, there was an accumulation of transcripts corresponding to the 5′-end of the gene and significant reduction of full-length transcripts (Figure 5D). These results are reminiscent of a gene that initiates transcription without completing elongation, thus supporting a possible role of Ikaros during productive huγ-gene transcription elongation.
Figure 5.
PIC assembly at huγ-genes in ln2 and ln2-Iknull yolk sac EryC. (A–C) ChIP on ln2 and ln2-Iknull yolk sac EryC carried out with TBP, Pol II and Pol II phospho Ser2 (PCTD) antibodies; fold enrichments (y-axis) were calculated as described in Figure 1E and are plotted as the mean ± SD of the measurements; a value of 1 (dashed line) indicates no enrichment; *P ≤ 0.05 by Student’s t-test; huε, huε-promoter; huγ, huγ-promoters; huγg, huγ-genes; huβ, huβ-promoter; Gpa, Glycophorin A promoter; dark gray bars: ln2 yolk sac EryC; light gray bars: ln2-Iknull yolk sac EryC; white bars: isotype-matched Ig (Ig ctl); (D) RT–qPCR of huγ-gene 5′- and 3′-primary transcripts performed on equal amounts of ln2 and ln2-Iknull yolk sac EryC using random primers for cDNA synthesis; transcript quantification was calculated according to Pfaffl (73) using the huγ-globin specific primer sets (Pr) depicted as colored bars and mouse Gapdh primary transcripts as internal control; ratios are plotted as the mean ± SD of the measurements; n ≥ 3; P ≤ 0.05; Ex: indicates the three exons of the huγ-gene.
Ikaros interacts with Cdk9
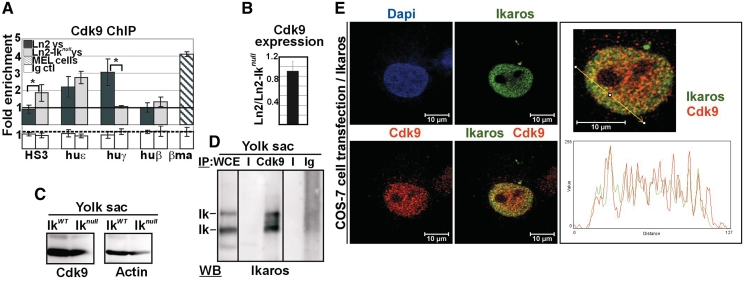
In higher eukaryotes, Pol II Ser2 phosphorylation is mediated primarily by the Cdk9 catalytic subunit of the P-TEFb complex (27). Considering that the Cdk9 inhibitor DRB is known to affect transcription elongation of huγ-genes (24,25), we investigated whether the P-TEFb complex is involved in huγ-gene transcription elongation and whether Ikaros influences the recruitment of P-TEFb to the huγ-genes. Interestingly, ChIP revealed that Cdk9 is recruited to huε- and huγ-promoters in ln2 yolk sac EryC, and that Cdk9 occupancy is significantly reduced at huγ- but not huε-promoters is in ln2-Iknull yolk sac EryC (Figure 6A). To evaluate the significance of the 3-fold enrichment observed in ln2 EryC, we performed ChIP with Cdk9 antibodies in DMSO-induced MEL cells since it has recently been shown that Cdk9 is recruited to the βmaj promoter in these cells (54). The level of Cdk9 enrichment at βmaj promoter in MEL cells is very similar to the occupancy levels of Cdk9 at huγ-promoters in ln2 yolk sac EryC, thus confirming the significance of the results obtained in ln2 yolk sac EryC. Importantly, loss of Ikaros does not modulate the level of Cdk9 transcripts (Figure 6B) or proteins (Figure 6C) in yolk sac EryC.
Figure 6.
Ikaros-mediated recruitment of Cdk9 to the huγ-genes in yolk sac EryC. (A) ChIP on ln2 and ln2-Iknull yolk sac EryC carried out with Cdk9 antibodies; fold enrichments (y-axis) were calculated as described in Figure 1E and are plotted as the mean ± SD of the measurements; a value of 1 (dashed line) indicates no enrichment; *P ≤ 0.05 by Student’s t-test; huε, huε-promoter; huγ, huγ-promoters; huβ, huβ-promoter; βma, βmajor promoter; dark gray bars: ln2 yolk sac EryC; light gray bars: ln2-Iknull yolk sac EryC; dark gray dashed bars: MEL cells; white bars: isotype-matched Ig (Ig ctl); (B) RT–qPCR of Cdk9 gene transcripts performed on equal amounts of ln2 and ln2-Iknull yolk sac EryC; transcript quantification was calculated according to Pfaffl (73) using mouse Actin cDNA as internal control; ln2/ln2-Iknull ratios are plotted as the mean ± SD of the measurements; n = 6; (C) WB performed on total cell lysates prepared from wild type or Iknull yolk sac EryC with Cdk9 or Actin antibodies; (D) protein co-IP of total cell lysates prepared from ln2 yolk sac EryC; cell lysates were immunoprecipitated with Cdk9 antibodies or isotype-matched Ig and WB were carried out with Ikaros antibodies; WCE: yolk sac EryC total cell lysate; (E) Confocal immunofluorescence of COS-7 cells expressing Flag/HA-Ikaros protein; cells were stained with mouse antibodies to the HA tag and with rabbit antibodies to Cdk9; the secondary staining was carried out with FITC-conjugated anti-mouse as well as TR-conjugated anti-rabbit antibodies; a single COS-7 cell is shown where Ikaros is detected as green signals, Cdk9 as red signals, and Ikaros-Cdk9 co-localization as yellow signals in the magnified merged image; line profile plots of Ikaros (in green) and Cdk9 (in red) were obtained by the WCIF ImageJ(®) program (NIH).
Cdk9 interacts with a number of transcription factors (27). To investigate whether Ikaros and Cdk9 interact in yolk sac EryC, we performed protein co-IP with Cdk9 antibodies or control Ig. Immunoblots probed with Ikaros antibodies revealed that Ikaros can be immunoprecipitated by Cdk9 antibodies but not by control Ig, hence suggesting that these proteins physically interact (Figure 6D). To assess Ikaros-Cdk9 interaction at single-cell level, we analyzed the subcellular localization of these proteins by IF and deconvolution microscopy (Figure 6E). COS-7 cells expressing Flag/HA-Ikaros protein were fixed with formaldehyde, permeabilized and immunolabeled with antibodies specific for HA (i.e. Ikaros detection) or Cdk9. Ikaros and Cdk9 were detected using secondary antibodies conjugated to FITC or TR fluorochromes, respectively. COS-7 cells transfected with the empty vector (mock control) showed undetectable levels of green stain (Supplementary Figure S2B). Confocal microscopy analysis revealed that both Ikaros (green signals) and Cdk9 (red signals) are distributed throughout the nucleus and that these proteins significantly overlap, even though they do not colocalize at each and every nuclear sites, in accordance with known evidence that Cdk9 is present in several other protein complexes (26).
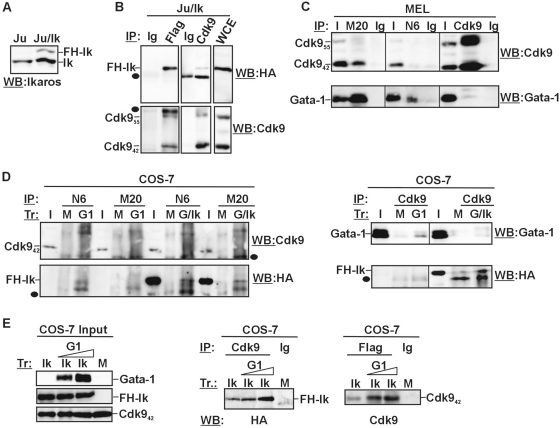
Since Ikaros is known to act as transcriptional repressor and activator of specific genes in T lymphocytes, it was intriguing to know whether Ikaros could possibly interact with Cdk9 in the T lymphocyte-like Jurkat cell line. Jurkat cells were stably transfected with the pOZN-FH-Ik expression vector that contains a bicistronic transcriptional unit, which allows expression of two proteins from a single transcript. This design ensures tight coupling between expression of the gene of interest and the selection marker, the interleukin-2 receptor α chain (IL2Rα) (18,38). Three rounds of magnetic affinity sorting with antibodies against the IL-2α receptor were sufficient to obtain a pure population of Jurkat cells (namely, Ju/Ik clone) that express Flag/HA-Ikaros at lower levels than endogenous Ikaros (Figure 7A). Total cell lysates were immunoprecipitated with Flag, Cdk9 antibodies or control Ig and WB performed with HA or Cdk9 antibodies. As shown in Figure 7B, Flag/HA-Ikaros is immunoprecipitated by both Flag and Cdk9 antibodies. As well, Cdk9 is immunoprecipitated by both Flag and Cdk9 antibodies, indicating that Ikaros and Cdk9 are part of the same protein complex. These results suggest that the Ikaros-Cdk9 interaction can also occur in lymphoid cells and does not require Ikaros overexpression because Flag/HA-Ikaros is expressed at lower levels than endogenous Ikaros.
Figure 7.
Ikaros interacts with Cdk9 in Jurkat and COS-7 cell lines. (A) WB performed on total cell lysates prepared from Jurkat cells (Ju) or Jurkat cells stably transfected with the pOZN-FH-Ik expression vector (Ju/Ik); endogenous (Ik) as well as double Flag/HA-Ikaros-specific bands (FH-Ik) are indicated; (B) Protein co-IP of total cell lysates prepared from Ju/Ik cells; filled circles represent Ig bands; WCE: Ju/Ik total cell lysate; (C) Protein co-IP of total cell lysates prepared from MEL cells; I, Input; M20, anti-Gata-1 antibodies, clone M20; N6, anti-Gata-1 antibodies, cloneN6; Ig, isotype-matched Ig; (D and E) Protein co-IP of total cell lysates prepared from COS-7 cells transfected (Tr) with empty vector (mock, M), Flag/HA-Ikaros (Ik), Gata-1 (G1) or Flag/HA-Ikaros together with Gata-1 (G/Ik); antibodies used for protein co-IP (IP) and WB are indicated on each panel; I, Input; M20, anti-Gata-1 antibodies, clone M20; N6, anti-Gata-1 antibodies, cloneN6; filled circles represent Ig bands.
Since Ikaros interacts with Gata-1 (18) (Figure 3B), and Gata-1 has been shown to interact with the P-TEFb component Cyclin T1 (CycT1) in megakaryocytes (55), we sought to investigate whether Gata-1 could interact with Cdk9 in mouse erythroleukemic MEL cells. Protein co-IP revealed that Cdk9 can be efficiently immunoprecipitated by one out of two antibodies tested directed against Gata-1 (clone M20) and that Gata-1 is immunoprecipitated, even though weakly, by Cdk9 antibodies (Figure 7C), thus indicating that Gata-1 and Cdk9 are part of the same protein complex in MEL cells. Next, we asked whether Gata-1 could either interfere with or enhance the interaction between Ikaros and Cdk9. To this end, COS-7 cells were transfected with pCMV-FH-Ik, pCMV-Gata-1 or pCMV-FH-Ik together with pCMV-Gata-1 expression plasmid. Cell lysates were immunoprecipitated with Gata-1 (N6 as well as M20 clone), Cdk9, Flag antibodies or control Ig and precipitated proteins were analyzed by WB with Cdk9, HA or Gata-1 antibodies. As expected, Ikaros and Gata-1 interact in COS-7 transfected cells that express both proteins, as revealed by Gata-1 co-IP and WB detection with anti-HA antibodies (Figure 7D, bottom panel). However, in COS-7 cells that express either Gata-1 or Gata-1 together with Flag/HA-Ikaros, Gata-1 is not detected upon Cdk9 co-IP nor is Cdk9 detected upon Gata-1 co-IP (Figure 7D), thus suggesting that Gata-1 and Cdk9 do not stably interact in non-hematopoietic, COS-7 cells. Using the same transfected cells and under the same stringency conditions, Flag/HA-Ikaros is detected upon Cdk9 co-IP and Cdk9 is detected upon Flag co-IP (Figure 7D and E), indicating that Ikaros and Cdk9 co-immunoprecipitate also in COS-7 cells and that additional hematopoietic-specific proteins are not required for this interaction. It is to notice that the interaction between Ikaros and Cdk9 can be maintained at higher stringency conditions, i.e. 200 mM NaCl or 1% NP-40 (Supplementary Figure S3A). Interestingly, Gata-1 appears to increase the degree of Ikaros-Cdk9 interaction, since the intensity of the immunoprecipitated bands is proportional to the levels of Gata-1 expression (Figure 7E). Altogether, these data indicate that (i) Ikaros and Gata-1 interact also in COS-7 transfected cells; (ii) Gata-1 interacts with Cdk9 in erythroid cells; (iii) Ikaros interacts with Cdk9 in erythroid, lymphoid as well as COS-7 transfected cells; and (iv) Ikaros-Cdk9 interaction does not require Gata-1 even though Gata-1 appears to enhance the strength of this interaction.
DISCUSSION
In the present study, we have shown that Ikaros acts as a stage-specific activator of huγ-genes in yolk sac EryC by (i) favoring Brg1 and Gata-1 recruitment to huγ-promoters and contributing to chromatin interaction between the βLCR and these promoters and (ii) supporting efficient PIC assembly and Cdk9 recruitment to huγ-promoters and hence, Pol II Ser2 phosphorylation which is required for transcription elongation. This suggests that through combined action with transcription factor and co-factor partners, Ikaros can exert control over both transcription initiation and elongation.
Ikaros and mouse embryonic development
Previous analysis of Iknull mice highlighted the importance of Ikaros for the regulation of hematopoietic genes (7). In particular Scl/Tal-1, Flk-2 receptor and c-kit receptor gene expression is affected in Iknull bone marrow hematopoietic stem cells (HSC) (56). However, in Iknull fetal liver EryC, Scl/Tal-1 expression levels are unchanged, whereas Eklf and Gata-2 expression levels are, respectively, decreased and increased (18,34). Finally, in Iknull yolk sac EryC, we report that Eklf, Gata-1, Fog-1 and Scl/Tal-1 expression levels are unchanged, whereas Gata-2 is overexpressed. Therefore, Ikaros, likely in association with specific chromatin regulators, can activate or repress a variety of hematopoietic-specific genes in a cell type- and developmental stage-specific manner.
Ikaros is essential for specification in lymphoid lineages, and inactivation of this protein exerts a profound effect on the production and differentiation of fetal lymphocyte progenitors (37). However, recent studies have identified Ikaros as an important regulator of lympho-myeloid lineage decisions in early hematopoietic progenitor cells (7). Ikaros can also influence erythroid cell differentiation. For example, adult Iknull mice display extramedullary hematopoiesis, bone marrow hypoplasia, and slight reduction in the number of BFU-E (Burst-Forming Unit Erythroid) and CFU-E (Colony-Forming Unit Erythroid) progenitors; however, reduced CFU-E and BFU-E activity is reported to be mainly due to a deficit in the HSC population rather than aberrant erythroid differentiation (56). Accordingly, erythropoiesis appears to be normal at earlier stages of development in fetal liver (18,37) and yolk sac (this work) isolated from Iknull mice where erythroid differentiation progresses to the point of producing Gpa transcripts, normal numbers of maturing proerythroblasts and typical levels of murine globin gene transcripts. Based on these findings, we conclude that reduced levels of huγ-gene expression in ln2-Iknull cells is not the consequence of impaired erythroid differentiation or maturation, and hence that Ikaros is recruited to, and directly contributes to regulation of, the huγ-genes in yolk sac EryC.
Ikaros, Brg1 and Gata-1 cooperate to control huγ-gene expression in yolk sac EryC
We have recently shown that Ikaros participates to huγ-gene silencing at the time of γ-to-β globin switching which takes place at ∼e12 in transgenic ln2 fetal liver by targeting a Gata-1/Fog-1/Hdac1/Mi-2-containing multiprotein complex to huγ-promoters (18). Here, we have shown that Ikaros, along with Gata-1 and Brg1, rather contributes to huγ-gene expression at an earlier developmental stage, i.e. in e10.5 yolk sac EryC. The observation that Ikaros can act as transcription activator and repressor for the same gene in a stage- and cell-specific manner is reminiscent of previous studies showing that Ikaros represses Il4 expression in mast cells (57), but activates that gene in Th2 cells, and also appears to exert distinct effects on Il4 and Ifnγ gene expression in the same cell type (Th1 and Th2) (58). The ability of Ikaros to preferentially interact with Mi-2, Brg1 or other known Ikaros co-regulators, and hence, to direct them to distinct gene regulatory regions, might depend on Ikaros post-translational modifications. Indeed, sumoylated Ikaros preferentially interacts with Brg1 versus Mi-2 (13). Thus, increased sumoylation levels in yolk sac relative to fetal liver EryC could account for the tendency of Ikaros to interact with Brg1, and possibly act as transcription activator. This could not be directly addressed since sumoylation is a highly dynamic process and a suitable cellular model for human primitive EryC is currently unavailable. A second, non-mutually exclusive explanation for different levels of Ikaros interaction with Mi-2 versus Brg1 is that expression of these co-regulators might fluctuate during development. In accordance with such hypothesis, we observed that Brg1 is expressed at higher levels in yolk sac than fetal liver EryC, whereas Mi-2 expression is higher in fetal liver EryC (Supplementary Figure S3B). Brg1 is the principal catalytic subunit of a SWI/SNF-like complex in Ter119+ EryC, and is essential for viability in mice (59). The use of hypomorphic (41) and conditional Brg1null mutations (42) have demonstrated that Brg1 is required for chromatin remodelling during both embryonic and adult β-globin gene transcription, and for maximal Pol II occupancy at the mouse βmaj promoter (60). A number of transcription factors are known to bind and recruit Brg1 to the globin locus (40,61–63); nonetheless using a Gata-1-inducible cell line, Kim et al. (22) reported that Gata-1 recruits Brg1 to the adult βmaj promoter more rapidly than other transcription factors tested.
The current results, together with previous published data (18), indicate that Gata-1 is a relevant Ikaros-interacting transcription factor and their combined effect is required for globin gene regulation in EryC. Indeed, Gata-1 and Ikaros are recruited in vivo to the huβ-globin locus, influence long-range chromatin interactions, contribute to huγ-gene regulation and co-immunoprecipitate in vivo in primary yolk sac (herein) and fetal liver (18) EryC. Gata-1 has been associated to both huγ-gene activation and repression, depending on the experimental model. For instance, Gata-1 contributes to huγ-gene silencing at the time of γ-to-β globin switching in mouse fetal liver (18) or in adult (32,33) EryC. By cooperating with BCL11a and SOX6, GATA-1 has recently been shown to contribute to huγ-gene silencing in adult human erythroid progenitors (19) and interestingly, Bcl11a is not expressed either in wild-type (30) or Iknull primitive EryC (Supplementary Figure S3C). However, GATA-1 has also been associated with huγ-gene activation (64). Finally GATA-1 overexpression in K562 cells does not change huγ-gene expression (65).
Despite the fact that Ikaros does not affect Gata-1 occupancy at βLCR HS2, recruitment of Gata-2 and Pol II to this region is significantly increased and decreased, respectively, in ln2-Iknull yolk sac EryC. It has recently been reported that Gata-1 occupancy at the well-defined −1.8 kb site of the Gata-2 locus is associated with Pol II expulsion and stable gene repression (66). Therefore, it is possible that, similarly to Gata-1 at the −1.8 kb site of the Gata-2 locus, increased Gata-2 occupancy at βLCR HS2 of the huβ-globin locus could partially display Pol II at this distal regulatory element.
In conclusion, even though we cannot exclude that Ikaros may also facilitate huγ-gene expression indirectly, e.g. by contributing to Gata-2 gene repression in yolk sac EryC, our data indicate that Ikaros directly contributes to huγ-gene expression mainly by conferring Gata-1 occupancy at huγ-promoters (see below) and, together with Brg1, by stabilizing huγ-promoter chromatin activation.
Ikaros promotes efficient huγ-gene transcription elongation
Here, we show that lack of Ikaros affects TBP and Pol II recruitment to huγ-promoters, and reduces Pol II Ser2 phosphorylation levels. Ikaros can interact with components of the basal transcription machinery such as TBP and TFIIB (67), and with Brg1, which is critical for long-range chromatin interaction and, such as reported (60), Pol II transfer. Even though transcription elongation has been regarded as an important regulatory step for β-like globin gene regulation (23,68,69), the severe reduction of Pol II Ser2 phosphorylation levels observed at huγ-promoters in ln2-Iknull yolk sac EryC was unanticipated in light of known properties of Ikaros. Our results indicate that Ikaros not only participates in PIC assembly but in addition contributes to Pol II Ser2 phosphorylation by recruiting the P-TEFb component Cdk9 to huγ-gene promoters.
Cdk9 expression levels increase in terminally differentiated cells (26). However, in the human erythroleukemic K562 cell line, Cdk9 knockdown induces erythroid differentiation (55), whereas erythroid differentiation of MEL cells does not lead to Cdk942 downregulation and in fact, engenders upregulation of Cdk955 (28). The reason for these discrepancies is not known, but it might depend on the intrinsic limitations of studying erythroid differentiation using transformed cell lines. Here we show that levels of Cdk9 expression are very similar in ln2 and ln2-Iknull yolk sac EryC and also in yolk sac EryC when compared to fetal liver EryC (Supplementary Figure S3D). We studied Ikaros-Cdk9 protein interaction in cells expressing predominantly Cdk942 (yolk sac EryC and COS-7 cells) or both isoforms (Jurkat cells) and based on our results we conclude that Ikaros preferentially interacts with Cdk942.
Lack of Ikaros leads to increased and reduced Cdk9 recruitment to HS3 and huγ-promoters respectively, and correlates with reduced long-range chromatin interactions between the βLCR and the huγ-promoters. It is therefore possible that, as proposed for Pol II (70), Cdk9 first accumulates at the βLCR and then, is transferred to the huγ-promoters when the two regulatory regions are in closer proximity. It is widely accepted that developmental control of gene expression can be associated with poised Pol II and the distribution of Pol II across Drosophila and human genomes have provided evidence that elongation control might direct the pattern of gene expression during development (71). In mammalian cells, the P-TEFb complex accounts for most of Pol II Ser2 phosphorylation activity (26) and therefore is the best-known activity capable of reactivating poised Pol II (27). Thus, Ikaros-dependent Cdk9 transfer to huγ-promoters might be important for the transition from poised Pol II to productive elongation at the huγ-genes.
Recruitment of P-TEFb to highly acetylated gene promoters can be mediated by the bromodomain protein Brd4, which interacts directly with CycT1 (27). However, certain transcription factors, e.g. c-Myc, CIITA, NF-kB, MyoD, STAT3 and the androgen receptor can recruit P-TEFb to defined promoter regions by interacting with either CycT1 or Cdk9 [(26,72) and references herein] and recently Ldb1 has been shown to recruit P-TEFb to multiple regions of the mouse β-globin locus (54). Interestingly, GATA-1 is reported to interact with CYCT1 in a megakaryocyte-like cell line whereas GATA-1 interaction with CDK9 in the same cells was not addressed (55). Gata-1 and Cdk9 co-immunoprecipitate in MEL cells but not in COS-7 cells expressing Gata-1 or Gata-1 together with Ikaros, under the same stringency conditions that allow co-IP of Ikaros and Cdk9. Thus, it appears that Ikaros can interact with Cdk9 in several cell lines, whereas Gata-1 interaction with Cdk9 would rather necessitate an erythroid environment. Interestingly, even in absence of a detectable interaction between Gata-1 and Cdk9 in COS-7 cells, the presence of Gata-1 can enhance the strength of the interaction between Ikaros and Cdk9. Whether the erythroid-specific association of Ikaros, Gata-1 and Cdk9 depends upon their relative level of expression or post-translational modifications or the presence of additional erythroid-specific proteins is thus far unknown. Nevertheless, it is tempting to assume that in vivo cooperation between Ikaros and Gata-1 is designed to facilitate Pol II Ser2 phosphorylation, possibly by targeting two distinct components of the P-TEFb complex to huγ-gene promoters.
Based on our results, we propose that the recruitment of Ikaros to huγ-promoters in yolk sac EryC, promotes stable PIC formation and contributes to Cdk9 recruitment, which leads to efficient transcription elongation of the huγ-genes. Therefore, unlike Ldb1-dependent recruitment of P-TEFb that occurs at multiple regions of the mouse β-globin locus (54), our results suggest that Ikaros contributes to Cdk9/P-TEFb recruitment specifically to promoters of the transcriptionally active huγ-genes. According to the observation that Ikaros and Cdk9 co-immunoprecipitate also in lymphoid-like cells (Figure 7B), it is likely that Ikaros influences transcription elongation in a similar manner at other transcriptionally active Ikaros target genes and in other hematopoietic cells.
In conclusion, we show that Ikaros contributes to efficient transcription initiation and elongation of the huγ-genes in primitive EryC. As such, this study contributes to a better understanding of the molecular mechanisms of Ikaros action to control gene expression in hematopoietic cells. Therefore, results presented inhere should provide a basis for understanding malignant hematopoietic processes characterized by Ikaros- or GATA-1-dependent gene dysregulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research (CIHR) (MOP 97738) held by E.M.; Fond de la recherche en santé du Québec (FRSQ to J.R. and E.M.). Funding for open access charge: Canadian Institutes of Health Research (CIHR) (MOP 97738).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Dean and E. Drobetsky for critical reading of the article, K. Georgopoulos for the Ikaros null mouse line, F. Grosveld for ln2 mouse line and G. D’Angelo for Wright-Giemsa staining analysis.
REFERENCES
- 1.Keys JR, Tallack MR, Zhan Y, Papathanasiou P, Goodnow CC, Gaensler KM, Crossley M, Dekker J, Perkins AC. A mechanism for Ikaros regulation of human globin gene switching. Br. J. Haematol. 2008;141:398–406. doi: 10.1111/j.1365-2141.2008.07065.x. [DOI] [PubMed] [Google Scholar]
- 2.Ng SY, Yoshida T, Georgopoulos K. Ikaros and chromatin regulation in early hematopoiesis. Curr. Opin. Immunol. 2007;19:116–122. doi: 10.1016/j.coi.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 3.Harker N, Naito T, Cortes M, Hostert A, Hirschberg S, Tolaini M, Roderick K, Georgopoulos K, Kioussis D. The CD8alpha gene locus is regulated by the Ikaros family of proteins. Mol. Cell. 2002;10:1403–1415. doi: 10.1016/s1097-2765(02)00711-6. [DOI] [PubMed] [Google Scholar]
- 4.Molnar A, Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol. Cell Biol. 1994;14:8292–8303. doi: 10.1128/mcb.14.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun L, Liu A, Georgopoulos K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 1996;15:5358–5369. [PMC free article] [PubMed] [Google Scholar]
- 6.Koipally J, Heller EJ, Seavitt JR, Georgopoulos K. Unconventional potentiation of gene expression by Ikaros. J. Biol. Chem. 2002;277:13007–13015. doi: 10.1074/jbc.M111371200. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida T, Ng SY, Georgopoulos K. Awakening lineage potential by Ikaros-mediated transcriptional priming. Curr. Opin. Immunol. 22:154–160. doi: 10.1016/j.coi.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koipally J, Renold A, Kim J, Georgopoulos K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 1999;18:3090–3100. doi: 10.1093/emboj/18.11.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabbattini P, Lundgren M, Georgiou A, Chow C, Warnes G, Dillon N. Binding of Ikaros to the lambda5 promoter silences transcription through a mechanism that does not require heterochromatin formation. EMBO J. 2001;20:2812–2822. doi: 10.1093/emboj/20.11.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 11.Sridharan R, Smale ST. Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes. J. Biol. Chem. 2007;282:30227–30238. doi: 10.1074/jbc.M702541200. [DOI] [PubMed] [Google Scholar]
- 12.Naito T, Gomez-Del Arco P, Williams CJ, Georgopoulos K. Antagonistic interactions between Ikaros and the chromatin remodeler Mi-2beta determine silencer activity and Cd4 gene expression. Immunity. 2007;27:723–734. doi: 10.1016/j.immuni.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Gomez-del Arco P, Koipally J, Georgopoulos K. Ikaros SUMOylation: switching out of repression. Mol. Cell Biol. 2005;25:2688–2697. doi: 10.1128/MCB.25.7.2688-2697.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Neill DW, Schoetz SS, Lopez RA, Castle M, Rabinowitz L, Shor E, Krawchuk D, Goll MG, Renz M, Seelig HP, et al. An ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol. Cell Biol. 2000;20:7572–7582. doi: 10.1128/mcb.20.20.7572-7582.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiefer CM, Hou C, Little JA, Dean A. Epigenetics of beta-globin gene regulation. Mutat. Res. 2008;647:68–76. doi: 10.1016/j.mrfmmm.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 17.Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bottardi S, Ross J, Bourgoin V, Fotouhi-Ardakani N, Affarel B, Trudel M, Milot E. Ikaros and GATA-1 combinatorial effect is required for silencing of human gamma-globin genes. Mol. Cell Biol. 2009;29:1526–1537. doi: 10.1128/MCB.01523-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu J, Sankaran VG, Ni M, Menne TF, Puram RV, Kim W, Orkin SH. Transcriptional silencing of {gamma}-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev. 24:783–798. doi: 10.1101/gad.1897310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 21.Song SH, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol. Cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SI, Bultman SJ, Kiefer CM, Dean A, Bresnick EH. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc. Natl Acad. Sci. USA. 2009;106:2259–2264. doi: 10.1073/pnas.0806420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawado T, Halow J, Bender MA, Groudine M. The beta -globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev. 2003;17:1009–1018. doi: 10.1101/gad.1072303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gribnau J, de Boer E, Trimborn T, Wijgerde M, Milot E, Grosveld F, Fraser P. Chromatin interaction mechanism of transcriptional control in vivo. EMBO J. 1998;17:6020–6027. doi: 10.1093/emboj/17.20.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson KD, Grass JA, Park C, Im H, Choi K, Bresnick EH. Highly restricted localization of RNA polymerase II within a locus control region of a tissue-specific chromatin domain. Mol. Cell Biol. 2003;23:6484–6493. doi: 10.1128/MCB.23.18.6484-6493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romano G, Giordano A. Role of the cyclin-dependent kinase 9-related pathway in mammalian gene expression and human diseases. Cell Cycle. 2008;7:3664–3668. doi: 10.4161/cc.7.23.7122. [DOI] [PubMed] [Google Scholar]
- 27.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 28.Meier N, Krpic S, Rodriguez P, Strouboulis J, Monti M, Krijgsveld J, Gering M, Patient R, Hostert A, Grosveld F. Novel binding partners of Ldb1 are required for haematopoietic development. Development. 2006;133:4913–4923. doi: 10.1242/dev.02656. [DOI] [PubMed] [Google Scholar]
- 29.Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- 30.Sankaran VG, Xu J, Ragoczy T, Ippolito GC, Walkley CR, Maika SD, Fujiwara Y, Ito M, Groudine M, Bender MA, et al. Developmental and species-divergent globin switching are driven by BCL11A. Nature. 2009;460:1093–1097. doi: 10.1038/nature08243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strouboulis J, Dillon N, Grosveld F. Developmental regulation of a complete 70-kb human beta-globin locus in transgenic mice. Genes Dev. 1992;6:1857–1864. doi: 10.1101/gad.6.10.1857. [DOI] [PubMed] [Google Scholar]
- 32.Harju-Baker S, Costa FC, Fedosyuk H, Neades R, Peterson KR. Silencing of Agamma-globin gene expression during adult definitive erythropoiesis mediated by GATA-1-FOG-1-Mi2 complex binding at the -566 GATA site. Mol. Cell Biol. 2008;28:3101–3113. doi: 10.1128/MCB.01858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu LR, Du ZW, Zhao HL, Liu XL, Huang XD, Shen J, Ju LM, Fang FD, Zhang JW. T to C substitution at -175 or -173 of the gamma-globin promoter affects GATA-1 and Oct-1 binding in vitro differently but can independently reproduce the hereditary persistence of fetal hemoglobin phenotype in transgenic mice. J. Biol. Chem. 2005;280:7452–7459. doi: 10.1074/jbc.M411407200. [DOI] [PubMed] [Google Scholar]
- 34.Lopez RA, Schoetz S, DeAngelis K, O'Neill D, Bank A. Multiple hematopoietic defects and delayed globin switching in Ikaros null mice. Proc. Natl Acad. Sci. USA. 2002;99:602–607. doi: 10.1073/pnas.022412699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanabe O, Katsuoka F, Campbell AD, Song W, Yamamoto M, Tanimoto K, Engel JD. An embryonic/fetal beta-type globin gene repressor contains a nuclear receptor TR2/TR4 heterodimer. EMBO J. 2002;21:3434–3442. doi: 10.1093/emboj/cdf340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sankaran VG, Menne TF, Xu J, Akie TE, Lettre G, Van Handel B, Mikkola HK, Hirschhorn JN, Cantor AB, Orkin SH. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 37.Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 38.Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- 39.Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol. Cell Biol. 2005;25:1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Neill D, Yang J, Erdjument-Bromage H, Bornschlegel K, Tempst P, Bank A. Tissue-specific and developmental stage-specific DNA binding by a mammalian SWI/SNF complex associated with human fetal-to-adult globin gene switching. Proc. Natl Acad. Sci. USA. 1999;96:349–354. doi: 10.1073/pnas.96.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bultman SJ, Gebuhr TC, Magnuson T. A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in {beta}-globin expression and erythroid development. Genes Dev. 2005;41:2849–2861. doi: 10.1101/gad.1364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffin CT, Brennan J, Magnuson T. The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development. 2008;135:493–500. doi: 10.1242/dev.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 44.Miccio A, Wang Y, Hong W, Gregory GD, Wang H, Yu X, Choi JK, Shelat S, Tong W, Poncz M, et al. NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. EMBO J. 29:442–456. doi: 10.1038/emboj.2009.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez P, Bonte E, Krijgsveld J, Kolodziej KE, Guyot B, Heck AJ, Vyas P, de Boer E, Grosveld F, Strouboulis J. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 2005;24:2354–2366. doi: 10.1038/sj.emboj.7600702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujiwara T, O'Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol. Cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ko LJ, Engel JD. DNA-binding specificities of the GATA transcription factor family. Mol. Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol. Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujiwara Y, Chang AN, Williams AM, Orkin SH. Functional overlap of GATA-1 and GATA-2 in primitive hematopoietic development. Blood. 2004;103:583–585. doi: 10.1182/blood-2003-08-2870. [DOI] [PubMed] [Google Scholar]
- 51.Johnson KD, Kim SI, Bresnick EH. Differential sensitivities of transcription factor target genes underlie cell type-specific gene expression profiles. Proc. Natl Acad. Sci. USA. 2006;103:15939–15944. doi: 10.1073/pnas.0604041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tevosian SG, Deconinck AE, Cantor AB, Rieff HI, Fujiwara Y, Corfas G, Orkin SH. FOG-2: A novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc. Natl Acad. Sci. USA. 1999;96:950–955. doi: 10.1073/pnas.96.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lahlil R, Lecuyer E, Herblot S, Hoang T. SCL assembles a multifactorial complex that determines glycophorin A expression. Mol. Cell Biol. 2004;24:1439–1452. doi: 10.1128/MCB.24.4.1439-1452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song SH, Kim A, Ragoczy T, Bender MA, Groudine M, Dean A. Multiple functions of Ldb1 required for {beta}-globin activation during erythroid differentiation. Blood. 2010;116:2356–2364. doi: 10.1182/blood-2010-03-272252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elagib KE, Mihaylov IS, Delehanty LL, Bullock GC, Ouma KD, Caronia JF, Gonias SL, Goldfarb AN. Cross-talk of GATA-1 and P-TEFb in megakaryocyte differentiation. Blood. 2008;112:4884–4894. doi: 10.1182/blood-2008-03-145722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nichogiannopoulou A, Trevisan M, Neben S, Friedrich C, Georgopoulos K. Defects in hemopoietic stem cell activity in Ikaros mutant mice. J. Exp. Med. 1999;190:1201–1214. doi: 10.1084/jem.190.9.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gregory GD, Raju SS, Winandy S, Brown MA. Mast cell IL-4 expression is regulated by Ikaros and influences encephalitogenic Th1 responses in EAE. J. Clin. Invest. 2006;116:1327–1336. doi: 10.1172/JCI27227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quirion MR, Gregory GD, Umetsu SE, Winandy S, Brown MA. Cutting edge: Ikaros is a regulator of Th2 cell differentiation. J. Immunol. 2009;182:741–745. doi: 10.4049/jimmunol.182.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 60.Kim SI, Bultman SJ, Jing H, Blobel GA, Bresnick EH. Dissecting molecular steps in chromatin domain activation during hematopoietic differentiation. Mol. Cell Biol. 2007;27:4551–4565. doi: 10.1128/MCB.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Armstrong JA, Bieker JJ, Emerson BM. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- 62.Brand M, Ranish JA, Kummer NT, Hamilton J, Igarashi K, Francastel C, Chi TH, Crabtree GR, Aebersold R, Groudine M. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat. Struct. Mol. Biol. 2004;11:73–80. doi: 10.1038/nsmb713. [DOI] [PubMed] [Google Scholar]
- 63.Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell. 2003;11:377–389. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 64.Yao X, Kodeboyina S, Liu L, Dzandu J, Sangerman J, Ofori-Acquah SF, Pace BS. Role of STAT3 and GATA-1 interactions in gamma-globin gene expression. Exp. Hematol. 2009;37:889–900. doi: 10.1016/j.exphem.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ikonomi P, Noguchi CT, Miller W, Kassahun H, Hardison R, Schechter AN. Levels of GATA-1/GATA-2 transcription factors modulate expression of embryonic and fetal hemoglobins. Gene. 2000;261:277–287. doi: 10.1016/s0378-1119(00)00510-2. [DOI] [PubMed] [Google Scholar]
- 66.Snow JW, Trowbridge JJ, Fujiwara T, Emambokus NE, Grass JA, Orkin SH, Bresnick EH. A single cis element maintains repression of the key developmental regulator Gata2. PLoS Genet. 2010;6:e1001103. doi: 10.1371/journal.pgen.1001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koipally J, Georgopoulos K. Ikaros-CtIP interactions do not require C-terminal binding protein and participate in a deacetylase-independent mode of repression. J. Biol. Chem. 2002;277:23143–23149. doi: 10.1074/jbc.M202079200. [DOI] [PubMed] [Google Scholar]
- 68.Bottardi S, Ross J, Pierre-Charles N, Blank V, Milot E. Lineage-specific activators affect beta-globin locus chromatin in multipotent hematopoietic progenitors. EMBO J. 2006;25:3586–3595. doi: 10.1038/sj.emboj.7601232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou Z, Li X, Deng C, Ney PA, Huang S, Bungert J. USF and NF-E2 cooperate to regulate the recruitment and activity of RNA polymerase II in the beta-globin gene locus. J. Biol. Chem. 285:15894–15905. doi: 10.1074/jbc.M109.098376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson KD, Grass JA, Boyer ME, Kiekhaefer CM, Blobel GA, Weiss MJ, Bresnick EH. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc. Natl Acad. Sci. USA. 2002;99:11760–11765. doi: 10.1073/pnas.192285999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Price DH. Poised polymerases: on your mark…get set…go! Mol. Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 72.Garriga J, Grana X. Cellular control of gene expression by T-type cyclin/CDK9 complexes. Gene. 2004;337:15–23. doi: 10.1016/j.gene.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 73.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.