Abstract
To explore the possibility of using restriction enzymes in a synthetic biology based on artificially expanded genetic information systems (AEGIS), 24 type-II restriction endonucleases (REases) were challenged to digest DNA duplexes containing recognition sites where individual Cs and Gs were replaced by the AEGIS nucleotides Z and P [respectively, 6-amino-5-nitro-3-(1′-β-d-2′-deoxyribofuranosyl)-2(1H)-pyridone and 2-amino-8-(1′-β-d-2′-deoxyribofuranosyl)-imidazo[1,2-a]-1,3,5-triazin-4(8H)-one]. These AEGIS nucleotides implement complementary hydrogen bond donor–donor–acceptor and acceptor–acceptor–donor patterns. Results allowed us to classify type-II REases into five groups based on their performance, and to infer some specifics of their interactions with functional groups in the major and minor grooves of the target DNA. For three enzymes among these 24 where crystal structures are available (BcnI, EcoO109I and NotI), these interactions were modeled. Further, we applied a type-II REase to quantitate the fidelity polymerases challenged to maintain in a DNA duplex C:G, T:A and Z:P pairs through repetitive PCR cycles. This work thus adds tools that are able to manipulate this expanded genetic alphabet in vitro, provides some structural insights into the working of restriction enzymes, and offers some preliminary data needed to take the next step in synthetic biology to use an artificial genetic system inside of living bacterial cells.
INTRODUCTION
Type II restriction endonucleases (REases) specifically recognize short, usually palindromic, sequences of DNA duplex 4–8 nucleobase pairs in length. In the presence of Mg2+, they cleave both strands of the duplex within or near the recognition sequence. Their enormous selectivity has been extremely valuable to biotechnology. Accordingly, many type II REases have been studied in detail (1,2).
Over the last two decades, we have been working to develop a synthetic biology based on artificially expanded genetic information systems (AEGIS) (3–12). These increase from 4 to 12 the number of nucleotides able to be independently replicated (Figure 1) by exploiting different hydrogen bonding patterns within a standard Watson–Crick geometry. As this system has now been developed to the point where it may be ready to be placed into living cells (10–12), it was appropriate to ask how REases might interact with DNA molecules that contain certain AEGIS non-standard nucleotides.
Figure 1.
Structure of the C:G and Z:P pairs and the C:P and Z:G mismatched pairs. 6-Amino-5-nitro-3-(1′-β-d-2′-deoxyribofuranosyl)-2(1H)-pyridone (Z) and its Watson–Crick complement, the purine analog 2-amino-8-(1′-β-d-2′-deoxyribofuranosyl)-imidazo[1,2-a]-1,3,5-triazin-4(8H)-one (P). Nucleobase pairing in this system conforms to the Watson–Crick geometry, with large purines (or purine analogs, both indicated by ‘pu’) pairing with small pyrimidines (or pyrimidine analogs, both indicated by ‘py’) joined by hydrogen bonds. The hydrogen-bonding donor (D) and acceptor (A) groups are listed from the major to the minor groove. The arrow indicates the hydrogen-bond between donor and acceptor. Unshared pairs of electrons (or ‘electron density’) presented to the minor groove are shown by the shaded lobes. The C:P and Z:G mismatches have one too few or one too many hydrogen atoms to form a stable base pair. (A) Perfectly matched pairs, C:G and Z:P, both following the size and hydrogen bond complementary. (B) Mismatches with electron density clashes (C:P) and with the middle hydrogen bond clashed (Z:G). (C) Conversion of mismatches into formal matches through protonation at low pH (protonated C:P) and deprotonation at high pH (deprotonated Z:G).
Here, we focus on two AEGIS components in particular, a pyrimidine analog that implements a hydrogen bond donor–donor–acceptor pattern [6-amino-5-nitro-3-(1′-β-d-2′-deoxyribofuranosyl)-2(1H)-pyridone, trivially called Z] and its complementary purine analog, which implements a hydrogen bond acceptor–acceptor–donor pattern [2-amino-8-(1′-β-d-2′-deoxyribofuranosyl)-imidazo[1,2-a]-1,3,5-triazin-4(8H)-one, trivially called P] (Figure 1) (11). This focus reflects our recent success implementing a six letter GACTZP-PCR with these AEGIS components (12).
If viewed from the major and minor grooves, the Z:P pair resembles the C:G pair in many respects, having the same ‘top’ and ‘bottom’ hydrogen bonds. However, the Z:P pair differs significantly from the C:G pair by not presenting an N-7 nitrogen on the purine to the major groove, and by having a bulky nitro group presented to the major groove by the pyrimidine analog (Figure 1).
With this focus, 24 type II REases whose recognition sequences contain one or more C:G pairs were chosen for detailed examination. Specific Cs and Gs in the recognition sequence were replaced by Z and P to determine whether the enzyme recognized the modified sequences as ‘foreign’. To assist in the analysis of the results, we exploited the three crystal structures of Bcn I, EcoO109I and NotI (13–15). The results were further analyzed by modeling AEGIS nucleotides and active site amino acids in contact with the recognition sequences. Finally, we used the PspOMI REase to quantify the fidelity of PCR reactions that incorporate the Z:P pair.
MATERIALS AND METHODS
Oligonucleotides and enzymes
Oligonucleotides, except those containing Z and P, were synthesized by Integrated DNA Technologies (Coralville, IA, USA). The oligonucleotides containing Z or P were synthesized in-house on an Expedite-8900 DNA synthesizer employing standard β-cyanoethylphosphoramidite chemistry using the Z and P protected phosphoramidites (5).
Bsp120I, Bme1390I, Cfr42I, Eco52I and BcnI were purchased from Fermentas (Glen Burnie, MD, USA). Dra II was purchased from Roche (Indianapolis, IN, USA). All the other REases were obtained from New England Biolabs (Beverly, MA, USA). Deep Vent (exo−), Deep Vent (exo+), Taq and Phusion DNA polymerases were purchased from New England Biolabs.
Digestion of the AEGIS duplexes by some type-II REases
In a 10 µl reaction volume, γ32P-labeled 51-mer or 58-mer templates (0.02 pmol, Table 1) were annealed to equimolecular complementary templates by heating at 95°C for 5 min followed by slow cooling to room temperature (over 1 h). REases (0.3 µl) (Table 2) were then added to the mixtures, which were incubated at various temperatures (Supplementary Figure S1) for 16 h. Since REases vary with respect to their ability to maintain activity in a reaction over an extended period of time (16), a second batch of enzymes (0.3 µl) was added into the reaction mixtures after 8 h. This was assumed to give ‘reaction to completion’, necessary for stringent tests of specificity; the amounts of enzymes added (never less than 1.5 U/assay) and these incubation times are far more than needed to completely digest standard DNA duplexes in the amounts added.
Table 1.
Oligonucleotides used in digestion tests
| Oligonucleotides | Sequence | Schematic |
|---|---|---|
| Z-51 | 5′–GCGTAATGGATGAGGATCGAGGGCCZGGCCGGATCGATCCGGTTAATTCGC-3′ | ——————Z—————— |
| P-51 | 3′-CGCATTACCTACTCCTAGCTCCCGGPCCGGCCTAGCTAGGCCAATTAAGCG-5′ | ——————P—————— |
| C-51 | 5′–GCGTAATGGATGAGGATCGAGGGCCCGGCCGGATCGATCCGGTTAATTCGC-3′ | ——————C—————— |
| G-51 | 3′-CGCATTACCTACTCCTAGCTCCCGGGCCGGCCTAGCTAGGCCAATTAAGCG-5′ | ——————G—————— |
| Z-58 | 5′-GCGAATTAACCCTCACTAAAGTACCGZGGCCGCTTATATACTGTCACTCGTGTTACTC-3′ | ——————Z—————— |
| P-58 | 3′-CGCTTAATTGGGAGTGATTTCATGGCPCCGGCGAATATATGACAGTGAGCACAATGAG-5′ | ——————P—————— |
| C-58 | 5′-GCGAATTAACCCTCACTAAAGTACCGCGGCCGCTTATATACTGTCACTCGTGTTACTC-3′ | ——————C—————— |
| G-58 | 3′-CGCTTAATTGGGAGTGATTTCATGGCGCCGGCGAATATATGACAGTGAGCACAATGAG-5′ | ——————G—————— |
58-meroligonucleotides were used in 11 REases digestion assays (EaeI, EagI, BsaJI, BsiEi, BstUI, BtgI, MspA1I, NotI, SacII, Cfr42I and Eco52I). 51-meroligonucleotides were used in the other REases digestion assays.
Table 2.
Recognition and cleavage of the DNA duplexes containing AEGIS nucleotides by some type-II REases
| Group | REase | Recognition sequence | Z | P | Z+P | Groove recognition |
|---|---|---|---|---|---|---|
| 1 | EaeI | Y^GGCCR | Majority block | Block | Block | Major(or both major and minor) |
| PspOMI | G^GGCCC | Block | Block | Block | ||
| ApaI | GGGCC^C | Block | Block | Block | ||
| Bsp120I | G^GGCCC | Block | Block | Block | ||
| 2 | BsaJI | C^CNNGG | Cut | Cut | Cut | Minor |
| ScrFI | CC^NGG | Cut | Cut | Cut | ||
| StyD4I | ^CCNGG | Majority cut | Majority cut | Majority cut | ||
| BsskI | ^CCNGG | Cut | Cut | Cut | ||
| Bme1390I | CC^NGG | Majority cut | Majority cut | Majority cut | ||
| 3 | EagI | C^GGCCG | Block | Cut | Block | C5 of dC in major groove (may include minor groove) |
| Eco52I | C^GGCCG | Block | Majority cut | Block | ||
| BsiEI | CG^RYCG | Block | Cut | Block | ||
| MspA1I | CMG^CKG | Block | Majority cut | Block | ||
| NotI | GC^GGCCGC | Majority block | Cut | Block | ||
| SacII | CCGC^GG | Block | Majority cut | P minority cut | ||
| Cfr42I | CCGC^GG | Block | Minority cut | Block | ||
| BcnI | CC^SGG | Block | Majority cut | P minority cut | ||
| NciI | CC^SGG | Block | Cut | Block | ||
| 4 | BanII | GRGCY^C | Cut | Block | Block | N7 of dG in major groove (may include minor groove) |
| Bsp1286I | GDGCH^C | Majority cut | Block | Block | ||
| DraII | RG^GNCCY | Cut | Block | Block | ||
| EcoO109I | RG^GNCCY | Cut | Block | Block | ||
| 5 | BstUI | CG^CG | Majority cut | Cut | Complicated | Complex |
| BtgI | C^CRYGG | Majority cut | Block | Complicated |
Assays for the REases in bold and italic type used 58-mertemplates, and the others used 51-mertemplates (Table 1). The underlined nucleotides were substituted with Z in ‘Z column’ reactions; the complementary nucleotides were accordingly substituted with P in ‘P column’ reactions; those were substituted with Z and P in ‘Z+P column’ reactions. All recognition sequences are written 5′–3′ using the single-letter code nomenclature with the point of cleavage indicated by a ‘^’ (B = C or G or T, D = A or G or T, H = A or C or T, K = G or T, M = A or C, N = A or C or G or T, R = A or G, S = C or G, V = A or C or G, W = A or T, Y = C or T).
Reactions were terminated by addition of quenching buffer (98% formamide, 10 mM EDTA). Products and substrates were then resolved on 10% denaturing PAGE gels.
Application of the REase (PspOMI) in testing the fidelity of six-letter PCRs
For each six-letter nucleotide system investigated, a PCR mixture containing four standard dNTPs (200 µM each) and two AEGIS nucleotides was cycled (30–40 rounds, 95°C for 30 s, then 55°C for 30 s, then 72°C for 1 min) with identical amounts of forward and reverse primers (0.25 µM each, the forward primer 5′-labeled with 32P using T4 polynucleotide kinase) and various concentrations of templates (see Tables 3 and 4 for sequences of oligonucleotides used in the misincorporation and retention tests, respectively). After PCR amplification, aliquots of the reaction mixtures (1 µl) were digested with PspOMI (in 10 µl of reaction volume) for 16 h. Products were resolved on 10% PAGE gel and visualized by autoradiography.
Table 3.
Oligonucleotides used in Figure 3
| R-17-Std | 32P-5′-CAGGAAGGAGCGAT*CGC-3′ |
| Temp-R-81 | 5′-CAGGAAGGAGCGATCGCAACGCGTATCGATGGTACCCGGCCGGGCCCACCGCGGTCTCCCATGGGCAGTCCGTCGTCCTAG-3′ |
| F-17-Std | 3′-CGTC*AGGCAGCAGGATC-5′ |
The position of phosphorothioate linkers are indicated by asterisk. The recognition sequence of the REase PspOMI is shown in underlined bold letters.
Table 4.
Oligonucleotides used in Figure 4
| R-24 | 32P-5′-TAGGACGACGGACTGCCTATGAG-3 |
| Temp-R-72-C | 5′-CTAGGACGACGGACTGCCTATGAGAGACATGAGGGCCCGGTACCATCGATACGTTGCGATCGCTCCTTCCTG-3 |
| Temp-R-72-Z: | 5′-CTAGGACGACGGACTGCCTATGAGAGACATGAGGGCCZGGTACCATCGATACGTTGCGATCGCTCCTTCCTG-3′ |
| F-24 | 3′-TATGCAACGCTAGCGAGGAAGGAC-5′ |
The recognition sequence of REase PspOMI is shown in underlined bold letters.
RESULTS
Recognition and cleavage of AEGIS duplexes by some type-II REases
A total of 24 type-II REases were challenged to digest duplex DNA containing AEGIS nucleotides Z and P at the sites indicated in Table 2. Three kinds of pairs involving AEGIS components were tested in these experiments: Z:P pairs (in duplexes 4 and 5), Z:G mismatched pairs (in duplex 2) and C:P mismatched pairs (duplex 3) (Figure 2). Results (Supplementary Figure S1) show that these REases had differing abilities to recognize and cleave the DNA duplexes. Based on those differences, the REases were classified into five groups (Table 2).
Figure 2.
Schematic models showing the digested DNA duplexes with AEGIS components. Duplex 1–5 denotes different annealing AEGIS double-strand DNAs of γ32P-labeled 51-mer or 58-mer templates and their complementary templates (see Table 1 for the sequence of oligonucleotides used). Duplex 1 is the control standard double-strand DNAs (C-58 and G-51 were radio-labeled, respectively); Duplexes 2 and 3 are AEGIS duplexes with one indicated Z–G and C–P mismatch base pair at cleavage sites; Duplexes 4 and 5 are Aegis duplexes with one Z–P base pair at cleavage sites, which have different radiolabeled DNA strand.
The Eae I, PspOMI, Apa I and Bsp 120I enzymes were placed in ‘Group 1’. These enzymes all refused to accept the Z:P, Z:G and C:P pairs at the selected C:G sites in their respective recognition sequences. Methylation on C also blocks the activity of these enzymes (Supplementary Table S1). Z- and P-containing sequences remained uncleaved (<10% cleavage), even after 16 h of digestion, even when perfectly matched as Z:P pairs.
The BsaJI, ScrFI, StyD4I, BsskI and Bme1390I enzymes all contain an unspecified base (‘N’ in Table 2) at a site in the middle of their recognition sequences. These enzymes were all able to accept Z:P, Z:G and C:P (and, of course, C:G) pairs in those sites and cleave both strands. These formed our ‘Group 2’.
EagI, Eco52I, BsiEI, MspA1I, NotI, SacII, Cfr42I, BcnI and NciI accepted P as a replacement for dG at selected sites, but not Z, and were classified as Group 3. They cleaved the P-containing strand of DNA duplexes with a P:C mismatch (duplex 3, Figure 2) but not a Z:G mismatch (duplex 2, Figure 2). When the Z:P pair occupied the site probed (duplexes 4 and 5), Group 3 REases other than BcnI and SacII failed to cleave the duplex entirely, while BcnI and SacII still retained the ability to cleave the strand with P and displayed ‘nickase’ activity (discussed below).
Group 4 REases, including BanII, Bsp1286I, DraII and EcoO109I, were able to accept Z but not P, cleaving DNA duplexes with Z:G mismatched pair (duplex 2) but not P:C mismatched pair (duplex 3). None of these enzymes cleaved at sites where a Z:P pair replaced a C:G pair.
The remaining enzymes, BtgI and BstUI were collected as ‘Group 5′. These two enzymes gave complicated cleavage patterns that did not fit into any of the other classes.
BtgI cut the DNA duplex with Z:G mismatched pair but not P:C mismatched pair. However, it was able to cleave both strands of DNA containing Z:P pair in the recognition site. BstUI cleaved both Z:G and P:C mismatches on the Z- and P-containing strands. Interestingly, when Z:P pair was located in the recognition site, its cleavage of the Z-containing strand was substantially reduced; but the cleavage of the P-containing strand almost remained unchanged (Supplementary Figure S1).
In attempting to understand these results, we noted that for the BtgI REase, the amount of cleavage of duplex 4 (Z:P, with the Z-containing strand labeled) appeared to correlate with the cleavage of duplex 5 (Z:P, with the P-containing strand labeled) (Supplementary Figure S1B). Thus, it appeared that in any individual duplex containing the Z:P pair, either the Z-containing strand was cleaved or the P-containing strand was cleaved, but not both strands in any single duplex. As the duplexes are short (58 nt), we considered the possibility that if the AEGIS substitution destroys the synchrony of strand cleavage, and if the enzyme fails to hold the duplex in its active site for a time sufficient following cleavage of the first strand, the nicked duplex might strand-separate to give single stranded DNA, which is no longer a substrate for the REase. This hypothesis can explain results from Group 5.
To explore this hypothesis, time courses were run on the cleavage of duplex substrate using BtgI and duplexes 4 and 5 (Supplementary Figure S2A). It is evident that BtgI cleaves the Z-containing strand far more rapidly than it cleaves the P-containing strand. Further, at ‘completion’, the amount of Z-containing product plus the amount of P-containing product sum approximately to the total amount of initial substrate.
A similar time course suggests that BstU1 (Supplementary Figure S2B) might be useful as a nickase when challenged with duplexes having the Z:P substitution at its operating temperature (60°C), given appropriate selections of incubation time and enzyme amount. Here, however, the strand containing P is cleaved more rapidly than the strand containing Z, the opposite of the behavior of BtgI.
Using REase PspOMI enzymes to the fidelity of six-letter (GACTZP) PCRs
These results deliver to synthetic biology a set of tools to manipulate DNA containing AEGIS nucleotides. These include REases that do not digest sites containing Z:P pairs, REases that generate nicks in the Z-strand of duplexes containing a Z:P-pair, REases that generate nicks in the P-strand of duplexes containing a Z:P-pair, REases that reject mismatches involving Z or P, and REases that selectively degrade a strand when it is mistmatched.
To illustrate the application of these tools, we used them to quantitate the rate at which DNA polymerases replace the Z:P pair by C:G pairs or introduce the Z:P pair as a replacement for the C:G pair during multiple cycles of GACTZP ‘six letter’ PCR. Here, we exploited the refusal of Group I REases to accept either Z or P in their recognition sites. This allows them to discriminate Z:P pairs and C:G pairs quantitatively. The PspOMI Group 1 REase was chosen because its cleavage was especially well blocked if Z or P appears in its recognition sequence.
First, we used PspOMI to estimate the rate of misincorporation by four thermophilic DNA polymerases (Taq, Deep Vent exo−, Deep Vent exo+, Phusion) of Z and/or P during the PCR amplification of a standard template containing the 5′-GGGCCC-3′ recognition sequence (Figure 3A and Table 3). As shown in Figures 3B and C, almost all of the PCR products were digested by PspOMI in control experiments having only dNTPs in the PCR mixtures (Figure 3B and C, lane 1). This was as expected for largely faithful PCR amplification, with the very small amount of undigested residual material being interpreted as evidence for single-stranded material or material that had suffered a mutation involving standard nucleotides.
Figure 3.
(A) Schematic showing the use of PspOMI digestion to evaluate the misincorporation rate of Z and (or) P pair during the PCR amplifications of the standard template (see Table 3 for the sequence of oligonucleotides used). First, the standard template (Tem-R-81) were amplified for 30–40 PCR cycles in the presence of dNTPs (200μM each), dZTP (25μM) and (or) dPTP (25μM). The final amplicon duplexes contained two kinds of products: one retaining the dC:dG pair (product 1), the other misincorporating Z:P pair (product 2). The product mixtures (including product 1 and 2) were then digested by PspOMI, and the ratio between the amount of radio-labeled 81-mer oligonucleotides (full-length product, FLP) and all the oligonucleotides [including 81-mer and 42-mer oligonucleotides (digested fragment)] represents the misincorporation rate of Z and (or) P in recognition sequence during PCR amplification. (B) Misincorporation rates of PCR amplification of the standard template in the presence of the AEGIS components using Deep Vent (exo+ and exo−) DNA polymerases at indicated pH values. Four parallel PCRs were performed to amply the standard template (Table 3) containing a recognition sequence (5′-GGGCCC-3′), followed by digestion with PspOMI for 16 h. The ratio between the amount of full-length product (FLP) and all the oligonucleotides indicate the misincorporation rate and shown on the figure. Lane 1: negative control PCR amplification of the standard template (Tem-R-81) in the presence of dNTPs (200 μM each), followed by digestion with PspOMI. Lane 2: five-letter PCR amplification of the standard template (Tem-R-81) in the presence of dNTPs (200 μM each) and dZTP (25 μM), followed by digestion with PspOMI. Lane 3: five-letter PCR amplification of the standard template (Tem-R-81) in the presence of dNTPs (200 μM each) and dPTP (25 μM), followed by digestion with PspOMI. Lane 4: six-letter PCR amplification of the standard template (Tem-R-81) in the presence of dNTPs (200 μM each), dZTP (25 μM) and dPTP (25 μM), followed by digestion with PspOMI. (C) Misincorporation rates of PCR amplification of the standard template in the presence of dZTP and (or) dPTP using Taq and Phusion DNA polymerases at indicated pH values. The reactions followed the same protocol as in Figure 3B except for the polymerases.
For analogous PCR experiments where dPTP (but no dZTP) was added (Figure 3B and C, lane 2), almost all of the amplicons were also digested by PspOMI. This indicated that the P:C mismatch was infrequently introduced by these polymerases under the conditions tested, so much so as to give essentially no nuclease-resistant products even after 30–40 cycles of PCR.
However, this was not the case for the PCRs containing dZTP (Figure 3B and C, lane 3). Here, after multiple PCR cycles, a small amount of the PCR products resisted digestion by PspOMI, suggesting that some C:G pairs were replaced by Z:G pairs in the recognition sequence during the amplification. Figures 3B and 3C also show that the misincorporation rate is pH-dependent, increasing with increasing pH. Since the deprotonated form of Z (pKa ∼7.8) is formally complementary to G (Figure 1), it is not surprising that Z:G mismatched pairs evidently form more frequently at higher pH. However, in the absence of dPTP, a DNA molecule containing a Z:G mismatch is not expected to be propagated efficiently.
Accordingly, when both dZTP and dPTP were present in the PCR, the digestion results (Figure 3B and C, lane 4) showed higher amounts of PspOM1-resistent products, again increasing with increasing pH. These results imply that after Z is first misincorporated into the PCR products as a Z:G mismatched pair, P is incorporated opposite Z in the next PCR cycle. As a result, the PCR products with Z:P pairs increase with increasing number of PCR cycles, as evidenced by greater amounts of PspOMI-resistant PCR products.
The PspOMI tool was also able to compare the relative infidelity of the four DNA polymerases tested (Figure 3). Taq DNA polymerase evidently had the best ability to avoid misincorporation, as PspOMI-resistant PCR products were generated the least, even at high pH. The Deep Vent (exo+) polymerase was second best. Phusion and Deep Vent (exo−) polymerases were then approximately equal as third best.
This assay could be applied in the reverse direction, to detect the loss of the Z:P pair to give a C:G pair as its replacement. To demonstrate this, three parallel PCRs using Taq polymerase at pH 8.0 were performed to amplify a synthetic oligonucleotide containing a Z in a sequence that, if it were replaced by C, would generate the recognition site for PspOMI (Temp-R-72-Z, Table 4). The PCR products were treated with PspOMI for 16 h. In products where the Z:P pair had been replaced by C:G pair, cleavage by PspOMI was expected. Thus, the retention of the Z:P pair during PCR amplification could be estimated from the ratio between the undigested full length product (FLP) and all of the products (including FLP and the digested fragments) (Figure 4A).
Figure 4.
(A) Schematic showing the use of PspOMI digestion to evaluate the retention rate of Z:P pair during the PCR amplification of DNA containing a single Aegis nucleoside (see Table 4 for the sequence of oligonucleotides used). First, the Z-template (Temp-R-72-Z) was amplified for 30 cycles in the presence of dNTPs, dZTP and dPTP using Taq DNA polymerase. The final amplicon duplexes contained two kinds of products: one retaining the Z:P pair (product 2), the other misincorporating dC:dG pair (product 1). The product mixtures (including product 1 and 2) were then digested by PspOMI, and the ratio between the amount of radio-labeled 72-meroligonucleotides (full-length product, FLP) and all the oligonucleotides [including 72-merand 39-mer(digested fragment)] reflects the retention rate of Z:P pair in recognition sequence during PCR amplification. (B) Retention rates of Z:P pair during the PCR amplification of DNA containing a single Aegis nucleoside with Taq DNA polymerase. The experiments were carried out according to the above schematic (Figure 4A). Lane 1 (control 1): PspOMI digestion of PCR product amplified by using the standard template (Temp-R-72-C). Final concentrations of PCR reaction mixture: dNTPs (200 μM each), forward and reverse primers (0.25 μM each), template (250 pM). Lane 2 (control 2): misincorporation rate of PCR amplification of the standard template (Temp-R-72-C) in the presence of dZTP and dPTP. Final concentrations of PCR reaction mixture: dNTPs (200 μM each), forward and reverse primers (0.25 μM each), template (250 pM), dZTP and dPTP (varying as indicated). Lanes 3–5: Retention rates of Z:P pair during the PCR amplification of Z-template (Temp-R-72-Z). Final concentrations of PCR reaction mixture: dNTPs (200μM each), dZTP and dPTP (varying as indicated). The concentration of the forward and reverse primers was fixed at 0.25 μM, while the concentration of templates were 250 pM (lane 3), 25 pM (lane 4) and 2.5 pM (lane 5), respectively.
In this PCR, the primer:template ratios were 103 (Figure 4B, lane 3), 104 (Figure 4B, lane 4) and 105 (Figure 4B, lane 5), requiring, respectively, 9.97, 13.29 and 16.61 theoretical rounds of PCR to consume the primers. The per cycle retention rates of Z:P pair were obtained by the equation y = (0.5 + f / 2)r where y is the fraction of full-length product, f is the fidelity (retention rate per cycle) per round and r is the number of theoretical rounds of PCR (17).
At high concentrations of dZTP and dPTP (200 μM each, Temp-R-72-Z, Table 4, Supplementary Figure S3, lanes 3–5), 99.1% of the Z:P pair is formally retained per cycle. This represents a lower limit, as the actual number of PCR cycles must be higher than the theoretical number to consume all of the primer.
PCR of a standard GACT template in the absence of dZTP and dPTP and in the presence of dZTP and dPTP at a high concentration (200 μM each; compare the experiment above at 25 μM each) served as controls, the second seeking misincorporation of dZTP and dPTP opposite G and C at high concentrations. Here with a primer:template ratio of 103, misincorporation (6.3%, lane 2, Supplementary Figure S3) was higher than that observed with low concentrations of dZTP and dPTP (3.2% 25 μM each) (Figure 3C). Misincorporation studies with just dZTP or dPTP (not both) showed again a small amount (∼4%) of misincorporation of Z but essentially no misincorporation of P, even with 200 μM dPTP (data not shown).
The PspOMI assay was then used to evaluate efforts to find conditions that increase the fidelity of PCR amplification of Z:P pairs. Here, the concentration of dZTP was kept low (50 μM) while the concentration of dPTP was varied [from 400 to 1200 μM (Figure 4B)]. The PspOMI assay estimated the retention rate per round to be 99.3, 99.6 and 99.8% (recognizing that these too are lower bounds) (Figure 4B). Under optimal conditions, the PspOMI assay found misincorporation after 9.97 theoretical cycles of standard template to be just 3% (Figure 4B, control 2: lane 2).
DISCUSSION
We reported here the performance of 24 type-II restriction endonucleases when challenged to digest duplexes where Z:P pairs replace selected C:G pairs within their recognition sites. This generated a series of REase tools that are now available to support the synthetic biology of a six letter GACTZP DNA alphabet. We applied one of these tools, PspOMI, to quantify and compare the fidelity of six-letter PCR amplification using four DNA polymerases. We report elsewhere how these tools were used to optimize the conditions of six-letter PCR to give almost 100% retention rate of Z:P pairs per round.
A structural comparison between the C:G and Z:P pairs suggested some general hypotheses to explain the different performance of the various REases when challenged to cleave AEGIS-containing duplexes. The minor groove of the Z:P pair is quite similar to that of the C:G pair. The major groove is different, however. In the major groove, Z has an additional exocyclic nitro group at C5. This is not only large, but also likely forms a hydrogen bond to the adjacent exocyclic amino group, hindering contact to this unit. Also differing, P replaces the nitrogen at position 7 by a CH unit (Figure 1).
If we lacked crystallographic information, we might infer from these experiments where contacts are made (and not made) between various REases and their substrates (Table 2). For example, the failure of Group 1REases to accept both Z and P would imply they inspect both C5 of C and N7 of G in major groove. Using analogous reasoning, we might infer that Group 2 REases do not make contacts to the nucleobases in the major groove, as they are able to accept both Z and P. Group 3 REases evidently inspect the C5 of cytosine but not N7 of the paired guanine, and Group 4 REases evidently inspect the N7 of G but not C5 of C. Isochizomers showed similar sensitivities to substitution of C and G by Z and P (Table 2), suggesting that they make similar contacts even though they have dissimilar sequences (18,19). Also generally (and as expected), it appears that enzymes best tolerate the Z:P substitution if it is made within the recognition sequence at a site where the exact nucleotide is not specified (in Table 2, N, S, Y). Of course, given the similarity of the Z:P and C:G pairs, none of these data exclude any inspection of the minor groove.
The nitro group of Z is, broadly speaking, analogous to the methyl group of 5-methylcytosine (m5dC), a methyl group that, at certain sites, prevents the cleavage by certain REases. The sensitivity of restriction endonucleases to C-methylation (from ‘REBASE’ database, http://rebase.neb.com/cgi-bin/mslist) is collected in Supplementary Table S1. In many cases, REases that are blocked by C-methylation also do not cleave sites where the C is replaced by Z. There are, however, six exceptions (marked in blue). These are puzzling and potentially important counterexamples to general strategies for using substrate analogs as probes.
Crystal structures have been determined for three of the REases studied here (BcnI, EcoO109I and NotI), allowing us to explore these hypotheses by modeling. To this end, we modeled Z and P and amino acids in contact with these portions from the active site of these REases. The C and G in the experimentally determined crystal structures were manually modified to Z and P in the model. Then the Z:P pair was subjected to an energy minimization within the side chains extracted from the active site using Macromodel 9.7 and Maestro 9.0 (Schrodinger, LLC, New York, NY, USA, 2009), while the other parts of DNA and protein were fixed. The figures were generated in Discovery Studio Visualizer 2.5 (Accelrys Inc., San Diego, CA, USA, 2009).
BcnI cleaves duplex DNA containing the sequence CC^SGG (S = C or G, ^ designates the cleavage position) to generate single nucleotide 5′-overhangs (13). When S was replaced by Z, cleavage stopped; an S to P replacement left cleavage activity (Figure 5). This implied that contacts were made in the vicinity of C5 of C, but not to N7 of G.
Figure 5.
Digestion of AEGIS duplexes by three REases with determined crystal structures (see Table 1 for the sequence of oligonucleotides used). In a 10 µl reaction volume, 1 µl of annealed duplex 1–5 (shown as Figure 2) was digested with 0.6 µl of REase BcnI, EcoO109I and NotI for 16 h, respectively. Reactions were terminated by addition of quenching buffer (20 µl, 98% formamide, 10 mM EDTA). An aliquot (4 µl) was then loaded on the wells of lane 1–5 of denaturing PAGE gels (10%) and resolved.
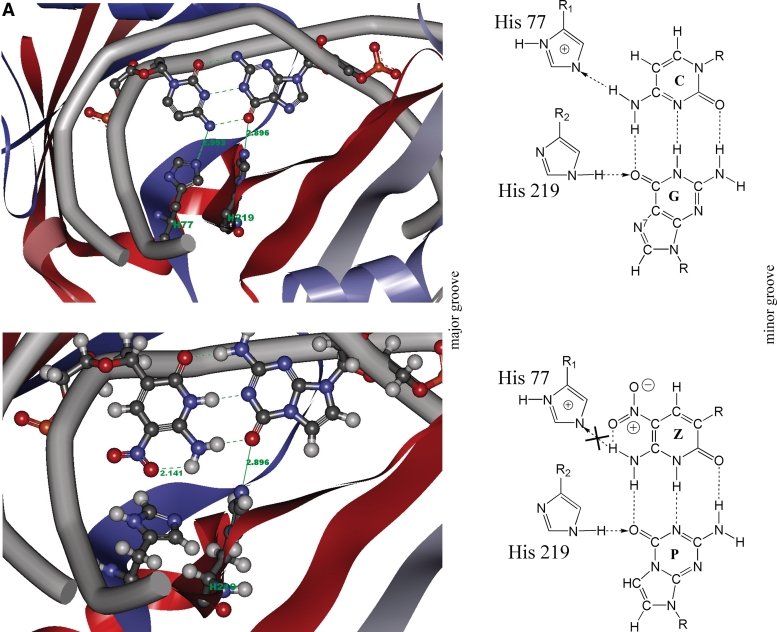
This is consistent with the BcnI crystal structure (Figure 6A). Since the minor grooves of Z:P and C:G pairs are essentially identical, we only discuss the amino acids contacting to the major groove. As shown in Figure 6A, the N4 atom of cytosine donates a hydrogen bond to the Nε atom of His77; the O6 atom of guanine accepts a hydrogen bond from the Nε atom of His219. When the central C:G pair was replaced by Z:P, the modeling found that the oxygen atom of the nitro group of Z formed an intramolecular hydrogen bond with its exocyclic NH. This breaks the intermolecular hydrogen bond between the N4 atom of Z and the Nε atom of His77, presumably disrupting the cleavage for Z. On the other hand, the nitro group of Z increased the distance between the N4 atom of Z and the Nε atom of His77 (to 3.962 Å). This also resulted in the breakage of this hydrogen bond. The structure shows that BcnI does not contact N7 of G, tolerating P at the central recognition base pair without destroying cleavage activity (Figure 5).
Figure 6.
(A1) Detailed diagram of the hydrogen bonding interactions between the central C–G (upper panel) or Z–P (lower panel) base pair (from major groove) and His 77, His 219 of BcnI (PDB: 2Q10). The hydrogen bonds are marked by green lines and their distances are labeled in green numbers. The atoms are colored by element. The lower panel shows that the oxygen atom of the nitro group of Z forms intramolecular hydrogen bond with its exocyclic NH, which disrupts the intermolecular hydrogen bond contacting to His 77. (A2) Schematic showing recognition of the central base pair from major groove with His 77 and His 219 of BcnI (recognition sequence: CCSGG). The upper panel shows the hydrogen bonding interactions between the central C–G base pair and His 77, 219 of BcnI. The arrow indicates the hydrogen-bond between donor and acceptor. The lower panel shows the hydrogen bonding interactions between the central Z–P base pair and His 77, 219 of BcnI. Here the nitro group of Z forms intramolecular hydrogen bond with its exocyclic NH. As a result, it disrupts the intermolecular hydrogen bond contacting to His 77 (indicated as cross). (B1) Detailed diagram of the hydrogen bonding interactions between the second (or sixth, by palindromy) C–G (upper panel) or Z–P (lower panel) base pair and Trp 130, Lys 173, Leu 134 of EcoO109I (PDB: 1WTE). The hydrogen bonds are marked by green lines and their distances are labeled in green numbers. H2O is presented as red sphere and the atoms are colored by element. In the lower panel, the hydrogen bond contacting to Trp 130 is retained, because the exocyclic NH of Z and the oxygen atom of acyl group of Trp 130 lie on one line. This is beneficial to the energy minimization and structural stability. On the contrary, the hydrogen bond contacting to Leu 134 is disrupted due to the substituent at 7 position of P. (B2) Schematic showing recognition of the second or sixth (by palindromy) base pair from major groove with Trp 130, Lys 173, Leu 134 of EcoO109I (recognition sequence: RGGNCCY). The upper panel shows the hydrogen bonding interactions between the second (or sixth) C–G base pair and EcoO109I. The arrow indicates the hydrogen-bond between donor and acceptor. The lower panel shows the hydrogen bonding interactions between the second (or sixth) Z–P base pair and EcoO109I. The hydrogen bond between Trp 130 and Z is retained, whereas the hydrogen bond between Leu 134 and P is disrupted (indicated as cross). (C1) Detailed diagram of the hydrogen bonding interactions between the second (or seventh) C–G (upper panel) or Z–P (lower panel) base pair and Asn 230, His 189, Gly 190 of NotI (PDB:3C25). The hydrogen bonds are marked by green lines and their distances are labeled in green numbers. The atoms are colored by element. Since the space steric hindrance of the nitro group of Z increases the distance between of the exocyclic amide-N and the oxygen of side chain carbonyl group of Asn 230, the hydrogen bond contacting to Asn 230 is disrupted. The hydrogen bond contacting to Gly 190 is also disrupted because of the substituent at 7 position of P. (C2) Schematic showing recognition of the second or seventh (symmetric) base pair from major groove with Asn 230, His 189, Gly 190 of NotI (recognition sequence: GCGGCCGC). The upper panel shows the hydrogen bonding interactions between the second or seventh C–G base pair and NotI. The arrow indicates the hydrogen-bond between donor and acceptor. The lower panel shows the hydrogen bonding interactions between the second or seventh Z–P base pair and NotI. The hydrogen bond contacting to Asn 230 and Gly 190 are destroyed (indicated as cross).
More interestingly, the Group 3 REase BcnI displayed nickase activity when challenged with a duplex replacing C:G by Z:P at the central base pair (Figure 5). This result is consistent with the crystal structure. BcnI is a monomer in solution that recognizes its target asymmetrically and nicks both DNA strands sequentially. Its crystal structure is more similar to a nickase MutH than any other structurally characterized restriction endonucleases (13). The Group 3 SacII also showed some kind of character of nickase when the target DNA duplexes contained Z:P base pairs, implying it may be a monomer in solution (Supplementary Figure S1).
EcoO109I recognizes double-stranded DNAs with a 7-bp motif, RG^GNCCY, and cleaves the phosphodiester bond between the second and third nucleotides to produce 5′-overhang DNA (14). Figure 5 showed that the C to Z replacement in the second (or sixth, by palindromy) nucleotide in the recognition sequence did not damage the cleavage activity; the structural model (Figure 6B) suggested that a hydrogen bond between the exocyclic amino group of Z and the oxygen atom of backbone C=O group of Trp130 in the active site is retained. Here, the exocyclic NH of Z presumably prefers to form an intermolecular hydrogen bond with Trp 130 over forming an intramolecular hydrogen bond with its oxygen atom of the nitro group because the exocyclic NH group of Z and the oxygen atom of Trp 130 lie in line; the resulting hydrogen bond is therefore presumably more stable than the bent intramolecular hydrogen bond. In regard to the replacement of G by P, the CH at position 7 of P cannot form a hydrogen bond the backbone NH of Leu134 (Figure 6B), leading to reduction of cleavage activity of EcoO109I (Figure 5).
NotI recognizes the eight base pair DNA sequence 5′-GC^GGCCGC-3′ and cleaves both strands of DNA to create 5′, 4-base cohesive overhangs (15). When C is replaced by Z in the second (or seventh, by palindromy) nucleotide, cleavage was significantly impaired (Figure 5). Modeling (Figure 6C) suggests that steric hindrance of the nitro group of Z increases the distance between the exocyclic amine-N of Z and the oxygen atom of side chain carbonyl group of Asn 230 (4.081 Å), destroying the intermolecular hydrogen bond and cleavage activity.
Although the C7 of P may disrupt the hydrogen bond between Gly190 and P, it does not damage the catalytic activity of NotI, indicating that this hydrogen bond may not be determinative (Figure 6C). The flexibility of NotI, which has a long eight base pair recognition site, may be related to a recent hypothesis (15) that it represents an evolutionary intermediate between mobile endonucleases (which recognize longer target sites, such as homing endonucleases) and canonical restriction endonucleases (whose recognition sites are generally only 4, 5 or 6 bp in length). Reflecting this hypothesis, NotI may have also acquired some of the lower sequence specificities of homing endonucleases, in that it tolerates one G to P replacement. Homing endonucleases do not have as stringently-defined recognition sequences as canonical type II restriction enzymes; single base changes usually do not abolish cleavage (20).
Z has an intramolecular hydrogen bond between one oxygen atom of its nitro group and exocyclic NH in the form of free-state (Supplementary Figure S4). However, this hydrogen bond is presumably weak because the N–H–O hydrogen bond is not linear. With NotI, the modeling showed that the amino group was twisted out of the plane of the pyridine ring, moving the amino group away from the nitro group (Figure 6C), weakening the intramolecular hydrogen bond further.
While these modeling results are subject to caveats appropriate for all modeling of this type, it is gratifying that they are ‘generally’ consistent with inferences that would be drawn from the cleavage data alone. This increases our confidence that inferences drawn about enzyme–substrate contacts drawn from cleavage data will be reliable to a similar extent. However, the failures of the performance of some REases with Z to correlated with their performance with methylated C are strong cautionary examples for the limitations of this approach.
These results both broaden our theoretical understanding of protein–nucleic acid interactions with these enzymes as well as our ability to manipulate this synthetic biological system in vitro. Looking forward, they should also be particularly helpful in taking the next step, moving this synthetic biology into living bacterial cells. In vivo, artificially expanded genetic information systems may well encounter restriction enzymes endogenous to many bacteria. An understanding of the outcome of such encounters will be important to predict how artificial GACTZP genetic systems behave in living cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Defense Threat Reduction Agency (DTRA-HDTRA1-08-1-0052); National Human Genome Research Institute (NHGRI-R01HG004831); National Institute of General Medical Sciences (NIGMS-R01GM081527). Funding for open access charge: NHGRI.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Stephen G. Chamberlin for helpful discussion.
REFERENCES
- 1.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010; 38(Database issue):D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piccirilli JA, Krauch T, Moroney SE, Benner SA. Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature. 1990;343:33–37. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]
- 4.Benner SA. Understanding nucleic acids using synthetic chemistry. Acc. Chem. Res. 2004;37:784–797. doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]
- 5.Sismour AM, Lutz S, Park JH, Lutz MJ, Boyer PL, Hughes SH, Benner SA. PCR amplification of DNA containing non-standard base pairs by variants of reverse transcriptase from human immunodeficiency virus-1. Nucleic Acids Res. 2004;32:728–735. doi: 10.1093/nar/gkh241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benner SA, Hutter D, Sismour AM. Synthetic biology with artificially expanded genetic information systems. From personalized medicine to extraterrestrial life. Nucleic Acids Res. 2003;3(Suppl.):125–126. doi: 10.1093/nass/3.1.125. [DOI] [PubMed] [Google Scholar]
- 7.Havemann SA, Hoshika S, Hutter D, Benner SA. Incorporation of multiple sequential pseudothymidines by DNA polymerases and their impact on DNA duplex structure. Nucleosides Nucleotides Nucleic Acids. 2008;27:261–278. doi: 10.1080/15257770701853679. [DOI] [PubMed] [Google Scholar]
- 8.Horlacher J, Hottiger M, Podust VN, Hübscher U, Benner SA. Recognition by viral and cellular DNA polymerases of nucleosides bearing bases with nonstandard hydrogen bonding patterns. Proc. Natl Acad. Sci. USA. 1995;92:6329–6333. doi: 10.1073/pnas.92.14.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z, Sismour AM, Sheng P, Puskar NL, Benner SA. Enzymatic incorporation of a third nucleobase pair. Nucleic Acids Res. 2007;35:4238–4249. doi: 10.1093/nar/gkm395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Z, Sismour AM, Benner SA. Nucleoside alpha-thiotriphosphates, polymerases and the exonuclease III analysis of oligonucleotides containing phosphorothioate linkages. Nucleic Acids Res. 2007;35:3118–3127. doi: 10.1093/nar/gkm168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Z, Hutter D, Sheng P, Sismour AM, Benner SA. Artificially expanded genetic information system: a new base pair with an alternative hydrogen bonding pattern. Nucleic Acids Res. 2006;34:6095–6101. doi: 10.1093/nar/gkl633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z, Chen F, Chamberlin SG, Benner SA. Expanded genetic alphabets in the polymerase chain reaction. Angew. Chem. Int. Ed. Engl. 2010;49:177–180. doi: 10.1002/anie.200905173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sokolowska M, Kaus-Drobek M, Czapinska H, Tamulaitis G, Szczepanowski RH, Urbanke C, Siksnys V, Bochtler M. Monomeric restriction endonuclease BcnI in the apo form and in an asymmetric complex with target DNA. J. Mol. Biol. 2007;369:722–734. doi: 10.1016/j.jmb.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto H, Shimizu T, Imasaki T, Kato M, Shichijo N, Kita K, Sato M. Crystal structures of type II restriction endonuclease EcoO109I and its complex with cognate DNA. J. Biol. Chem. 2005;280:5605–5610. doi: 10.1074/jbc.M411684200. [DOI] [PubMed] [Google Scholar]
- 15.Lambert AR, Sussman D, Shen B, Maunus R, Nix J, Samuelson J, Xu SY, Stoddard BL. Structures of the rare-cutting restriction endonuclease NotI reveal a unique metal binding fold involved in DNA binding. Structure. 2008;16:558–569. doi: 10.1016/j.str.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. New England BioLabs. http://www.neb.com/nebecomm/tech_reference/restriction_enzymes/survival_restriction_endonucleases_in_reaction.asp (8 December 2010, date last accessed)
- 17.Sismour AM, Benner SA. The use of thymidine analogs to improve the replication of an extra DNA base pair: a synthetic biological system. Nucleic Acids Res. 2005;33:5640–5646. doi: 10.1093/nar/gki873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovall RA, Matthews BW. Type II restriction endonucleases: structural, functional and evolutionary relationships. Curr. Opin. Chem. Biol. 1999;3:578–583. doi: 10.1016/s1367-5931(99)00012-5. [DOI] [PubMed] [Google Scholar]
- 19.Niv MY, Ripoll DR, Vila JA, Liwo A, Vanamee ES, Aggarwal AK, Weinstein H, Scheraga HA. Topology of Type II REases revisited; structural classes and the common conserved core. Nucleic Acids Res. 2007;35:2227–2237. doi: 10.1093/nar/gkm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.