Abstract
Many eukaryotic genes are regulated at the level of transcript elongation. Nucleosomes are likely targets for this regulation. Previously, we have shown that nucleosomes formed on very strong positioning sequences (601 and 603), present a high, orientation-dependent barrier to transcription by RNA polymerase II in vitro. The existence of this polar barrier correlates with the interaction of a 16-bp polar barrier signal (PBS) with the promoter-distal histone H3–H4 dimer. Here, we show that the polar barrier is relieved by ISW2, an ATP-dependent chromatin remodeler, which translocates the nucleosome over a short distance, such that the PBS no longer interacts with the distal H3–H4 dimer, although it remains within the nucleosome. In vivo, insertion of the 603 positioning sequence into the yeast CUP1 gene results in a modest reduction in transcription, but this reduction is orientation-independent, indicating that the polar barrier can be circumvented. However, the 603-nucleosome is present at the expected position in only a small fraction of cells. Thus, the polar barrier is probably non-functional in vivo because the nucleosome is not positioned appropriately, presumably due to nucleosome sliding activities. We suggest that interactions between PBSs and chromatin remodelers might have significant regulatory potential.
INTRODUCTION
Eukaryotic genomes are organized into chromatin in order to pack DNA efficiently into the nucleus and to regulate gene expression. The structural repeat unit of chromatin is the nucleosome, a complex containing two molecules of each core histone (H2A, H2B, H3 and H4) and 147 bp of DNA (1). The question of how transcription occurs in the presence of nucleosomes has been addressed extensively (2). In vivo, histones undergo displacement and exchange during intense transcription (3–6), but at the moderate levels of transcription typical of most genes, nucleosomes survive (5,7,8) and influence the rate of transcript elongation. Indeed, RNA polymerase II (Pol II) tends to stall at promoter-proximal sites (9–11), where it is often associated with the first nucleosome on the gene (12).
In vitro, the nucleosome is a potent inhibitor of transcription by Pol II (13,14). This can be ascribed to several factors, including: (i) intrinsic pausing signals specified by the underlying DNA sequence, which are observed even in the absence of nucleosomes, but tend to be greatly amplified when assembled into nucleosomes (15,16); (ii) nucleosome structure, which directs pausing at two specific sites within the nucleosome, at +15 and +45 with respect to the proximal nucleosome border (14); (iii) nucleosome stability, defined by the affinity of the DNA sequence for the histone octamer (14).
Nucleosomes assembled on the synthetic high-affinity 601, 603 and 605 positioning sequences (17), present a polar barrier to transcription in vitro (14). That is, Pol II transcribes through the nucleosome with much more difficulty when approaching from one side than when approaching from the other side, referred to as the ‘non-permissive’ and ‘permissive’ nucleosome orientations, respectively. Our recent studies suggest that the intra-nucleosomal location of a 16-bp sequence common to the 601, 603 and 605 positioning sequences [‘R(2–3)’] might act as a polar barrier signal (PBS) (18). Mutations in the PBS decrease the strength of the polar barrier and reduce the affinity of the DNA for histones, without affecting nucleosome positioning. It was proposed that the location of the PBS in the distal half of the non-permissive nucleosome is critical for barrier function.
Here, we show that ISW2-mediated translocation of a non-permissive nucleosome over a short distance, such that the PBS no longer interacts with the distal half of the nucleosome, is sufficient to nullify the polar barrier. In vivo, we find that insertion of 603 into the yeast CUP1 gene does not result in a strong polar barrier to transcription by Pol II, probably because the precise nucleosome position necessary for polar barrier function is not maintained.
MATERIALS AND METHODS
See Supplementary Data for more details.
Nucleosome assembly and remodeling in vitro
In the nomenclature used previously (18), the permissive orientations were 601R, 603 and 605; the non-permissive orientations were 601, 603R and 605R, where R indicates the reverse-complement of the published sequence (17). The 601, 603 and 605 templates were constructed using pGEM-3Z/601, pGEM-3Z/603 and pGEM-3Z/605 (17) and PCR primers listed in Supplementary Table S1. One primer was 5′-end-labeled. The resulting DNA fragments were digested with TspRI and reconstituted into nucleosomes by octamer transfer from long chicken erythrocyte chromatin (19). Remodeling was performed with purified ISW2 as described (20). Briefly, nucleosomal templates (5 ng/µl) were incubated with 35 ng ISW2 per 370 ng total DNA at 30°C for 30 min in 25 mM HEPES–KOH (pH 7.6), 5 mM MgCl2, 40 mM KCl, 0.1 mg/ml BSA, 6 mM Tris–HCl (pH 8.0), 5% glycerol, 30 mM NaCl, 0.3 mM ATP. Nucleosomal templates were analyzed by non-denaturing 4.5% PAGE (19).
Assembly of elongation complexes and transcription
Pol II elongation complexes were assembled using purified His-tagged yeast Pol II, DNA templates and 9 nt RNA, and immobilized on Ni-NTA resin (21). Nucleosomal templates, before or after remodeling, were annealed to immobilized EC9 complexes through their TspRI sticky ends at 16°C for 30 min in TB300 (20 mM Tris–HCl (pH 7.9), 5 mM MgCl2, 300 mM KCl, 2 mM 2-mercaptoethanol). ISW2 dissociates from DNA in 300 mM KCl, preventing further remodeling. Annealed complexes were washed twice with TB300 and once with TB40 (as TB300, but with 40 mM KCl) and ligated in TB40 containing 0.5 mM ATP, 1% PEG8000, 0.2 mg/ml BSA and 400 U T4 DNA ligase (NEB) at 16°C for 30 min. Ligated EC9 complexes were washed once with TB40 and twice with TB300. Limited transcription was initiated in TB300 with 20 µM ATP, 20 µM CTP and 20 µCi [α-32P]-GTP (3000 Ci/mmol) for 15 min at room temperature. GTP was added to 10 µM for 5 min to form EC45. Immobilized labeled EC45 complexes were washed twice with TB300, twice with TB40 and eluted with 0.1 M imidazole. Transcription was continued in TB with 0.4 mM NTPs at different KCl concentrations. Transcripts were resolved by denaturing 8% PAGE. Phosphorimages were quantified using ImageQuant (GE). The procedure for quantitation of the amounts of the run-off transcripts is designed to take into account possible variations in the efficiency of ligation and/or in the amount of material loaded on the denaturing gel. First, total amounts of transcripts that are longer than 45 nt (and therefore can be produced only on ligated templates) in each lane were quantified. Second, the fraction of the run-off transcripts to total amount of >45-nt transcripts was quantified. Finally, the yields of the run-off transcript were normalized to the amount of run-off transcripts produced in 1 M KCl (was set at 100%).
Preparation of core particles
Yeast strains were constructed as described in Supplementary Data. Cells were grown in synthetic complete medium at 30°C to A600 ∼0.5. Copper induction was for 20 min in 50 µM CuSO4. Nuclei were prepared from ∼250 A600 units of cells as described (22), resuspended in 2.4 ml 10 mM HEPES-K pH 7.5, 35 mM NaCl, 0.5 mM MgCl2, 0.5 mM CaCl2, divided into six aliquots of 400 µl and digested with micrococcal nuclease (Worthington) at 100, 150, 225, 340, 500 or 750 U/ml for 2 min at room temperature. The reaction was stopped with 10 mM EDTA and 1% SDS. DNA was purified and dissolved in 50 µl 50 mM Tris–HCl pH 8.0, 10 mM EDTA, 100 µg/ml RNase A. The extent of digestion was determined by analysis in agarose gels stained with ethidium bromide. Core particle DNA from samples in which almost all of the chromatin had been digested to core particles was gel-purified and repaired using the PreCR DNA repair kit (NEB). The quality of the core particle DNA was verified in 8% native and 10% denaturing polyacrylamide gels. Core particle DNA was fully trimmed to 147–150 bp.
RESULTS
ISW2-induced nucleosome translocation relieves the polar barrier
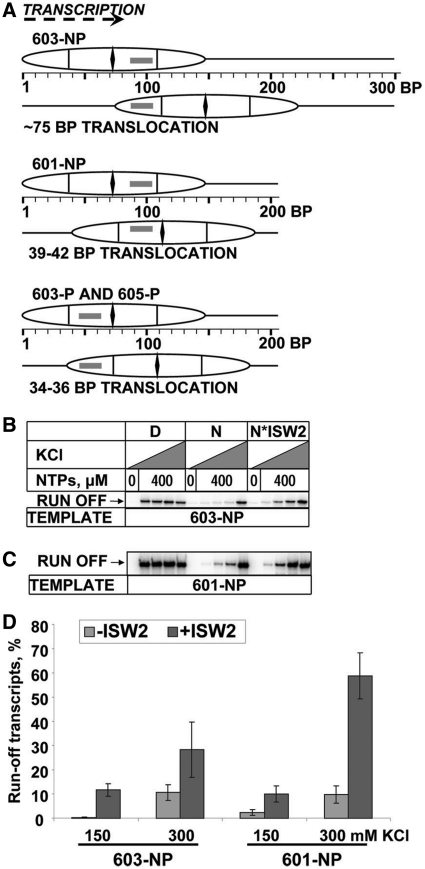
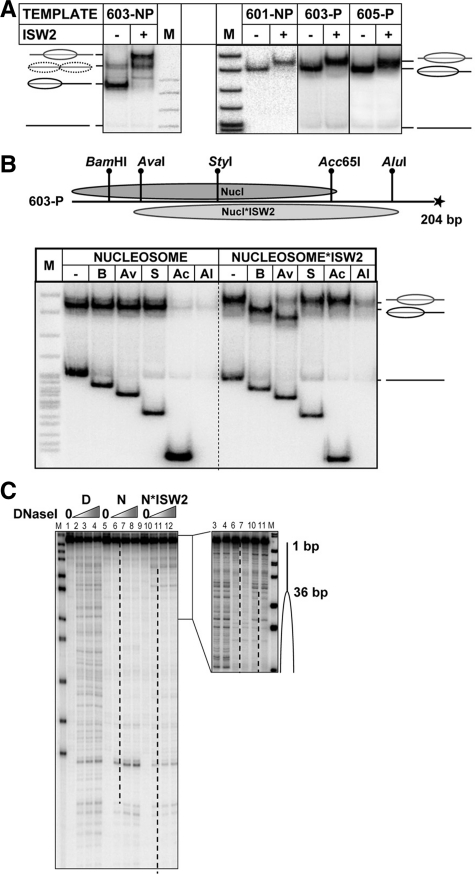
To evaluate the effect of re-location of the PBS on transcription by Pol II, an ATP-dependent chromatin remodeler was employed to translocate nucleosomes from the original 601, 603 or 605 positioning sequences to alternative positions. ISW2 was selected because it minimally perturbs nucleosome structure and moves nucleosomes over predictable distances, depending on the DNA length and the original nucleosome position (23,24). Templates were designed for translocation over short distances, placing the PBS in different locations with respect to the nucleosome, such that it interacts with either the promoter-proximal H2A–H2B dimer, the promoter-proximal H3–H4 dimer, or the promoter-distal H3–H4 dimer (Figure 1A). Nucleosome positions before and after ISW2 remodeling were verified by analysis in a native gel (Figure 2A), restriction enzyme sensitivity (Figure 2B) and DNase I footprinting (Figure 2C).
Figure 1.
ISW2-induced nucleosome translocation relieves the polar barrier. (A) Nucleosome positions and translocation distances on permissive (603-P and 605-P) and non-permissive (603-NP and 601-NP) templates before and after remodeling by ISW2. The PBS is indicated by a rectangle. (B) 603-NP or (C) 601-NP nucleosomes (N) or nucleosomes remodeled with ISW2 (N*ISW2), or free DNA (D) were transcribed by Pol II in 40 mM KCl without NTPs, or in 40, 150, 300 or 1000 mM KCl with 0.4 mM NTPs. Pulse-labeled RNA was analyzed by denaturing PAGE. Only the run-off products are shown (see Supplementary Figure S2 for the entire gel). (D) Quantitation of run-off transcripts in (B) and (C). The amount of run-off transcript produced in 1 M KCl was set at 100%. The data represent means of three independent experiments, with standard errors.
Figure 2.
Analysis of ISW2-induced nucleosome translocation. (A) Nucleosomes were reconstituted on 210- or 300-bp end-labeled DNA fragments containing the 601, 603 or 605 nucleosome positioning sequences (Figure 1A), incubated with ISW2 and analyzed by native PAGE. Nucleosome positions are indicated. M, end-labeled pBR322-MspI digest. (B) Mapping of 603-P nucleosome positions: restriction enzyme sensitivity assay. Top: Expected positions of nucleosomes on permissive 603-P template before (Nucl.) and after ISW2 remodeling (Nucl*ISW2). Unique sites for restriction enzymes are indicated. Bottom: Analysis of end-labeled 603-P nucleosomes by native PAGE before and after remodeling and digestion with a restriction enzyme. Histone-free DNA was added to the nucleosomal templates as an internal control for restriction enzyme activity. (C) Mapping of 603-P nucleosome positions before and after remodeling by ISW2: DNase I footprinting. ISW2 induces translocation of the 603-P nucleosome by 36 bp. Footprinting of DNA (D) and the nucleosome (N) was conducted before and after remodeling. Analysis by denaturing PAGE. Nucleosome positions are indicated (dashed lines and ovals).
The translocated nucleosomes were transcribed using pre-assembled yeast Pol II elongation complexes (EC). Briefly, EC9 complexes (where the number indicates the length of the transcript) were pre-assembled using synthetic oligonucleotides. The properties of these elongation complexes are very similar to those of promoter-initiated elongation complexes (14,25). The EC9 complexes were then annealed to the nucleosomal templates before or after ISW2 treatment, ISW2 was removed by washing, and the template was ligated. The nascent RNA was pulse-labeled with ATP, CTP and radioactive GTP to produce EC45 complexes, with Pol II stalled upstream of the promoter-proximal nucleosome boundary, awaiting the addition of UTP. The nucleosomal templates were transcribed in the presence of all four NTPs and at different concentrations of KCl to determine the strength and intranucleosomal location of the barrier to Pol II (Supplementary Figure S1).
The effect of ISW2-induced nucleosome translocation on the polar barrier in non-permissive 601 and 603 nucleosomes (601-NP and 603-NP, Figure 1A) was analyzed. In these nucleosomes, the PBS is located at +87 to +102 with respect to the promoter-proximal nucleosome boundary, where it interacts with the promoter-distal H3–H4 dimer (18). ISW2 induced the translocation of the 603-NP nucleosome to a new position ∼75 bp downstream of the original position, such that the PBS now interacts with the promoter-proximal H2A–H2B dimer (Figure 1A). Transcription of the 603-NP template before remodeling was characterized by strong pausing at +45 (Supplementary Figure S2): >99% of transcribing Pol II failed to yield run-off transcript in 150 mM KCl (Figure 1B and D). The 603-NP nucleosome barrier was relieved only in 1 M KCl, when nucleosome structure is disrupted. However, after nucleosome translocation by ISW2, the strength of the barrier was dramatically reduced, by ∼30-fold in 150 mM KCl (Figures 1B and D, and 3D). In 150 mM KCl, only ∼0.4% of Pol II transcribed through the 603-NP nucleosome, whereas ∼12% was able to transcribe through the nucleosome after translocation. In 300 mM KCl, the fraction of Pol II able to transcribe through the 603-NP nucleosome increased from ∼10 to ∼30% (a ∼3-fold decrease in barrier strength) (Figure 1D).
Figure 3.
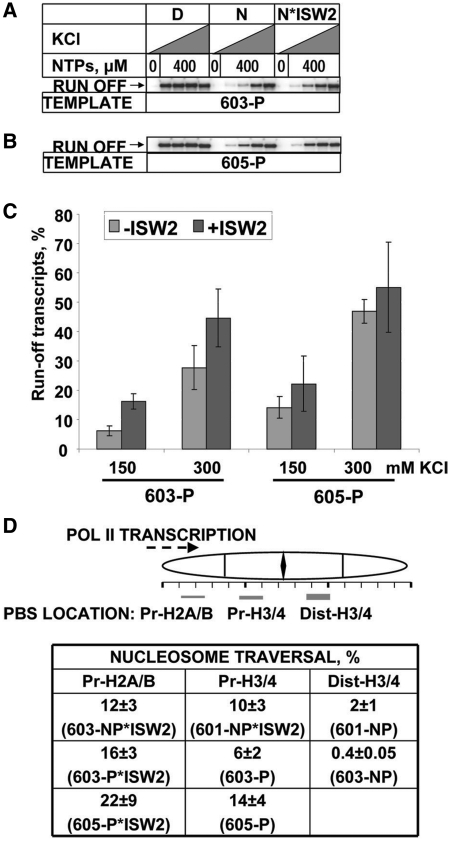
The efficiency of transcription through permissive nucleosomes is minimally affected by ISW2. (A) 603-P or (B) 605-P nucleosomes (N) or nucleosomes remodeled with ISW2 (N*ISW2), or free DNA (D) were transcribed by Pol II in 40 mM KCl without NTPs, or in 40, 150, 300 or 1000 mM KCl with 0.4 mM NTPs. Pulse-labeled RNA was analyzed by denaturing PAGE. Only the run-off products are shown (see Supplementary Figure S3 for the entire gel). Pulse-labeled RNA was analyzed by denaturing PAGE. (C) Quantitation of run-off transcripts in (A) and (B). (D) Intra-nucleosomal location of the PBS and the height of the nucleosomal barrier to transcription. Top: location of the PBS within different nucleosomes; the heights of the polar barrier to Pol II are indicated by the height of the rectangle which indicates the location of the PBS. Pr-H2A/B, Pr-H3/4 and Dist-H3/4: Pr = promoter-proximal; Dist = promoter-distal. Bottom: yields of run-off transcripts from the various nucleosomes in 150 mM KCl. The data represent the means of three independent experiments, with standard errors.
ISW2 induced translocation of the 601-NP nucleosome over a short distance to a new position only 38–42 bp away from the original position (Figure 1A) (26). This resulted in a large increase in run-off transcript produced at 150 and 300 mM KCl (Figure 1C and D). The strength of the barrier was reduced ∼5-fold in 150 mM KCl (Figure 1D): only ∼2% of Pol II transcribed through the 601-NP nucleosome, whereas ∼10% was able to transcribe through the nucleosome after translocation. In 300 mM KCl, the fraction of Pol II able to transcribe through the 601-NP nucleosome increased from ∼10 to ∼60% (a ∼6-fold decrease in barrier strength). Thus, the effect of translocation on the 603-NP nucleosome was greater than on the 601-NP nucleosome, probably because the 603-NP nucleosome is intrinsically more difficult to transcribe. In conclusion, ISW2-induced translocation of the non-permissive 601 and 603 nucleosomes over distances of 38–75 bp relieved the polar barrier to transcription, even though the translocated nucleosomes still contain the PBS.
ISW2-induced nucleosome translocation has little effect on the non-polar barrier
To determine whether translocation has any effect on the non-polar barrier to transcription that is exhibited by all nucleosomes, permissive 603 and 605 nucleosomes were remodeled by ISW2 and transcribed (Figure 3). In the 603-P and 605-P nucleosomes, the PBS interacts with the promoter-proximal H3–H4 dimer. ISW2 induced nucleosome translocation to new positions 34 and 36 bp downstream of the original positions, respectively (Figure 1A). In both cases, the PBS was re-located such that it interacts with the promoter-proximal H2A–H2B dimer. In 150 mM KCl, there was a relatively modest increase in transcription of both the 603-P and 605-P nucleosomes (∼2.5- and 1.5-fold, respectively) as a result of translocation (Figure 3). In 300 mM KCl, the increase in transcription due to translocation was slight for both nucleosomes (Figure 3C, Supplementary Figure S3). Thus, the strength of the barrier posed by the permissive nucleosomes decreased only slightly in response to translocation. The strength of the barrier was characteristic of nucleosomes formed on natural DNA, which allow progression of 10–40 and 45–90% of transcribing Pol II in 150 and 300 mM KCl, respectively (14). In conclusion, ISW2-induced nucleosome translocation over a short distance (34–75 bp) strongly relieved the polar barrier to transcription, by 5-fold (601) or 30-fold (603) (Figure 3D). The effect on the non-polar barrier was only 1.6–2.7-fold.
The 603-positioning sequence does not form a polar barrier in vivo
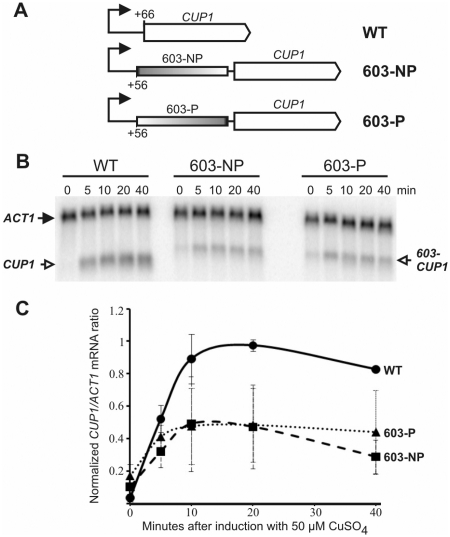
Is the PBS functional in vivo? The 603-sequence was inserted into the yeast CUP1 gene at +56, between the major transcription start site at +1 and the start codon at +66, in the permissive or the non-permissive orientation (Figure 4A). CUP1 encodes an inducible metallothionein which protects cells against the toxic effects of copper (27). Sequence-603 was chosen because it has a more polar barrier than 601 (Figures 1 and 3C). Haploid strains (WT, 603-NP and 603-P) were constructed in which a CUP1 gene, with or without the 603-insert, was integrated into the chromosome, replacing the original CUP1 locus (Figure 4A). These strains have only one copy of CUP1 per cell. Growth assays showed that WT cells were resistant to 50 µM copper, whereas both 603 strains grew moderately well only in 10 µM CuSO4. Thus, the 603-insert had an adverse effect on CUP1 function.
Figure 4.
The 603-nucleosome positioning sequence does not form a polar barrier to transcription in vivo. (A) Yeast strains with 603 inserted at +56 in the 5′-untranslated region of CUP1. The major transcription start site (+1) is indicated by the arrow. The 603-NP and 603-P strains have 603 inserted in the non-permissive or permissive orientations, respectively. (B) Time courses of induction with 50 µM CuSO4. The phosphorimager scan of a northern blot probed for CUP1 and ACT1 transcripts is shown. (C) Time courses of induction: WT (circles); 603-P (triangles); 603-NP (squares). The data represent the means of two independent experiments, with standard deviations.
The effect of 603 on copper-induction was determined (Figure 4B). CUP1 was strongly induced in WT cells, increasing rapidly, peaking between 10 and 20 min, and then falling off somewhat (Figure 4C), as expected (28). The presence of the 603-insert was confirmed by the larger size of the 603-CUP1 transcripts. Both 603-CUP1 strains showed significantly elevated basal levels of CUP1 expression and exhibited a robust induction, similar to that of wild-type cells, but reduced by ∼2-fold, probably accounting for the increased copper-sensitivity of the 603-CUP1 strains. Most importantly, 603-CUP1 transcription was independent of the orientation of 603 (Figure 4C). Thus, 603 does not dictate a polar barrier to transcription in vivo. This could be due to nucleosome translocation by remodeling complexes, as shown above for ISW2 in vitro. Consequently, we asked whether the nucleosome is correctly positioned on 603 in vivo.
The 603-nucleosome is positioned correctly in only a small fraction of cells
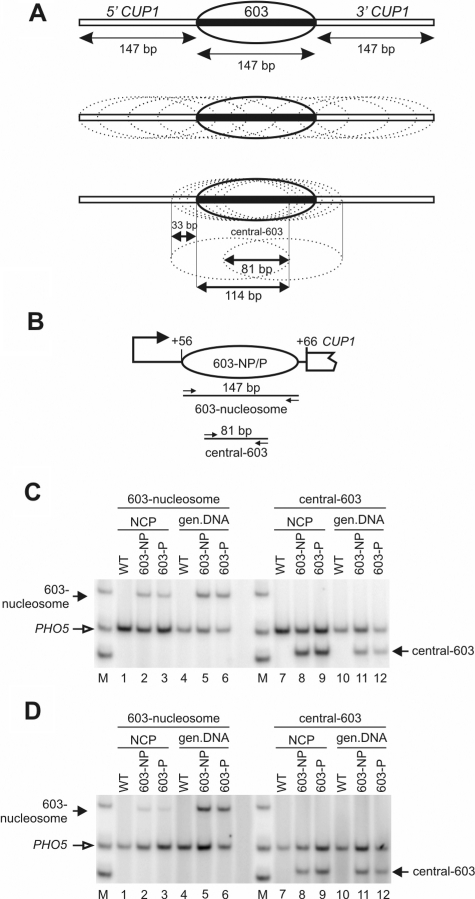
A quantitative PCR assay was used to measure the nucleosome occupancy of 603 in vivo. The assay depends on the fact that the 603-nucleosome position has been determined with high precision (29), corresponding exactly to the 147-bp inserted into CUP1. Thus, a pair of primers corresponding to the ends of the 603-insert yield the positioning sequence as a 147-bp PCR product. Using nucleosome core particle DNA trimmed to 145–150 bp by MNase as template (Supplementary Figure S4), this PCR product will be observed only if the nucleosome is located within a few base pairs of the expected position (Figure 5A). If trimming is incomplete, the occupancy of the 603-position would tend to be over-estimated.
Figure 5.
The occupancy of the 603 nucleosome positioning sequence is low in vivo. (A) Nucleosome occupancy is defined as the fraction of nucleosomes precisely positioned on the 147-bp 603-insert (solid oval) of all nucleosomes containing any part of 603 (dotted ovals). Any nucleosome including part of 603 will physically prevent the formation of the correctly positioned 603-nucleosome. An approximate occupancy is measured by determining the fraction of nucleosomes correctly positioned on 603, of all nucleosomes containing the central 81 bp of 603 (lower panel). (B) The 603-CUP1 promoter showing a nucleosome correctly positioned on 603. The locations of the 603-nucleosome and central 603 primer pairs are indicated (small arrows) with their respective amplicon sizes. (C) Measurement of 603 occupancy in non-induced cells. NCP: nucleosome core particles; gen. DNA: genomic DNA; M: marker (75, 100 and 150 bp). Lanes 1–6: multiplex PCR with the 603-nucleosome and PHO5 primers (internal control). Lanes 7–12: PCR with the central-603 and PHO5 primers. (D) Measurement of 603-nucleosome occupancy in cells induced with 50 µM CuSO4 for 20 min. Phosphorimages are shown.
The occupancy can be defined as the fraction of nucleosomes correctly positioned on 603 of all nucleosomes overlapping any part of 603 (Figure 5A). If the 603-nucleosome is present in all cells, the occupancy would be 1. Measuring the occupancy precisely is difficult, because all nucleosomes containing 1 bp or more of the 603-insert must be measured accurately and summed to provide the denominator. Instead, an approximate occupancy was determined by measuring all nucleosomes containing the central 81 bp of the 603-insert and using this as the denominator. This represents a significant over-estimate of 603 occupancy because many potential alternative positions are not included (Figure 5A). Primers corresponding to the central 101 bp of the −1 nucleosome in the PHO5 promoter (30) were used for normalization.
Multiplex PCR was performed using core particle DNA purified from the WT, 603-NP and 603-P strains, grown without copper induction (Figure 5B). The signals for core DNA were normalized using the signals from genomic DNA, since the amounts of 603 and PHO5 must be equal in genomic DNA. Core particles from the 603-NP and 603-P strains gave rise to some 147 bp product (Figure 5C, lanes 2 and 3), proving that at least some 603-NP and 603-P cells contain a correctly positioned 603-nucleosome. However, for core particles from both strains, the intensity of the 603-nucleosome band was much lower than that of the PHO5 internal control band (Figure 5C, lanes 2 and 3; ∼5- and ∼8-fold lower, respectively), whereas the ratio of the 603-nucleosome band to the PHO5 internal control was close to 1 for genomic DNA (Figure 5C, lanes 5 and 6).
The occupancy of 603 was measured by normalizing the 603-nucleosome band and the central-603 band to their respective PHO5 internal controls and then to their respective genomic 603/PHO5 ratios, and dividing the 603-nucleosome ratio by the central-603 ratio. The occupancy of 603 was 20 ± 7% in 603-NP cells and 14 ± 11% in 603-P cells (averages of three independent experiments). Copper-induction had little effect on occupancies (Figure 5D): these were 12 ± 4% in 603-NP cells and 12 ± 9% in 603-P cells (averages of two independent experiments). In both cases, the orientation of 603 had no significant effect on occupancy. In conclusion, the large majority of cells do not have a correctly positioned 603-nucleosome.
Evidence for nucleosomes positioned over the CUP1-603 junctions
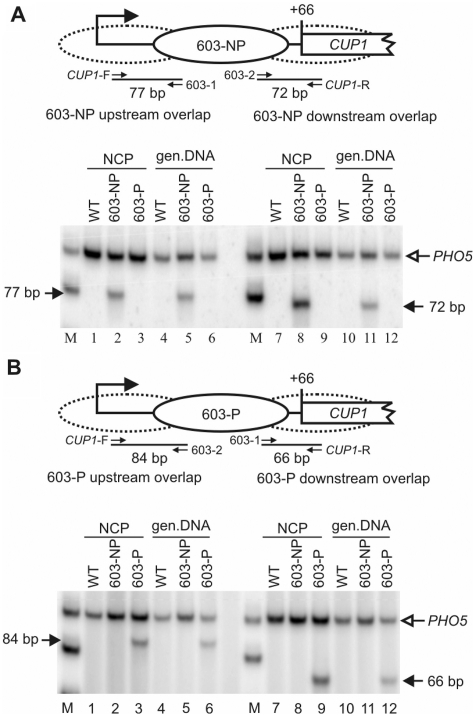
Given that nucleosomes are not positioned on 603 as expected, it should be possible to detect overlapping nucleosomes containing part of the 603-insert and some flanking up or downstream CUP1 DNA. To verify the presence of such nucleosomes, different primer pairs had to be used for the 603-NP and 603-P strains, because of the orientation of 603 (Figure 6A). PCR performed with the upstream junction primers gave the expected 77 bp product using core particle and genomic DNA from the 603-NP strain (Figure 6A, lanes 2 and 5). The downstream junction primers gave a similar result. The results confirm the presence of nucleosomes in positions overlapping the junctions between CUP1 and 603 in 603-NP cells. The same was shown to be true in 603-P cells using different primer combinations (Figure 6B).
Figure 6.
Evidence for nucleosomes positioned over the CUP1-603 junction sequences in non-induced cells. Detection of nucleosomes positioned over the CUP1-603 junctions in (A) 603-NP cells and (B) 603-P cells. The schematics show a nucleosome correctly positioned on 603. The dotted ovals represent nucleosomes in positions overlapping the junctions of 603 with CUP1. The locations of the upstream and downstream CUP1 primers and of internal 603 primers 1 and 2 are indicated (small arrows). Phosphorimages of PCR assays are shown. NCP: nucleosome core particles; gen. DNA: genomic DNA; M: marker (75 and 100 bp). Lanes 1–6: multiplex PCR with the upstream junction primer pair and PHO5 primers (internal control). Lanes 7–12: PCR with the downstream junction primer pair and PHO5 primers. Similar results were obtained with copper-induced cells (Supplementary Figure S5). The data are quantified in Supplementary Table S2.
A quantitative measure of the junction nucleosomes was obtained by comparison with nucleosomes containing the central 603-sequence (Supplementary Table S2). In non-induced 603-NP cells, nucleosomes were more likely to include the central 603-sequence or the downstream 603-CUP1 junction DNA than the upstream CUP1-603 junction. In 603-P cells, the central 603 sequence was more likely to be nucleosomal than either junction sequence. However, these differences were small. Induction with copper had only minor effects on nucleosome distribution (Supplementary Figure S5 and Table S2). Thus, overall, 603 was not strongly preferred over neighboring CUP1 sequences for nucleosome assembly in vivo. The fact that the fraction of junction nucleosomes is high is consistent with the low occupancy of 603, because the presence of a junction nucleosome precludes the presence of the 603-nucleosome, as they both cannot be present on the same DNA molecule and therefore in the same cell.
DISCUSSION
In summary, the presence of a strong polar barrier to transcription correlates with the interaction of the PBS with the promoter-distal H3–H4 dimer (Figure 3D). The polar barrier is therefore dependent on precise nucleosome positioning: even a small change in position (35–75 bp) results in re-location of the PBS and loss of the barrier. Thus, ISW2, an ATP-dependent chromatin remodeler, can strongly affect the transcription rate by moving the nucleosome such that the PBS is now in a different location, even within the same nucleosome (Figure 3D). The PBS within 603, one of the strongest nucleosome positioning sequences known, does not act as a polar barrier in vivo, probably because in most cells, the nucleosome is not positioned as required.
Circumvention of the polar barrier by ISW2-induced nucleosome translocation in vitro
Our data are consistent with the proposal that the polar barrier is negated by re-location of the PBS within the nucleosome. When ISW2 moves the PBS away from the distal H3–H4 dimer, the sequence contained within the nucleosome is changed and the histone octamer is likely to be bound at a less energetically favorable location. Although the sequence context has changed, there is a good correlation between the location of the PBS sequence within the nucleosome and the height of the barrier to transcribing Pol II (Figure 3D). The relative de-stabilization of the nucleosome per se is unlikely to account for the loss of the polar barrier, because the polar barrier does not depend on the overall stability of the nucleosome, but rather on its orientation with respect to the approaching polymerase (14). In the 601-NP and 603-NP nucleosomes, the PBS is located such that it interacts with the promoter-distal half of the H3–H4 tetramer (Figure 3D), resulting in a very high barrier to transcription. Translocation by ISW2, resulting in re-location of the PBS such that it interacts with the proximal H3–H4 dimer or with the proximal H2A–H2B dimer, greatly reduces the strength of the polar barrier. In the 603-P and 605-P nucleosomes, the PBS interacts with the promoter-proximal H3–H4 dimer and the barrier is relatively weak, similar to that exhibited by nucleosomes formed with natural DNA. After translocation, the PBS interacts with the promoter-proximal H2A–H2B dimer, with little effect on the weak barrier. Thus, there is a strong correlation between the presence of a strong polar barrier and the interaction of the PBS with the promoter-distal H3–H4 dimer.
The absence of a polar barrier in vivo
How is the polar barrier circumvented in vivo? A likely explanation is the low occupancy of 603; most cells do not contain a nucleosome correctly positioned on 603, which is necessary for the PBS to function. In the 10–20% of 603-NP cells which do contain the 603-nucleosome, the polar barrier might be present but undetected, given the error in the measurements. In these cells, the polar barrier might cause Pol II to stall inside the 603-nucleosome. An alternative view is that nucleosomes on CUP1 and 603 might be in flux, due to various nucleosome sliding activities, spending only a short time in the 603-position (31,32). If this is the case, a transient polar barrier might be formed in all 603-NP cells, active only when the nucleosome is in the 603-position. We propose that the polar barrier is absent in vivo because nucleosome sliding activities drive translocation of the 603-nucleosome from its critical position, preventing interaction of the PBS with the promoter-distal H3–H4 dimer.
Direct support for translocation of the 603-nucleosome by remodeling complexes could be obtained by examining nucleosome sliding mutants. However, in addition to ISW2, there are several chromatin remodeling complexes capable of mobilizing nucleosomes in yeast, including ISW1, SWI/SNF and RSC. This level of redundancy makes it difficult to eliminate nucleosome-sliding activity in yeast cells.
The 603-insert does not position a nucleosome strongly in vivo
We have shown that most cells do not have the 603-nucleosome at the correct position. It is perhaps surprising that the occupancy of 603 is so low in vivo, given that it is one of the strongest positioning sequences known, and that DNA sequence is a major determinant of positioning in yeast (33–35). Perhaps 603 is not such a strong positioning sequence in vivo, as the related 601 sequence is a relatively poor competitor for histones under some conditions in vitro (36). The 5S RNA gene is a natural, strong positioning sequence and it also fails to position uniquely in vivo (37), although this is also true in vitro (38), unlike 601. More generally, these observations imply that the nucleosome positioning information [the ‘nucleosome code’ (39)] that is undoubtedly present in DNA is insufficient to predict the locations of all nucleosomes unambiguously in vivo. Other factors likely to be of critical importance include the binding of transcription factors, which may act as nucleosome phasing signals (31,32) and the formation of nucleosomal arrays in vivo, which imposes restrictions on the locations of neighboring nucleosomes once the first nucleosome has been assembled (e.g. during DNA replication) (28). The promoters of yeast genes tend to be relatively low-affinity sites for nucleosome formation in vitro and tend to exclude nucleosomes in vivo (31,32). However, like the PHO5 promoter (26), the CUP1 promoter is an exception: there is a nucleosome on the CUP1 promoter in non-induced cells, which adopts one of several alternative positions in vivo (29) and in vitro (30). Insertion of 603 just downstream of the major transcription start site would necessarily interrupt these positioning signals, perhaps interfering with the positioning of the nucleosome on the 603-insert just downstream.
However, probably most important is the fact that ISW2 (Figure 1A) and RSC (40,41) can move the nucleosome quantitatively from 601 or 603 to a much less energetically favorable position in vitro. Thus, remodeling complexes can overcome the very high affinity of the histone octamer for 601 or 603, shifting the nucleosome to a higher free-energy state. This is also likely to be the case in vivo: nucleosome sliding activities use the free-energy gained from ATP hydrolysis to move nucleosomes to positions that are not necessarily those of the lowest free energy. Thus, the insertion of 603 elsewhere in the yeast genome, where there is much less sliding activity, perhaps in a silenced region, might result in higher occupancies.
Nucleosomal barriers and remodeling complexes
The polar barrier to yeast Pol II cannot be relieved by the elongation factor TFIIS or by the histone chaperone FACT, although IIS can facilitate transcription though the polar barrier by human Pol II (14). ISW2 is the first yeast factor to be identified that can relieve the polar barrier in vitro. Although the existence of polar nucleosomal barriers in natural DNA remains to be demonstrated, it seems likely that such sequences exist in vivo. Polar barriers might possess significant regulatory potential if combined with nucleosome sliding activities capable of controlling the occupancy of the critical nucleosome position relative to other positions.
More generally, our data suggest that nucleosome re-positioning could be involved in regulation of transcription. In particular, elongating Pol II is paused immediately downstream from the transcription start site (TSS) on thousands of genes in Drosophila and man (9,10). The first nucleosome downstream of the TSS (the +1 nucleosome) is positioned relatively well and is translocated during activation of transcription (12,42). Furthermore, elongating RNA polymerase II co-maps with the promoter-proximal boundary of the +1 nucleosome on many Drosophila genes (12). Finally, the +1 nucleosome can form a strong, activator-sensitive barrier to transcribing human Pol II in vitro (13). However, a causal relationship between +1 nucleosome translocation and transcription activation remains to be established.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institutes of Health (NICHD); National Institutes of Health (GM58650 to V.M.S.); National Institutes of Health (GM70864 to B.B.). Funding for open access charge: Intramural Research Program of the National Institutes of Health (to D.J.C.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Donal Luse for helpful comments on the article.
REFERENCES
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Kulaeva OI, Gaykalova DA, Studitsky VM. Transcription through chromatin by RNA polymerase II: Histone displacement and exchange. Mutat. Res. 2007;618:116–129. doi: 10.1016/j.mrfmmm.2006.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kristjuhan A, Svejstrup JQ. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 6.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Thiriet C, Hayes JJ. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 2005;19:677–682. doi: 10.1101/gad.1265205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown SA, Imbalzano AN, Kingston RE. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- 14.Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell. 2006;24:469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 15.Izban MG, Luse DS. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991;5:683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- 16.Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M. Nature of the nucleosomal barrier to RNA polymerase II. Mol. Cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 17.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 18.Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, Studitsky VM. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat. Struct. Mol. Biol. 2009;16:1272–1278. doi: 10.1038/nsmb.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaykalova DA, Kulaeva OI, Bondarenko VA, Studitsky VM. Preparation and analysis of uniquely positioned mononucleosomes. Methods Mol. Biol. 2009;523:109–123. doi: 10.1007/978-1-59745-190-1_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kassabov SR, Henry NM, Zofall M, Tsukiyama T, Bartholomew B. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol. Cell. Biol. 2002;22:7524–7534. doi: 10.1128/MCB.22.21.7524-7534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 22.Kim Y, Clark DJ. SWI/SNF-dependent long-range remodeling of yeast HIS3 chromatin. Proc. Natl Acad. Sci. USA. 2002;99:15381–15386. doi: 10.1073/pnas.242536699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gangaraju VK, Prasad P, Srour A, Kagalwala MN, Bartholomew B. Conformational changes associated with template commitment in ATP-dependent chromatin remodeling by ISW2. Mol. Cell. 2009;35:58–69. doi: 10.1016/j.molcel.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat. Struct. Mol. Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 25.Walter W, Kireeva ML, Studitsky VM, Kashlev M. Bacterial polymerase and yeast polymerase II use similar mechanisms for transcription through nucleosomes. J. Biol. Chem. 2003;278:36148–36156. doi: 10.1074/jbc.M305647200. [DOI] [PubMed] [Google Scholar]
- 26.Kagalwala MN, Glaus BJ, Dang W, Zofall M, Bartholomew B. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J. 2004;23:2092–2104. doi: 10.1038/sj.emboj.7600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karin M, Najarian R, Haslinger A, Valenzuela P, Welch J, Fogel S. Primary structure and transcription of an amplified genetic locus: the CUP1 locus of yeast. Proc. Natl Acad. Sci. USA. 1984;81:337–341. doi: 10.1073/pnas.81.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen CH, Leblanc BP, Neal C, Akhavan R, Clark DJ. Targeted histone acetylation at the yeast CUP1 promoter requires the transcriptional activator, the TATA boxes, and the putative histone acetylase encoded by SPT10. Mol. Cell. Biol. 2002;22:6406–6416. doi: 10.1128/MCB.22.18.6406-6416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morozov AV, Fortney K, Gaykalova DA, Studitsky VM, Widom J, Siggia ED. Using DNA mechanics to predict in vitro nucleosome positions and formation energies. Nucleic Acids Res. 2009;37:4707–4722. doi: 10.1093/nar/gkp475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svaren J, Horz W. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci. 1997;22:93–97. doi: 10.1016/s0968-0004(97)01001-3. [DOI] [PubMed] [Google Scholar]
- 31.Kim Y, McLaughlin N, Lindstrom K, Tsukiyama T, Clark DJ. Activation of Saccharomyces cerevisiae HIS3 results in Gcn4p-dependent, SWI/SNF-dependent mobilization of nucleosomes over the entire gene. Mol. Cell. Biol. 2006;26:8607–8622. doi: 10.1128/MCB.00678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark DJ. Nucleosome positioning, nucleosome spacing and the nucleosome code. J. Biomol. Struct. Dyn. 2010;27:781–793. doi: 10.1080/073911010010524945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen CH, Clark DJ. DNA sequence plays a major role in determining nucleosome positions in yeast CUP1 chromatin. J. Biol. Chem. 2001;276:35209–35216. doi: 10.1074/jbc.M104733200. [DOI] [PubMed] [Google Scholar]
- 34.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat. Struct. Mol. Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu C, Travers A. Relative affinities of DNA sequences for the histone octamer depend strongly upon both the temperature and octamer concentration. Biochemistry. 2005;44:14329–14334. doi: 10.1021/bi050915w. [DOI] [PubMed] [Google Scholar]
- 37.Chipev CC, Wolffe AP. Chromosomal organization of Xenopus laevis oocyte and somatic 5S rRNA genes in vivo. Mol. Cell. Biol. 1992;12:45–55. doi: 10.1128/mcb.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Donohue MF, Duband-Goulet I, Hamiche A, Prunell A. Octamer displacement and redistribution in transcription of single nucleosomes. Nucleic Acids Res. 1994;22:937–945. doi: 10.1093/nar/22.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol. Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shukla MS, Syed SH, Montel F, Faivre-Moskalenko C, Bednar J, Travers A, Angelov D, Dimitrov S. Remosomes: RSC generated non-mobilized particles with approximately 180 bp DNA loosely associated with the histone octamer. Proc. Natl Acad. Sci. USA. 2010;107:1936–1941. doi: 10.1073/pnas.0904497107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koerber RT, Rhee HS, Jiang C, Pugh BF. Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol. Cell. 2009;35:889–902. doi: 10.1016/j.molcel.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.