Abstract
Using a 10-species oral biofilm consortium and defined mutants, we show that high-level capacity to generate ammonia from a common salivary substrate is needed to maintain community diversity. This model appears to be suitable for the study of the effects of individual genetic determinants on the ecology of oral biofilms.
Dental plaque is a biofilm with a highly diverse microbial composition that functions as a true microbial community. The stability of oral biofilms represents a highly dynamic balance achieved by a range of synergistic and antagonistic interactions among the constituent species. Generally, the stable climax communities that establish on different surfaces in the mouth are associated with oral health and provide benefit by preventing colonization by pathogenic microorganisms. On occasion, however, this natural balance can break down, usually as a result of sustained and severe environmental stresses, leading to overgrowth by previously minor components of the microflora. A consequence of this ecological upset can be the development of caries or periodontal diseases (3, 4, 10, 21, 22, 31).
In the case of dental caries, increased consumption of dietary carbohydrates results in repeated and sustained acidification of oral biofilms, which leads to the emergence of a flora that is enriched for highly acidogenic and acid-tolerant (aciduric) organisms, such as mutans streptococci, other aciduric streptococci, and lactobacilli (7, 17, 18, 20, 32, 33). However, it is also established that some bacteria, such as Streptococcus sanguis, Streptococcus gordonii, and Actinomyces naeslundii (2, 16, 18), show a negative correlation with caries. One possible explanation for the association of these species with health is their ability to generate ammonia from salivary and dietary substrates, resulting in neutralization of acids within the biofilms (9). When cariogenic species begin to become numerically significant, it appears to be, at least in part, at the expense of organisms capable of alkali generation. Thus, a reasonable hypothesis that has been put forth is that the production of alkali-generating systems by selected oral bacteria is a critical component in oral biofilm stability (9).
Urea hydrolysis by urease enzymes and catabolism of arginine via the arginine deiminase pathway are the two primary ammonia-producing pathways in the oral cavity (9). The most active of these enzymes is urease, which converts the urea that is supplied in saliva (concentrations of roughly 1 to 10 mM in healthy subjects) into two molecules of ammonia and one of carbon dioxide. A variety of studies have indicated that ammonia generation from urea is negatively correlated with initiation and progression of caries (9). For example, the application of urea to oral bacterial populations results in a rapid increase in pH (27, 28). Renal dialysis patients, who have highly elevated levels of salivary urea, have diminished plaque acidification and lower caries rates (25, 26). More direct demonstration of the roles of urea metabolism in moderation of plaque pH and caries resistance include the use of genetically engineered plaque streptococci to demonstrate a nearly direct correlation between the amount of urease produced and the pH-moderating capacity of ureolytic bacteria (14). In a subsequent report, Clancy et al. demonstrated that rats infected with strains of Streptococcus mutans that were genetically engineered to produce high levels of urease had a dramatically decreased rate and severity of caries development compared with animals that were infected with the wild-type S. mutans (15). Collectively, these experiments lend credence to the idea that the alkali-generating capacity of oral biofilms can strongly influence caries development and progression.
One inadequately resolved issue related to oral alkali generation is whether ammonia production exerts its influence primarily by inhibiting enamel demineralization or whether ammonia generation can actually result in stabilization of a healthy flora by suppressing the emergence of aciduric and acidogenic bacteria. To begin to approach this question, and to establish whether in vitro mixed biofilm models could be utilized to test the effects of specific gene products on oral biofilm composition and activity, we modified a 10-species biofilm consortium model to include genetically manipulated bacteria with altered ureolytic capacities. Here, we demonstrate the importance of urease enzymes in fostering community stability and preventing the emergence of a cariogenic microflora by using an in vitro 10-species biofilm model.
Model.
For this study, we adopted the 10-species consortium originally described by Marsh and coworkers (5, 19). Stable biofilm communities form in this system, with all 10 species being well represented unless a significant stress is placed on the populations in the form of lowered pH or repeated carbohydrate pulses without pH control (5, 6, 19). In these cases, community diversity is lost, and aciduric lactobacilli and streptococci and lactate-metabolizing Veillonella dispar dominate the biofilms. The original 10-species consortium was modified for this particular study by replacing Streptococcus sanguis with the urease-producing Streptococcus salivarius 57.I (12, 29) and Actinomyces viscosus with the urease-producing Actinomyces naeslundii WVU45 (23). Streptococcus mutans UA159, a strongly cariogenic organism that can be genetically manipulated (8, 24) and which was the focus of a genome-sequencing project (1), was used to replace the original S. mutans strain in the consortium used by Marsh et al. Thus, our consortium contained eight strains of nonureolytic bacteria (Streptococcus mutans UA159, Streptococcus oralis SK193, Lactobacillus rhamnosus ATCC 7469, Neisseria subflava A1078, Veillonella dispar ATCC 17745, Prevotella nigrescens T588, Fusobacterium nucleatum ATCC 10953, and Porphyromonas gingivalis W50) as well as two urease-producing strains (Streptococcus salivarius 57.I and Actinomyces naeslundii WVU 45). For evaluation of the role of urease in community stability, two otherwise isogenic, urease-negative derivatives of the input Streptococcus salivarius (13) and Actinomyces naeslundii (23) strains were used.
We elected to cultivate the 10-species consortium in a constant-depth film fermenter (CDFF) because this thin-film model has been shown to be of significant value for oral biofilm ecology studies (19, 34). Each strain was grown individually in liquid medium to late exponential phase, and 5 ml of each culture was combined, gently mixed, and inoculated into the CDFF through a pump over a 6-h period. Concentrations of input organisms were carefully controlled for each run, with bacteria counts ranging from 6.0 × 107 to 8.3 × 108 CFU ml−1. During the inoculation period and thereafter, a mucin-containing medium, BMM (6), containing 2.5 g of porcine gastric mucin liter−1, 2 g of proteose peptone liter−1, 2.5 g of KCl liter−1, 1 g of yeast extract liter−1, 1 g of Trypticase peptone liter−1, 0.1 g of cysteine hydrochloride liter−1, and 0.001 g of hemin liter−1 that was supplemented with 10 mM glucose and 10 mM urea (BMMUG) was used to cultivate the biofilms. Urea concentrations in the saliva of healthy subjects range from about 3 to 10 mM, and the glucose concentration provided in BMMUG is similar to what would be experienced during a significant carbohydrate intake (11). In all cases, the pHs of the media were adjusted to pH 7.5 prior to autoclaving. Subsequent to the inoculation period, BMMUG was pumped into the CDFF at a rate of 100 ml h−1 for 7 days. Then, depending on the experiment, addition of BMMUG or BMM base medium supplemented with 10 mM glucose but no urea (BMMG) was continued until day 11 at the same flow rate. The CDFF was housed in a 37°C warm room under aerobic conditions.
Biofilms were grown on 4.75-mm-diameter polytetrafluoroethylene (PTFE) plugs located in 15 PTFE sample pans, which were inserted into a rotating stainless-steel turntable (19). Each pan contained five plugs that were recessed to a depth of 300 μm, and the thickness of the biofilms was maintained by a scraper bar with the turntable rotating at 3 rpm. Samples were taken for culture analysis using the inocula and then from the biofilms formed on PTFE plugs at 7, 9, 10, and 11 days postinoculation. To sample the biofilms, a sample pan was aseptically removed from the fermenter, and using sterile forceps, plugs were carefully extruded with the biofilm still intact on the surface. The biofilms on the plugs were dispersed in 5 ml of 0.5% proteose peptone-0.25% KCl by vortexing for 10 s and sonicating for 10 s at low power (Sonic Dismembrator, model 100; Fisher Scientific). The cell suspensions were serially diluted in the same buffer and plated on a range of selective and nonselective media as previously described (5, 19). After sampling of each of the five plugs, the PTFE sample pan was placed into 10 ml of sterile deionized water and vortexed and the pH of the suspension was recorded. The experiments consisted of two phases. The first phase involved evaluation of the importance of urea in the medium in maintaining community diversity and preventing dominance by an aciduric flora as well as determination that urea could be used as a major substrate for moderation of acidification. The second phase involved the use of defined mutants to directly assess the contribution of urease enzymes to community diversity and prevention of dominance by aciduric bacteria.
Role of urea.
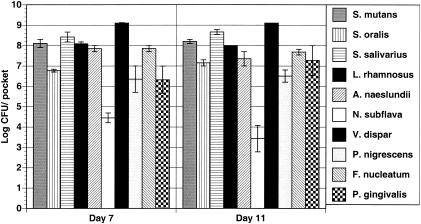
The goal of the first portion of the study was to determine whether urea could be used as a primary substrate to induce stability of the 10-species model. As described above, the CDFF was inoculated with all 10 species and biofilms were allowed to form for 7 days. At that time, sample plugs were removed, biofilm constituents were quantified by plating as previously described, and the pH of the biofilms was determined. When the biofilms were allowed to form in BMMUG medium, all 10 species were well represented in the consortium (Fig. 1). The bulk of the biofilms were constituted by the streptococci, Actinomyces, Lactobacillus, and Veillonella spp., which is probably a reflection of the effects of the relatively high concentration of glucose added to the base medium. The anaerobes, P. gingivalis, F. nucleatum, and P. nigrescens, and the obligate aerobe N. subflava were present but at levels about 2 and 4 logs lower than those of the most abundant organisms, respectively. After sampling again at day 11, it was found that all 10 species thrived and that the composition of the community remained stable (Fig. 1). The pH of the biofilms on day 7 was 6.06, and it was 5.91 on day 11.
FIG. 1.
Wild-type biofilms formed in the presence of urea. Biofilms were cultivated in complete BMMUG medium for the entire 11-day period. Organisms were recovered from the CDFF and enumerated by viable counting on selective media as detailed in the text. Viable counts are expressed as the average of the log CFU obtained per pocket, which provides the number of organisms that could be recovered from each of the five recessed pockets in the PTFE pans. Error bars indicate standard deviation. The data represent results from at least two complete runs of 11 days, and all platings were done in triplicate using three separate PTFE pans.
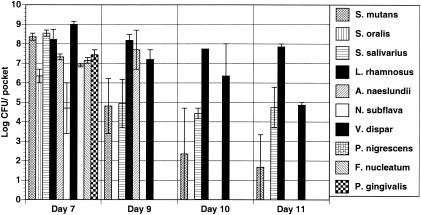
These results were in stark contrast to those obtained when the biofilms were formed in the presence of urea (BMMUG) for 7 days, followed by removal of urea from the growth medium (Fig. 2). In this case, biofilm composition after 7 days was identical to that seen in the initial experiments. However, after 2 days of incubation with BMMG as the nutrient source, community diversity was dramatically affected and P. gingivalis, P nigrescens, F. nucleatum, N. subflava, and S. oralis were no longer cultivable from the biofilms. By day 10, all that remained were aciduric streptococci, L. rhamnosus, and the lactic acid-consuming organism V. dispar. The pH values of the biofilms were 6.20 on day 7, 4.12 on day 9, and 4.45 on day 11. The overall numbers of organisms in the biofilms were reduced, likely due to growth inhibition from acid conditions.
FIG. 2.
Wild-type biofilms before and after removal of urea from the medium. Biofilms were cultivated in complete BMMUG medium for 7 days and then in BMMG for days 8 through 11. Organisms were recovered from the CDFF and enumerated as detailed in the text and in the legend to Fig. 1.
These experiments demonstrated the essential role of urea in maintaining community stability and the feasibility of phase two of the study. It is likely that the primary mechanism by which ureolysis conferred protection to the community is the neutralizing acids produced from catabolism of the glucose and the carbohydrates provided in mucin. In fact, profound effects of urea on the pH profiles of microcosm biofilms cultivated from plaque samples from human volunteers have been noted (30). It is also interesting that a concentration of urea similar to that found in saliva (10 mM) is sufficient to offset a fairly strong and sustained challenge with over 10 mM carbohydrate, or roughly 1,000-fold that found in resting plaque and comparable to what can be measured after ingestion of heavily sweetened beverages (11). In the oral cavity, urea is delivered almost continually to oral biofilms, whereas carbohydrate is presented only intermittently. Consequently, it seems that the level of urea present in saliva may be adequate to offset even a strong cariogenic challenge and that it is the amount of urease enzyme in the biofilms which is a critical factor in pH homeostasis (14). Clearly, in this in vitro model, the presence of the strongly ureolytic S. salivarius and the weakly ureolytic A. naeslundii was sufficient to provide enough alkali generation to maintain the pH of the biofilms near neutrality, preventing the domination by aciduric streptococci.
Importance of high levels of urease enzymes.
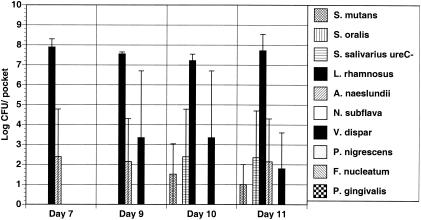
The purpose of this study's second phase was to utilize the biofilm model to determine the importance of the contribution of the individual urease enzymes of S. salivarius and A. naeslundii to community stability. To accomplish this, the 10-species consortium was first constituted with nine of the wild-type organisms and a urease-deficient S. salivarius strain that was constructed by allelic exchange of the ureC gene (large subunit) of urease with a polar antibiotic-resistance determinant (13). The results obtained here were dramatic (Fig. 3). In the absence of ureolytic S. salivarius, the 7-day biofilms were dominated primarily by lactobacilli and ureolytic A. naeslundii. Other organisms were present in numbers too small to detect. After cultivation for an additional 2, 3, or 4 days in the absence of urea, the biofilms contained the aciduric streptococci and lactobacilli, the lactate-metabolizing Veillonella, and the urease-positive A. naeslundii. The pH values of the biofilms were 4.12 on day 7, 4.38 on day 9, and 4.31 on day 11. These results demonstrate that ammonia production by S. salivarius is an essential determinant for the establishment and persistence of diverse biofilms and prevents dominance of biofilms by aciduric species. It is also of interest that the presence of the urease-producing A. naeslundii was not sufficient to offset the impact of the loss of S. salivarius. This is not entirely surprising since A. naeslundii produces about 50-fold less urease than S. salivarius. Clancy and Burne have previously shown that when the level of urease drops below a certain threshold, the rate of urea breakdown becomes insufficient to offset glycolytic acidification of the environment (14). Thus, it seems that the low level of urease produced by A. naeslundii was unable to provide enough ammonia from urea to avoid loss of community diversity, although urea breakdown may enhance survival of A. naeslundii (23). Similarly, when biofilms were formed with urease-deficient strains of both S. salivarius and A. naeslundii, the results were similar to what was seen with omission of the S. salivarius urease (data not shown).
FIG. 3.
Biofilms formed with the urease-deficient Streptococcus salivarius. Biofilms were cultivated in complete BMMUG medium for 7 days and then in BMMG for days 8 through 11. Organisms were recovered from the CDFF and enumerated as detailed in the text and in the legend to Fig. 1.
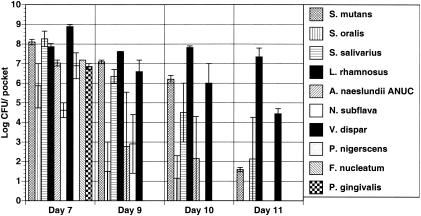
In the converse experiment, when a wild-type S. salivarius strain and urease-deficient A. naeslundii strain were utilized, community diversity after 7 days in BMMUG was essentially identical to that of the biofilms formed with 10 wild-type species (Fig. 4), indicating that the amount of ammonia generated by the S. salivarius urease was adequate to create an environment conducive to the persistence of all species. However, removal of urea from the medium resulted in a rapid loss of diversity, similar to what was observed with the wild-type, 10-species consortium, confirming that urea was required for the effect on the communities. It is also noteworthy that the pH of the biofilms on day 7 was 6.6, which was higher than when the wild-type A. naeslundii was included with the ureolytic S. salivarius (pH = 6.1 to 6.2). This difference may indicate that there is competition for urea between the two organisms and that A. naeslundii incorporates more of the amino nitrogen of urea into proteins than is released to alkalinize the environment.
FIG. 4.
Biofilms formed with the urease-deficient Actinomyces naeslundii. Biofilms were cultivated in complete BMMUG medium for 7 days and then in BMMG for days 8 through 11. Organisms were recovered from the CDFF and enumerated as detailed in the text and in the legend to Fig. 1.
Summary.
The production of an adequate amount of ammonia-generating capacity in a mixed-species model of oral biofilms is essential for the stabilization of diverse microbial communities in the face of a strong carbohydrate challenge. Loss of sufficient quantities of urease resulted in acidification of the biofilms, leading to loss of community diversity and the emergence of an aciduric flora. Extrapolating these results to supragingival biofilms in the human oral cavity would suggest that strategies that enhance the alkali-generating capacity of oral biofilms as a mechanism to inhibit caries formation may be highly effective. As our knowledge of the genetics and biochemistry of alkali-generating pathways is expanded, a clearer picture of how to design and deliver rational therapeutics that enhance the expression of ammonia-producing systems, or that enhance the persistence of alkali-producing organisms, may come into focus. Finally, this study demonstrates that the 10-species model can be adapted to the use of genetically modified organisms, allowing for the testing of a wide variety of mutants and engineered strains. Marsh and coworkers have effectively used the 10-species model for pH studies and studies on oxygen metabolism and for monitoring the effects of antimicrobials. Future studies with recombinant organisms could be performed to test a wide range of theories on interactions of microbes and effects of individual genetic determinants on community ecology.
Acknowledgments
We thank José Lemos for critical evaluation of the manuscript and Phil Marsh and David Bradshaw for providing strains and helpful advice in establishing the 10-species consortium.
This work was supported by DE13239 and DE10362.
Editor: V. J. DiRita
REFERENCES
- 1.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowden, G. H., J. Ekstrand, B. McNaughton, and S. J. Challacombe. 1990. Association of selected bacteria with the lesions of root surface caries. Oral Microbiol. Immunol. 5:346-351. [DOI] [PubMed] [Google Scholar]
- 3.Bowden, G. H., and I. R. Hamilton. 1998. Survival of oral bacteria. Crit. Rev. Oral Biol. Med. 9:54-85. [DOI] [PubMed] [Google Scholar]
- 4.Bowden, G. H. W., D. C. Ellwood, and I. R. Hamilton. 1979. Microbial ecology of the oral cavity, p. 135-217. In M. Alexander (ed.), Advances in microbial ecology. Plenum Press, New York, N.Y.
- 5.Bradshaw, D. J., P. D. Marsh, K. M. Schilling, and D. Cummins. 1996. A modified chemostat system to study the ecology of oral biofilms. J. Appl. Bacteriol. 80:124-130. [DOI] [PubMed] [Google Scholar]
- 6.Bradshaw, D. J., A. S. McKee, and P. D. Marsh. 1989. Effects of carbohydrate pulses and pH on population shifts within oral microbial communities in vitro. J. Dent. Res. 68:1298-1302. [DOI] [PubMed] [Google Scholar]
- 7.Burne, R. A. 1998. Oral streptococci: products of their environment. J. Dent. Res. 77:445-452. [DOI] [PubMed] [Google Scholar]
- 8.Burne, R. A., Y. M. Chen, D. W. Wexler, H. Kuramitsu, and W. H. Bowen. 1996. Cariogenicity of Streptococcus mutans strains with defects in fructan metabolism assessed in a program-fed specific pathogen free rat model. J. Dent. Res. 75:1572-1577. [DOI] [PubMed] [Google Scholar]
- 9.Burne, R. A., and R. E. Marquis. 2000. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol. Lett. 193:1-6. [DOI] [PubMed] [Google Scholar]
- 10.Burne, R. A., R. G. Quivey, Jr., and R. E. Marquis. 1999. Physiologic homeostasis and stress responses in oral biofilms. Methods Enzymol. 310:441-460. [DOI] [PubMed] [Google Scholar]
- 11.Carlsson, J. 1984. Regulation of sugar metabolism in relation to the “feast-and-famine” existence of plaque, p. 205-211. In B. Guggenheim (ed.), Cariology. Karger, Basel, Switzerland.
- 12.Chen, Y., K. A. Clancy, and R. A. Burne. 1996. Streptococcus salivarius urease: genetic and biochemical characterization and expression in a dental plaque streptococcus. Infect. Immun. 64:585-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, Y. Y., C. A. Weaver, and R. A. Burne. 2000. Dual functions of Streptococcus salivarius urease. J. Bacteriol. 182:4667-4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clancy, A., and R. A. Burne. 1997. Construction and characterization of a recombinant ureolytic Streptococcus mutans and its use to demonstrate the relationship of urease activity to pH modulating capacity. FEMS Microbiol. Lett. 151:205-211. [DOI] [PubMed] [Google Scholar]
- 15.Clancy, K. A., S. Pearson, W. H. Bowen, and R. A. Burne. 2000. Characterization of recombinant, ureolytic Streptococcus mutans demonstrates an inverse relationship between dental plaque ureolytic capacity and cariogenicity. Infect. Immun. 68:2621-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emanuelsson, I. R., and E. Thornqvist. 2000. Genotypes of mutans streptococci tend to persist in their host for several years. Caries Res. 34:133-139. [DOI] [PubMed] [Google Scholar]
- 17.Hamada, S., and H. D. Slade. 1980. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44:331-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keltjens, H. M., M. J. Schaeken, J. S. van der Hoeven, and J. C. Hendriks. 1987. Microflora of plaque from sound and carious root surfaces. Caries Res. 21:193-199. [DOI] [PubMed] [Google Scholar]
- 19.Kinniment, S. L., J. W. Wimpenny, D. Adams, and P. D. Marsh. 1996. Development of a steady-state oral microbial biofilm community using the constant-depth film fermenter. Microbiology 142:631-638. [DOI] [PubMed] [Google Scholar]
- 20.Loesche, W. J., and S. A. Syed. 1973. The predominant cultivable flora of carious plaque and carious dentine. Caries Res. 7:201-216. [DOI] [PubMed] [Google Scholar]
- 21.Marsh, P. D. 2003. Are dental diseases examples of ecological catastrophes? Microbiology 149:279-294. [DOI] [PubMed] [Google Scholar]
- 22.Marsh, P. D., and D. J. Bradshaw. 1997. Physiological approaches to the control of oral biofilms. Adv. Dent. Res. 11:176-185. [DOI] [PubMed] [Google Scholar]
- 23.Morou-Bermudez, E., and R. A. Burne. 1999. Genetic and physiologic characterization of urease of Actinomyces naeslundii. Infect. Immun. 67:504-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murchison, H. H., J. F. Barrett, G. A. Cardineau, and R. Curtiss III. 1986. Transformation of Streptococcus mutans with chromosomal and shuttle plasmid (pYA629) DNAs. Infect. Immun. 54:273-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterson, S., J. Woodhead, and J. Crall. 1985. Caries resistance in children with chronic renal failure: plaque pH, salivary pH, and salivary composition. Pediatr. Res. 19:796-799. [DOI] [PubMed] [Google Scholar]
- 26.Sirrakou, E. 1994. Plaque pH and plaque organic acid production in end-stage renal dialysis patients on hemodialysis. M.S. thesis. University of Rochester, Rochester, N.Y.
- 27.Sissons, C. H., and T. W. Cutress. 1987. In vitro urea-dependent pH-changes by human salivary bacteria and dispersed, artificial-mouth, bacterial plaques. Arch. Oral Biol. 32:181-189. [DOI] [PubMed] [Google Scholar]
- 28.Sissons, C. H., T. W. Cutress, and E. I. Pearce. 1985. Kinetics and product stoichiometry of ureolysis by human salivary bacteria and artificial mouth plaques. Arch. Oral Biol. 30:781-790. [DOI] [PubMed] [Google Scholar]
- 29.Sissons, C. H., E. M. Hancock, H. E. R. Perinpanayagam, and T. W. Cutress. 1988. The bacteria responsible for ureolysis in artificial dental plaque. Arch. Oral Biol. 33:727-734. [DOI] [PubMed] [Google Scholar]
- 30.Sissons, C. H., L. Wong, and M. Shu. 1998. Factors affecting the resting pH of in vitro human microcosm dental plaque and Streptococcus mutans biofilms. Arch. Oral Biol. 43:93-102. [DOI] [PubMed] [Google Scholar]
- 31.Theilade, E. 1990. Factors controlling the microflora of the healthy human mouth, p. 1-56. In M. J. Hill and P. D. Marsh (ed.), Human microbial ecology. CRC Press, Boca Raton, Fla.
- 32.van Houte, J., J. Lopman, and R. Kent. 1994. The predominant cultivable flora of sound and carious human root surfaces. J. Dent. Res. 73:1727-1734. [DOI] [PubMed] [Google Scholar]
- 33.van Ruyven, F. O. J., P. Lingstrom, J. van Houte, and R. Kent. 2000. Relationship among mutans streptococci, “low-pH” bacteria, and iodophilic polysaccharide-producing bacteria in dental plaque and early enamel caries in humans. J. Dent. Res. 79:778-784. [DOI] [PubMed] [Google Scholar]
- 34.Wimpenny, J. W. 1997. The validity of models. Adv. Dent. Res. 11:150-159. [DOI] [PubMed] [Google Scholar]