Abstract
The vertebrate 2-5A system is part of the innate immune system and central to cellular antiviral defense. Upon activation by viral double-stranded RNA, 5′-triphosphorylated, 2′–5′-linked oligoadenylate polyribonucleotides (2-5As) are synthesized by one of several 2′–5′-oligoadenylate synthetases. These unusual oligonucleotides activate RNase L, an unspecific endoribonuclease that mediates viral and cellular RNA breakdown. Subsequently, the 2-5As are removed by a 2′-phosphodiesterase (2′-PDE), an enzyme that apart from breaking 2′–5′ bonds also degrades regular, 3′–5′-linked oligoadenylates. Interestingly, 2′-PDE shares both functionally and structurally characteristics with the CCR4-type exonuclease–endonuclease–phosphatase family of deadenylases. Here we show that 2′-PDE locates to the mitochondrial matrix of human cells, and comprise an active 3′–5′ exoribonuclease exhibiting a preference for oligo-adenosine RNA like canonical cytoplasmic deadenylases. Furthermore, we document a marked negative association between 2′-PDE and mitochondrial mRNA levels following siRNA-directed knockdown and plasmid-mediated overexpression, respectively. The results indicate that 2′-PDE, apart from playing a role in the cellular immune system, may also function in mitochondrial RNA turnover.
INTRODUCTION
Interferons (IFNs) are vertebrate-specific cytokines with key roles in innate immunity (1,2). They induce a range of biological responses, of which their antiviral, antitumor and immuno-modulatory effects are of major medical significance (3,4). Upon viral infection, cells begin synthesizing and secreting the IFNs, which then interact with specific receptors at the cell surface to activate an intracellular signal cascade, eventually leading to the changes in gene expression that dictate the biological outcome.
The 2-5A system forms a major part of the cellular antiviral defense in which IFNs initially induce expression of a set of double-stranded RNA-dependent 2′–5′-oligoadenlyate synthetases (dsRNA-dependent 2′–5′-OASs). Upon activation by viral dsRNA, these enzymes begin synthesizing a set of unusual 5′-triphosphorylated, 2′–5′-linked oligoadenylate polyribonucleotides (2-5As) from ATP with the general structure, pppA(2′p5 ′A)n (5). Accumulation of 2-5As then causes activation of RNase L, an otherwise latent endoribonuclease, leading to the degradation of viral RNA. However, RNase L is only moderately specific towards viral RNA and will degrade cellular RNA if not deactivated (6,7). Consequently, the intracellular 2-5A level is quickly downregulated by a nuclease, which, in contrast to canonical exonucleases, degrades 2′–5′-linked RNA (8,9).
The removal of 2-5As is thought to initially involve dephosphorylation by a 5′-phosphatase, as activation of the pathway by viral infection and IFN stimulation in mice and in cell cultures have been shown to lead to accumulation of 5′-dephosphorylated 2-5As (2-5A core molecules) rather than their 5′-triphosphorylated counterparts (10,11). Although less active in stimulating RNase L (12), the dephosphorylated 2-5A core molecules still have significant antiviral and anti-growth properties and need to be fully removed to shut down the response (13,14). Both phosphorylated and dephosphorylated 2-5As can be degraded by a 2′–5′-specific exoribonuclease known as 2′-phosphodiesterase [2′-PDE (also named phosphodiesterase 12 (PDE12))], which is thus a key regulator of the 2-5A system (9).
2′-PDE is also able to degrade 3′–5′-linked oligoadenylate RNAs in vitro (9,15,16), and has therefore been classified as a putative RNA deadenylase (17). Deadenylases are a diverse group of enzymes responsible for removing the poly(A) tail during early mRNA turnover in the eukaryotic cytoplasm. Most deadenylases are 3′–5′ exonucleases that produce AMP by hydrolyzing the phosphodiester bonds of RNA in a divalent metal ion-dependent fashion (17). The process represents the first and rate-limiting step during mRNA turnover and is therefore highly regulated (17,18). All deadenylases characterized to date belong to either the DEDD or the exonuclease–endonuclease–phosphatase (EEP) superfamily (17). In DEDD-type nucleases, the active site consists of three aspartate residues and one glutamate that coordinate two essential divalent metal ions (19–21). In humans, members of this group include the POP2, CAF1Z, PARN and PAN2 proteins. The EEP-type nucleases, to which 2′-PDE belongs, also depend on divalent metal ions and contain an active site composed of aspartate and histidine residues (22,23). This group comprises enzymes active on both DNA and RNA, and includes the CCR4, Nocturnin and ANGEL proteins.
Here we show that human 2′-PDE, unlike other components constituting the 2–5A system (24–29), is located in the mitochondrial matrix compartment, and comprises a general 3′–5′ exoribonuclease exhibiting a preference for oligo-adenosine RNA like canonical cytoplasmic deadenylases (21,22,30–33). Furthermore, we document the presence of a marked negative association between 2′-PDE and mitochondrial mRNA levels, following siRNA-directed knockdown and plasmid-mediated overexpression, respectively. Collectively, the results suggest that 2′-PDE may play a dual role in cells, being involved in mitochondrial RNA turnover as well as the 2-5A system.
MATERIALS AND METHODS
2′-PDE expression constructs
The cDNA clone DKFZp667B1218 (GenBank accession no. BX647169) was used as template for PCR amplification of the 2′-PDE fragments, inserted into either pcDNA3 (Invitrogen) or pTriEx-3 Neo (Novagen).
Cell culturing and transfection
HeLa, HEK293 or COS-7 cells were grown in DMEM (Gibco BRL/Life Technology) containing 10% FBS and 1% Penicillin/Streptomycin (Gibco BRL/Life Technology). HEK293 and HeLa cells were transfected using the PolyFect Transfection Reagent (Qiagen), and COS-7 cells were transfected using Lipofectamine (Invitrogen). Cells were lysed with 1% NP40, 0.5 M CH3COOK containing 2 × Complete protease inhibitor cocktail (Roche). The cell lysates were centrifuged at 20 000g for 3 min and the supernatants recovered as protein extracts. Protein concentrations were estimated using the BCA Protein Assay Kit (Pierce).
Escherichia coli expression and protein purification
Escherichia coli BL21 (DE3) cells were transformed with 2′-PDE615-His, 2′-PDEΔmTP600-His and OAS1-p42 in pTriEx-3 Neo, and as a control also with empty vector pTriEx-3 Neo. Then the bacteria were grown in LB medium containing 50 µg/ml ampicillin before induction at OD600 = 0.8 with 1 mM IPTG for 10 h at 25°C. Bacteria were pelleted at 10 000g for 20 min and resuspended in 300 mM KCl, 15% glycerol, 5 mM MgCl2, 5 mM β-mercaptoethanol, 1 mM PMSF, 1× protease inhibitor cocktail and 50 mM Tris–HCl, pH 7.5. This suspension was sonicated and lysed by high-pressure homogenization at 15 000–20 000 psi. Cell debris were spun down at 30 000g for 1 h and discarded. For purification, the supernatant was filtered through a 0.45-μm filter and applied to a Ni-NTA column (QIAGEN) pre-equilibrated in 300 mM KCl, 20 mM imidazole, 5 mM β-mercaptoethanol, 1 mM PMSF, 1× protease inhibitor cocktail and 50 mM Tris–HCl, pH 7.5. Proteins were eluted by a step-gradient using 300 mM imidazole. The Ni-NTA eluate was diluted and applied to a MonoQ HR 16/10 column (GE Healthcare) pre-equlibrated in 50 mM KCl, 5 mM β-mercaptoethanol and 50 mM Tris–HCl, pH 7.5, and eluted with a linear KCl gradient of 0.2%/ml going from 50 to 500 mM. Fractions were pooled and concentrated on a 5-kDa MWCO Vivaspin filter (Sartorius), and stored in 50% glycerol.
SDS–PAGE and immunoblotting
Proteins were denatured at 95°C for 5 min and applied to 10% SDS–PAGE. Gels were stained in Coomassie Brilliant Blue R-250 (Pierce). For immunoblotting, proteins were electro-blotted onto Immobilon-P PVDF membranes (Millipore). Membranes were blocked in 5% skimmed milk powder in PBS-T (PBS with 0.05% Tween-20), washed in PBS-T and incubated with anti-His antibodies (Rockland or GenScript Corporation). Membranes were washed in PBS-T and incubated with either of two HRP-conjugated IgGs, goat anti-rabbit antibody (DakoCytomation) or goat anti-mouse antibody (GE Healthcare). Proteins were detected with the ECL reagent (Amersham ECL PlusTM, GE Healthcare).
Bioinformatics
The following online bioinformatic tools were used: WoLF PSORT (http://wolfpsort.org/), pTARGET (http://bioapps.rit.albany.edu/pTARGET/), MitoProt II (http://mips.helmholtz-muenchen.de/cgi-bin/proj/medgen/mitofilter), PSORT (http://psort.ims.u-tokyo.ac.jp/form.html), GPMAW (http://www.gpmaw.com/Downloads/downloads.html), Phobius (http://phobius.sbc.su.se/), TopPred (http://www.cbib.u-bordeaux2.fr/pise/toppred.html) and MultiAlin (http://bioinfo.genotoul.fr/multalin/multalin.html).
Immunocytochemistry
HeLa and HEK293 cells were seeded on coverslips, transfected with PolyFect and incubated for 36 h. The growth media was supplemented with 100 nM Mitotracker (Red CMXRos, Invitrogen) for 30 min. Cells were fixed with 4% PFA, washed in PBS, permeabilized with 0.2% Triton-X 100, washed again and finally blocked with 0.1% Tween-20 in PBS. Cells were then incubated with an anti-His antibody (Rockland or Genscript), washed and incubated with a FITC-conjugated secondary antibody (Sigma). Cells were finally washed, air-dried and the coverslips mounted using Vectashield (Vector Laboratories). Images were acquired with a Zeiss LSM510 MetaConfocal microscope. The brightness and contrast of the resulting images were adjusted using the LSM Image Software.
Synthesis of 2′–5′ oligoadenylate tetramer core, ApApApA
In 200 ml, 17 μg/ml of a highly active 2′–5′-OAS1 was incubated with 2 mM ATP in 4 mM Mg(OAc)2, 0.2 mM DTT, 40 µM EDTA, 0.2 mg/ml Poly(I)-Poly(C), 0.1 mg/ml BSA, 2% glycerol and 4 mM Tris–HCl, pH 7.8 for 3 h at 37°C. The ATP concentration was increased from 2 to 10 mM by addition of 1 mM ATP every 10 min. The sample was heat-inactivated at 85°C for 15 min, filtered (0.45 µm) and then applied 100 U/ml alkaline phosphatase (Roche Applied Science) overnight at 37°C. The sample was then divided into 10 ml aliquots and each aliquot loaded onto a MonoQ HR 16/10 column pre-equilibrated in 20 mM Tris–HCl, pH 7.5, and eluted with 1 M NaCl using a step-wise linear gradient of 0–10% over 3.6-column volumes and 10–30% over 24-column volumes. The tetramer oligoadenylate core fractions were then pooled, diluted in 20 mM Tris–HCl, pH 7.5, again applied to the MonoQ HR 16/10 column and finally eluted with a step-gradient to 0.5 M NaCl. The identity of the tetramer core was determined by MS and a concentration estimate was obtained by MonoQ Sepharose using ATP as an internal control.
Degradation assay and determination of Km and Vmax
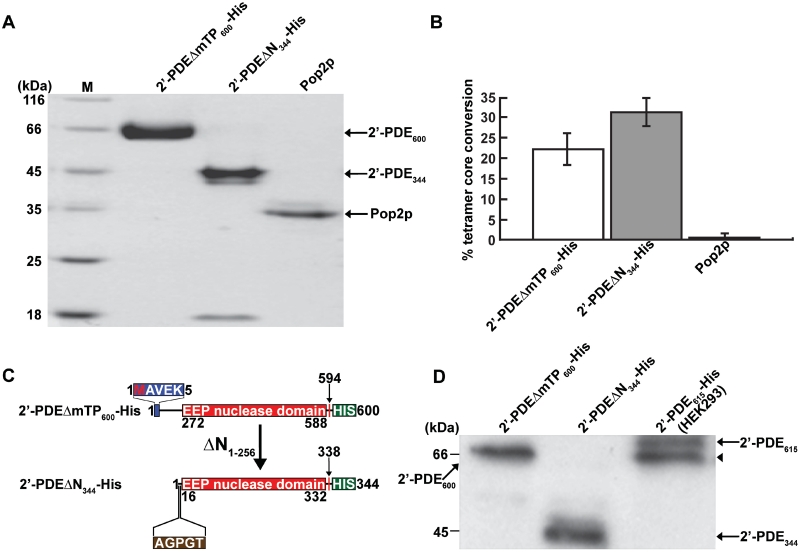
For the 2′–5′-linked tetramer, ApApApA, reactions were carried out with 1 pmol/μl 2′-PDE and 0.5 mM substrate in 5 mM MgCl2, 1 mM DTT, 1 mg/ml BSA, 1x protease inhibitor cocktail and 20 mM Hepes, pH 7.0 for 1 h at 37°C. The samples were diluted in 20 mM Tris–HCl, pH 7.5 and filtered on AcroPrepTM 10K Filter Plates (Pall Corporation). The filtrates were applied to a MonoQ HR 5/5 column (GE Healthcare) pre-equilibrated in 20 mM Tris–HCl, pH 7.5, and eluted with 1 M NaCl using a step-wise linear gradient of 0–12% and 12–16% over 18-column volumes and 20-column volumes, respectively. The percentage tetramer core conversion was calculated by integrating the individual peaks.
The 2′–5′ oligoadenylate degradation assay was repeated with the 3′–5′-linked tetramer substrates, ApApApA and CpCpCpC (Dharmacon), using 1 pmol/µl 2′-PDE and varying substrate concentrations (25 µM–1.5 mM). Reactions were incubated 20 min at 37°C and processed as above by filtration, MonoQ Sepharose fractionation and peak integration. Km and Vmax kinetic parameters were determined by non-linear regression and Michaelis–Menten enzyme kinetics using the GraphPad Prism Software.
For the 30-mer RNA- and DNA-degradation assay, the RNA–Poly(A), the RNA Stemloop–Poly(A) and the DNA–Poly(A) substrates were synthesized with a 5′-fluorescein label whereas the RNA–Poly(C) substrate was synthesized with a 5′-TAMRA label (Invitrogen). All substrates were gel-purified. In vitro reactions contained a 100-fold excess of enzyme (1 μM) over RNA (10 nM) and were carried out in 50 mM KCl, 7.1 mM MgCl2 and 10 mM Tris–HCl, pH 8.0. Reactions were incubated at 30°C for up to 2 h, quenched with RNA-loading buffer (8 M urea, 0.5% (w/v) bromophenol blue, 0.5% (w/v) xylene cyanol FF, 20 mM EDTA and 5 mM Tris–HCl, pH 8.0), separated by denaturing gel electrophoresis on 20% urea-acrylamide gels running at 15 W and the bands visualized with a Typhoon 9400 and Typhoon Trio scanner (GE Healthcare). The RNA and DNA ladders were constructed by limited alkaline hydrolysis of the respective substrates in 50 mM NaCO3, pH 8.9 for 10 min at 96°C followed by neutralisation with 300 mM Na(CH3COO), pH 4.5.
Differential cell processing
Cells were harvested in PBS using a cell scraper, pelleted at 1000g for 10 min, resuspended in 220 mM mannitol, 70 mM sucrose, 1 mM EDTA, 2 mg/ml BSA, 2× protease inhibitor cocktail, 0.5 mM PMSF and 20 mM Hepes, pH 7.6, homogenized with a Dounce homogenizer, and the cell debris pelleted at 1000g for 10 min. Recovered supernatants were centrifuged for 20 min at 10 000g, and the pellet (mitochondria) and supernatant (cytosol and microsomes) split up. Mitochondria were resuspended as above but without BSA, and any remaining cell debris pelleted at 1000g for 10 min. The supernatants were centrifuged, again for 20 min at 10 000g, and the resulting mitochondrial pellets resuspended in urea buffer (6 M urea and 50 mM Tris–HCl, pH 8.0) for MS and gel-fractionation, or 1x OFFGEL Solution (6.7 M urea, 2 M thiourea, 60 mM DTT and 1.2% ampholytes, pH 3–10) for off-gel fractionation, or BSA Buffer (3 mg/ml fatty acid-free BSA, 250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 2 mM KH2PO4, 5 mM methionine and 10 mM MOPS–KOH, pH 7.2) for in vitro mitochondrial import. The cytosol and microsomes were segregated by centrifugation at 330 000g for 30 min and the supernatant (cytosol) TCA-precipitated and resuspended in urea buffer. Mitochondrial protein concentration was estimated using the Bradford assay reagent (Pierce) and the cytosolic protein concentration using the BCA method.
35S-labeling and in vitro mitochondrial import
35S-labeled 2′-PDE615-His was synthesized at 30°C for 1.5 h using the TNT T7 Quick Coupled Transcription/Translation system (Promega). Import reactions were carried out in 1% ethanol, 0.45 mg/ml mitochondria, 1.25 μl 35S-labeled 2′-PDE, 5 mM ATP, pH 7.0 and 10 mM sodium succinate, pH 7.5 in BSA buffer. The membrane potential (Δψ) was disrupted by substituting ethanol with 80 μM antimycin A, 10 μM valinomycin and 20 μM oligomycin and leaving out sodium succinate. Reactions were carried out at 30°C for different time-points and stopped with 2 mM valinomycin. Non-imported protein was digested for 10 min at 4°C using 50 μg/ml proteinase K followed by inhibition of the protease with 2 mM PMSF at 4°C for 10 min. Mitochondria were pelleted as above and washed by resuspension in SEM Buffer (250 mM sucrose, 1 mM EDTA, 2 mM PMSF and 10 mM MOPS–KOH, pH 7.2). The pellets were finally dissolved in sample buffer containing 2 mM PMSF and applied to SDS–PAGE. For the carbonate extraction experiment, mitochondria were exposed to 0.1 M sodium carbonate (pH 11.5) for 30 min on ice after protein import. Samples were then ultracentrifuged at 100 000g for 30 min at 4°C and the supernatants (soluble and peripheral membrane proteins) exposed to TCA-precipitation in 125 pg/µl sodium deoxycholate. Proteins were finally redissolved in PMSF-containing sample buffer and allowed to solubilize by incubation for 10 min at RT. Ultracentrifuged pellets (integral membrane proteins) were resuspended in sample buffer containing PMSF. For the mitochondrial swelling experiment, mitochondria were resuspended in SM Buffer (250 mM sucrose with 10 mM MOPS–KOH, pH 7.2) after protein import and treated with nine volumes 10 mM MOPS–KOH, pH 7.2 for 15 min on ice, or as a control, kept intact by soaking in nine volumes SM Buffer. Accessible proteins were removed by proteinase K treatment or left untreated as control. The mitochondria/mitoplasts were then pelleted by centrifugation, washed in SEM Buffer and redissolved in sample buffer.
For autoradiography, samples were applied to SDS–PAGE and the gel fixed and dried. Proteins were visualized by phosphorimaging using the Typhoon 9400 and Typhoon Trio scanner.
Tryptic digest of crude mitochondria
Mitochondrial proteins (∼1 mg) dissolved in urea buffer were reduced in 10 mM DTT for 18 h at 37°C, then alkylated in 30 mM iodoacetamide for 1 h at room temperature. The reaction was stopped by adding additional DTT to 40 mM. The proteins were diluted 10 times with 1 mM CaCl2 in 10 mM Tris–HCl, pH 8.0, and digested using 5 ng/µl trypsin for a period of 18 h at 37°C. The pH of samples for MS analysis was adjusted to approximately 2–3 with 0.1% TFA.
Off-gel fractionation and preparation for MS
Proteins were off-gel fractionated at 200 V for 25 h (5 kVh) and 50 µA (3100 OFFGEL fractionators, Agilent) into 12 fractions using immobilized pH gradient gel strips covering the pH interval 3–10. The fractions were digested with trypsin and the peptides purified by Zip-Tips (0.2 µl C18 resin, Millipore). Peptides were finally dried down by SpeedVac centrifugation and redissolved in 0.1% TFA.
RP-HPLC and MS
Samples were analyzed using the UltiMate 3000 LC system (Dionex) linked to a micrOTOF-Q ESI-Qq-TOF mass spectrometer (Bruker Daltonics, Bremen). The software Bruker Compass HyStar (Version 3.2-SR2, Bruker Daltonics) was used to link the RP-HPLC and the mass spectrometer controlling software packages. The HPLC used a C18 RP-column (PepMap 100 nanocolumn, Dionex) to fractionate the peptides. Peptides were applied to the column, the column washed in 2% acetonitrile + 0.1% formic acid and peptides eluted with a linear gradient of 98% acetonitrile in 0.1% aqueous formic acid. The gradient was run from 0% to 2%, from 2% to 42% and from 42% to 99% over, respectively, 20, 160 and 76 µl with a flowrate of 4 µl/min. The peptides MS and MS/MS spectra were recorded online by the mass spectrometer.
MS sample files were processed, denoted and deconvoluted using DataAnalysis (Version 4.0 SP1, Bruker Daltonics, Germany). The peptide spectra were exported to BioTools (Version 3.1, Bruker Daltonics, Germany) and matched against the peptides resulting from a virtual tryptic digest of 2′-PDE. To estimate mitochondrial purity, MS/MS data were applied to the Mascot search engine (Version 2.2, Matrix Science, United Kingdom) and probed against the full repertoire of human proteins contained in the Swiss-Prot protein database. Purities were deduced based on the known cellular localization of the detected proteins.
siRNA knockdown and plasmid-mediated overexpression of 2′-PDE in HeLa cells, for qRT-PCR
A pool of four siRNAs (target sequences: 5′-GAAGAUUGAAUUAGCGUGU-3′, 5′-AGUACAAGGUGGAGCGCAA-3′, 5′-CGGCAUCUCUACACGAAGA-3′ and 5′-CAAACUCAGCCUCGAAUUU-3′) (Dharmacon) was employed to knockdown the 2′-PDE gene expression level (2′-PDE ON-TARGET plus SMARTpool siRNA). A negative control siRNA was targeting GFP (target sequence: 5′-GACGTAAACGGCCACAAGTC-3′) (Dharmacon). Plasmid-mediated protein overexpression was done using the 2′-PDE615-His construct cloned in pcDNA3.
siRNA and plasmid transfections of HeLa cells were carried out in 6-well plates using the DharmaFECT 1 (Dharmacon) or PolyFect (Qiagen) Transfection Reagents, respectively, according to the manufacturer’s instructions. 72 hours (siRNA) and 36 h (plasmid) post-transfections, cells were harvested and the total RNA isolated using the RNeasy Mini Kit (Qiagen).
Quantitative reverse transcriptase PCR analysis
First-strand cDNA was prepared using 1.5 µg total RNA, oligo(dT)18 primer and the MMLV Reverse Transcriptase (Epicenter Biotechnologies) in a final volume of 50 µl. Each quantitative reverse transcriptase (qRT)-PCR reaction contained 1 µl first-strand cDNA and 200 nM of both forward and reverse primers in a final volume of 20 µl, and was carried out using the Platinum SYBR Green qPCR SuperMix UDG Kit supplemented with ROX Reference Dye (Invitrogen). Gene-specific primer sets were as follows: 2′-PDE forward, 5′-GTCATCAATGGCAGCATTCCAGAG-3′; 2′-PDE reverse, 5′-CTATTTCCATTTTAAATCACATACAAGTGC-3′; CoxI forward, 5′-ACCCTAGACCAAACCTACGCCAAA-3′; CoxI reverse, 5′-TAGGCCGAGAAAGTGTTGTGGGAA-3′; CoxII forward, 5′-ACAGATGCAATTCCCGGACGTCTA-3′; CoxII reverse, 5′-GGCATGAAACTGTGGTTTGCTCCA-3′; CoxIII forward, 5′-ACTTCCACTCCATAACGCTCCTCA-3′; CoxIII reverse, 5′-TGGCCTTGGTATGTGCTTTCTCGT-3′; CytB forward, 5′-TCCTCCCGTGAGGCCAAATATCAT-3′; CytB reverse, 5′-AAAGAATCGTGTGAGGGTGGGACT-3′; ND4 forward, 5′-ACAAGCTCCATCTGCCTACGACAA-3′; ND4 reverse, 5′-TTATGAGAATGACTGCGCCGGTGA-3′; ND6 forward, 5′-AGGATTGGTGCTGTGGGTGAAAGA-3′; ND6 reverse, 5′-ATAGGATCCTCCCGAATCAACCCT-3′; RPL13A forward, 5′-CCTGGAGGAGAAGAGGAAAGAGA-3′; RPL13A reverse, 5′-TTGAGGACCTCTGTGTATTTGTCAA-3′. The results were obtained with the Mx3005P System (Stratagene), and the PCR thermocycling conditions were: 95°C for 10 min, and 40 cycles of 95°C for 10 s, 58°C for 20 s and 72°C for 20 s. Band sizes of PCR products were verified by standard agarose gel electrophoresis, and the melting-curve analysis of all PCR products displayed single melting peaks. Signal levels were normalized to the ROX reference dye for correction of errors due to differences in plastic-ware transparency and reflectivity, or aliquoting errors. The outcome Ct values were converted to quantities using the comparative Ct method, incorporating the efficiencies of the reaction with each primer set, which were determined as described previously (34). Relative gene expression levels were normalized to values of the RPL13A gene, encoding a stably expressed ribosomal protein (35).
RESULTS
2′-PDE contains an N-terminal mitochondrial signal peptide
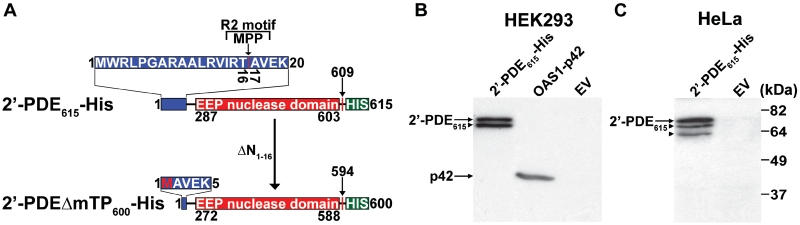
Initially, full-length 2′-PDE (609 amino acids) was cloned from human cDNA into the eukaryotic expression vector pcDNA3, to encode a 615 amino acids (68 kDa) fusion protein containing a C-terminal 6х His-tag (2′-PDE615-His) (Figure 1A). After transient transfections, two (HEK293 cells, Figure 1B) or three (HeLa cells, Figure 1C) specific bands were observed corresponding to the full-length protein and either degraded or post-translationally processed products. No degradation was observed upon expression of a control construct containing the 2′–5′-oligoadenylate synthetase, OAS1-p42, suggesting that protein processing could be involved in generating the multiple bands.
Figure 1.
Constructs and expression of 2′-PDE in human cells. (A) The 2′-PDE constructs. 2′-PDE615-His encodes the full-length protein added a C-terminal His-tag, whereas 2′-PDEΔmTP600-His makes up an N-terminal truncated form of the 2′-PDE615-His devoid of the first 16 residues (vertical arrow in top row) and containing an artificial initiator methionine (M). The EEP-nuclease domain is boxed in red, and the mitochondrial matrix-targeting peptide, mTP in blue. The matrix-targeting R2 motif, xRx↓(S/x) and associated proteolytic-cleavage site (MPP) are indicated. Vertical red lines specify the final residue occurring in the natural protein. The initiator methionine in 2′-PDEΔmTP600-His, not present in the native protein, is highlighted in red. MPP, mitochondrial processing peptidase. (B) and (C) 2′-PDE615-His was transfected into the human HEK293 and HeLa cell lines and protein expression allowed to proceed for 36 h upon which cells were lysed and analyzed by immunoblotting using an antibody against the His-tag (Rockland). Extracts from cells transfected with a construct encoding His-tagged OAS1-p42 or an empty-pcDNA3 vector (EV) were used as positive and negative controls, respectively. Arrows indicate the full-length proteins whereas arrowheads specify N-terminally processed forms. Of total protein, 20 µg were loaded into each lane.
To further analyze the likelihood of protein processing and possibly a sub-cellular localization other than the cytosol, the algorithms WoLF PSORT and pTARGET were used to predict the cellular localization of 2′-PDE based on sequence (36). Both clearly identified a mitochondrial signal peptide, also referred to as a matrix-targeting peptide, mTP, in the N-terminus of 2′-PDE (37,38). MitoProtII (39), a bioinformatic tool designed specifically for the identification of mTPs as well as the most likely N-terminal processing site confirmed the putative mitochondrial localization with a probability of 95% with the mTP predicted to constitute residues 1–16 of the precursor (Figure 1A). Specifically, the N-terminus of 2′-PDE contains an R2 motif, xRx↓x(S/x), where the arrow marks a proteolytic cleavage site, in this case between Thr16 and Ala17. The presence of an R2 motif in 2′-PDE suggests that mitochondrial import proceeds through a single cleavage event mediated by a matrix-localized protease known as mitochondrial processing peptidase, MPP (40). A characteristic feature of mTPs is the presence of an amphipathic α-helix at the N-terminus of the protein, which gets recognized by membrane-bound import receptors in the mitochondrion (41–43). To explore this possibility, GPMAW (44) was used to predict the secondary structure in the N-terminus of 2′-PDE and a likely helical element was found covering the initial 27 residues. Helical-wheel analysis subsequently confirmed that the amino acid residues 7–24 would be consistent with an amphipathic helix (Supplementary Figure S1).
Recombinantly expressed 2′-PDE co-localizes with mitochondria in an mTP-dependent fashion
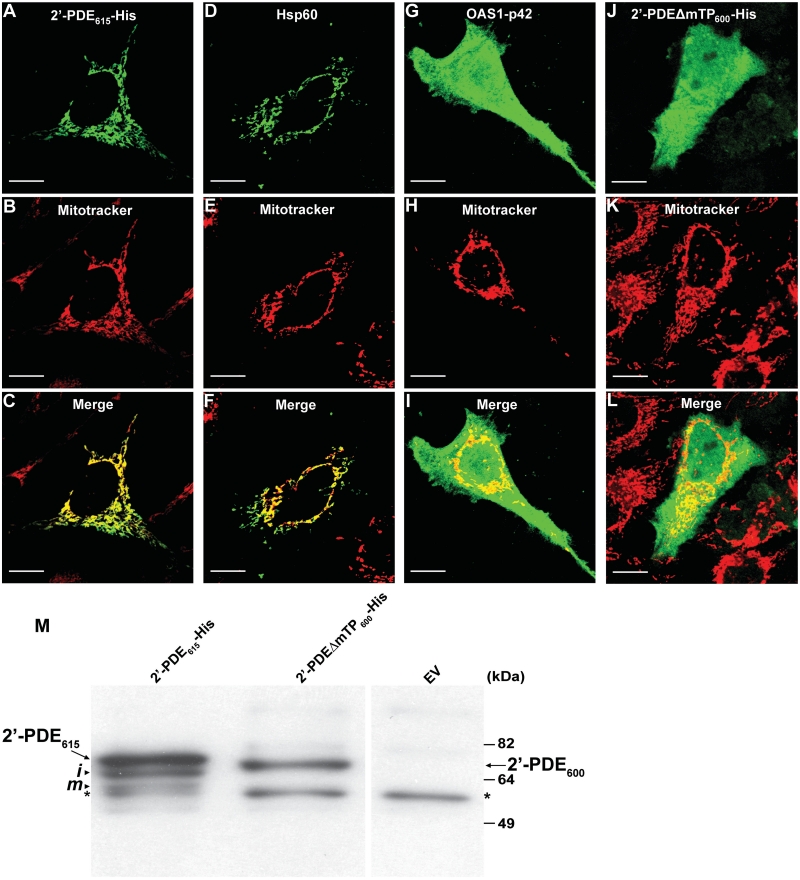
Prompted by our bioinformatic discoveries, we next wanted to experimentally confirm the sub-cellular localization of recombinant 2′-PDE expressed in human cell lines. Accordingly, the 2′-PDE615-His construct was transfected into HeLa cells, and the cells then analyzed by immuno-fluorescence microscopy in which mitochondria were tracked by an organelle-specific stain (Mitotracker) and the 2′-PDE by an antibody toward the C-terminal His-tag (Figure 2). The results clearly demonstrated an almost complete co-localization of 2′-PDE with mitochondria under these conditions (Figure 2A–C). His-tagged constructs encoding the known matrix-localized heat shock protein 60, Hsp60 (45) and the non-mitochondrial protein, OAS1-p42 (26) were included as positive and negative controls, respectively. To confirm the significance of the predicted mTP, a variant lacking the expected signal peptide, 2′-PDEΔmTP600-His (Figure 1A) was analyzed in a similar way. This deletion variant showed a uniform distribution throughout the cell, and furthermore was found not to undergo post-translational processing to the same extent as the full-length protein (Figure 2M). Given the detection of an unspecific band in the immunoblot in Figure 2M, it was important to ascertain that cross-reactivity of the anti-His antibody did not hamper the interpretation of the results described in Figure 2A–L. This was confirmed by conducting parallel immuno-fluorescence analyses on HeLa cells not transfected, in addition to HeLa cells transfected with empty vector pcDNA3, which did reveal no background staining whatsoever (data not shown). Finally, these data defining 2′-PDE as a mitochondrial protein were affirmed by similar stainings done on 2′-PDE-transfected HEK293 cells, which also showed a mitochondrial distribution of the recombinant protein (Supplementary Figure S2).
Figure 2.
Cellular localization and post-translational processing of 2′-PDE. (A–L) Immuno-fluorescence microscopy images of HeLa cells transiently transfected with constructs encoding 2′-PDE615-His (A–C), the His-tagged Hsp60 (D–F) and OAS1-p42 controls (G–I) or the 2′-PDEΔmTP600-His deletion variant (J–L). After 36 h, cells were probed with a His-tag specific antibody (Genscript) and stained with a FITC-conjugated secondary antibody (green). Mitchondria were visualized by treating live cells with Mitotracker (red). Scale bars; 10 μm. (M) Immunoblotting on HeLa protein extracts obtained 36 h post-transfection with 2′-PDE615-His and 2′-PDEΔmTP600-His using a His-tag specific antibody (Genscript). EV (empty-pcDNA3 vector) was used as a negative control. Arrows specify full-length 2′-PDEs, whereas arrowheads mark the ‘intermediate’ (i) and ‘mature’ (m) processed forms of 2′-PDE615-His. The asterisks indicate an unspecific band. Of total cell lysate, 20 μg were loaded into each lane. Note that due to practical reasons the results in Figure 2 were obtained with a different anti-His antibody as compared with the one utilized in Figure 1.
In summary, 2′-PDE appears to locate to the mitochondria via an mTP-dependent mechanism. The presence of at least three forms of 2′-PDE when expressed in HeLa cells suggests that a minimum of two processing events are required to generate the mature, mitochondrial form of 2′-PDE, which is most likely represented by the band of lowest molecular mass. The protein forms are therefore denoted ‘precursor’, ‘intermediate’ and ‘mature’, by decreasing size.
Recombinant 2′-PDE associates with the mitochondrial sub-cellular fraction
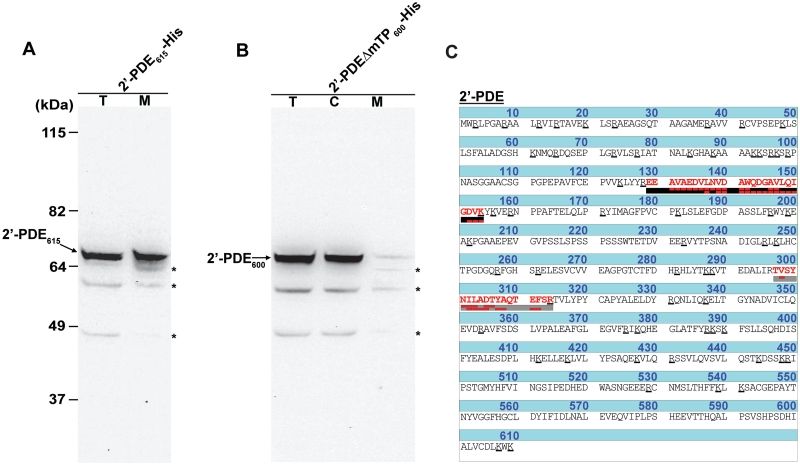
To further analyze the complex processing and localization pathway of 2′-PDE, COS-7 cells transiently expressing the protein were separated into cytosolic and mitochondrial fractions through differential centrifugation of cellular compartments. COS-7 cells were used in this case for practical reasons and are generally assumed to maintain a very homologous mitochondrial import and processing function to the one found in human cells. In this set-up, the full-length form encoded by 2′-PDE615-His and the construct lacking the mTP, 2′-PDEΔmTP600-His, were used. If the predicted mTP is indeed functional, the truncated construct should be retained in the cytosol and thus represent an appropriate control that would also validate the purity of the mitochondrial fractions utilized in these experiments. Following a 36 h transient expression, His-tag immunoblotting on the total cell lysate and a mitochondrial fraction clearly showed that the 2′-PDE615-His associates with mitochondria (Figure 3A). In contrast, the 2′-PDEΔmTP600-His is not targeted to this organelle, but is exclusively found in the cytosolic fraction (Figure 3B).
Figure 3.
Human recombinant and endogenous 2′-PDE targets the mitochondria of cells. (A) Immunoblotting on total cell lysate (T) and mitochondrial (M) fractions isolated upon transient expression of 2′-PDE615-His for 36 h in COS-7 cells using a His-tag specific antibody (Genscript). (B) Immunoblotting as in (A) on 2′-PDEΔmTP600-His-specific total cell lysate (T), mitchondrial (M) and cytoplasmic (C) samples obtained following 36 h of recombinant expression in COS-7 cells. In (A) and (B), arrows indicate 2′-PDEs and asterisks mark unspecific bands. Twenty micrograms of total cell lysate/cytoplasm and 5 µg of mitochondrial protein were loaded into the respective lanes. (C) Intrinsic and mitochondrial-specific 2′-PDE peptides (red) in HEK293 cells as determined by LC-MS/MS of a tryptic digest prepared from all proteins in off-gel fraction 6 (emphasized in red in Supplementary Figure S4). The detected y-ions (upper red boxes) and b-ions (lower red boxes) are shown, and the Arg(R)/Lys(K) residues underscored in the protein sequence.
The identity of the 2′-PDE band associated with mitochondria was subsequently confirmed by LC-MS/MS of tryptic peptides prepared from all proteins contained in the mitochondrial fractions. Accordingly, within the 2′-PDE615-His-specific sample, twelve peptides were found matching 2′-PDE (Supplementary Figure S3), whereas none were identified in the mitochondrial fraction from cells expressing 2′-PDEΔmTP600-His. This is despite a relatively low-estimated mitochondrial purity of ∼39% based on the known localization of a wide range of other identified proteins in the samples.
Endogenous 2′-PDE associates with mitochondria
As protein expression by transient transfection could potentially produce localization artifacts (46), we decided to check if endogenous 2′-PDE also localizes to mitochondria using a similar MS-based approach. In this case, human HEK293 cells were used due to a known high expression level of endogenous 2′-PDE (9). Mitochondrial fractions were isolated as before and protein complexity reduced by off-gel isoelectric focusing in the pH interval 3–10. In this set-up, a single mitochondrial preparation was split into 12 individual samples distinguished by a difference in pH (Supplementary Figure S4). Individual samples were then treated with trypsin and analyzed by LC-MS/MS. Two peptides specific for 2′-PDE were identified in the sample corresponding to a pI of approximately 6, consistent with the predicted pI of 6.11 for full-length 2′-PDE (Figure 3C). No 2′-PDE-specific peptides were found in the remaining samples. In total, 545 proteins were identified of which 225 represented known mitochondrial proteins yielding a mitochondrial purity of ∼41%. Note that in comparison to this HEK293-specific enzymatic identification, no peptides derived from endogenous 2′-PDE were identified in the 2′-PDEΔmTP600-His mitochondrial preparation isolated from COS-7 cells (described in the previous section). Specifically, this disparity is likely to be the result of either (1) the fact that the COS-7 sample was not off-gel fractionated prior to LC-MS/MS analyses, or (2) simply that the 2′-PDE exhibits differential expression between COS-7 and HEK293 cells.
2′-PDE synthesized in vitro targets the mitochondrial matrix
We next wanted to analyze the functional significance of the observed mitochondrial localization of 2′-PDE by identifying in which mitochondrial sub-compartment, i.e. the outer membrane, the intermembrane space, the inner membrane or the matrix, the protein is located. The presence of the R2-sequence motif immediately suggests that the protein would localize to the matrix, a destination also supported by the sub-cellular prediction tool PSORT (47). In addition, the trans-membrane prediction servers, Phobius (48) and TopPred (49), did not locate any membrane-associated elements, suggesting that 2′-PDE most likely is a soluble matrix protein.
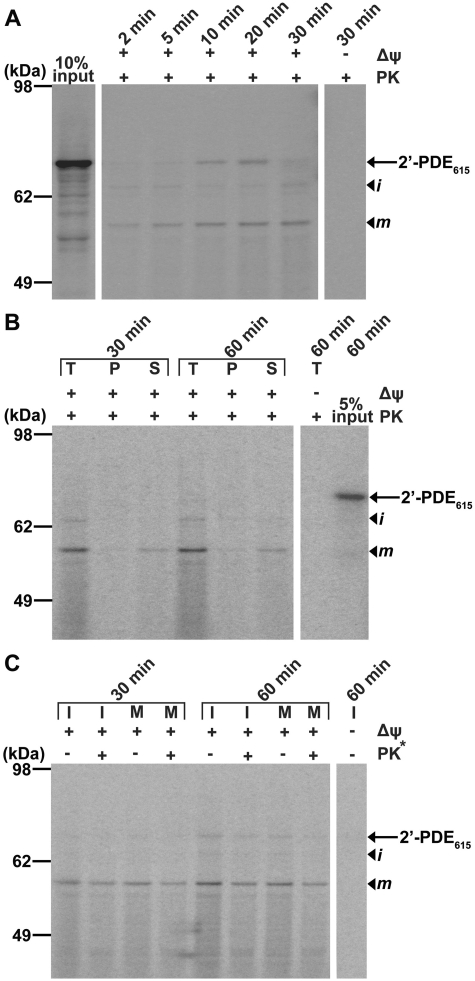
To confirm this hypothesis, the import of in vitro synthesized, 35S-labeled 2′-PDE615-His into mitochondria was studied by incubation of the radioactive protein with organelles obtained from HeLa cells. First, a control time-course experiment was conducted whereby components, i.e. 35S-labeled 2′-PDE and mitochondria, were mixed for 2–30 min followed by gel-electrophoretic analysis of the mitochondria pre-treated with proteinase K, to reveal the efficiency of import (Figure 4A). In this case, protein bands corresponding to the ‘precursor’, ‘intermediate’ and ‘mature’ forms of 2′-PDE appeared over time, showing that 2′-PDE615-His is both imported into mitochondria and at the same time undergoes N-terminal processing like seen for the recombinantly expressed protein. Furthermore, mitochondrial import was found to be dependent on the membrane potential across the inner membrane (Δψ), which is consistent with a localization to the matrix as only precursor proteins destined for the inner membrane or matrix and in rare cases the intermembrane space, show Δψ-dependence during import (50).
Figure 4.
Mitochondrial import of radioactively labeled 2′-PDE. (A) Time-course experiment in which 35S-labeled 2′-PDE615-His and HeLa mitochondria were incubated for 2–30 min followed by proteolysis of non-imported proteins by treatment with proteinase K (+PK). The sequestered 35S-labeled proteins were subsequently separated by SDS–PAGE and visualized by phosphorimaging. Δψ indicates whether the mitochondrial membrane potential has been disrupted (−) or not (+). The 10% input lane shows the result of in vitro transcription/translation prior to import, and is relative to the total quantity of 35S-labeled 2′-PDE added into each import reaction (100%). The arrow marks the position of 2′-PDE615-His and arrowheads the location of the processed ‘intermediate’ (i) and ‘mature’ (m) forms. (B). Carbonate extraction experiment in which import was performed as in (A) for 30 and 60 min, after which the mitochondria were split by carbonate extraction into protein fractions containing the integral membrane (P) and the soluble and peripheral membrane parts (S). Specifically, whereas the T lanes provide the total quantity and pattern of 35S-labeled protein after mitochondrial import but prior to carbonate extraction, the 5% input lane presents the 35S-2′-PDE615-His-starting material prior to import, and is relative to the amount utilized for each reaction (100%). (C) Osmotic-induced swelling experiment in which mitoplasts (M) were obtained after import for 30 and 60 min by osmotic-induced rupturing of the outer membrane followed by proteolysis (where indicated). The asterisk marks that this PK treatment constitutes the second treatment used to remove accessible proteins upon stripping of the outer membrane. Shown are also the control experiments where the membrane was kept intact (I).
We then wanted to clarify whether 2′-PDE is part of the soluble mitochondrial proteome represented by the matrix and the intermembrane space, or alternatively, associates with the inner mitochondrial membrane. To analyze this, mitochondria were subjected to carbonate extraction after import of the pre-protein, a technique that allows the separation of soluble and peripheral membrane proteins (S) from integral membrane proteins (P) (51). Specifically, the 35S-labeled 2′-PDE615-His was firstly incubated with the mitochondria, this time for 30–60 min, upon which the organelles pre-treated with proteinase K were carbonate extracted, and the obtained fractions analyzed by gel electrophoresis (Figure 4B). The results clearly indicated that 2′-PDE is present in the soluble phase, i.e. either in the intermembrane space or in the matrix.
Osmotic-induced swelling of mitochondria, whereby the outer membrane is burst by lowering the external solute concentration in a mitochondrial suspension while leaving the inner membrane intact, can be used to distinguish matrix proteins from proteins located in the intermembrane space. In this technique, proteins within or facing the intermembrane space become accessible to an external protease, while matrix-located proteins remain shielded. Accordingly, the subsequent isolation and analysis of mitoplasts, the remaining particles comprising the matrix enclosed by the inner membrane, allow for discrimination between intermembrane space and matrix proteins (52). Thus, using mitoplasts isolated from HeLa mitochondria incubated with 35S-labeled 2′-PDE615-His for 30–60 min, the sub-organellar localization of 2′-PDE could be deduced from gel analysis (Figure 4C). The ‘mature’ protein seemed unaffected by protease treatment of the isolated mitoplasts [lanes marked M (mitoplasts) with or without proteinase K treatment], suggesting that 2′-PDE is a matrix protein. Although this does not formally exclude that 2′-PDE might be partly embedded in the inner membrane with no parts protruding into the intermembrane space, we have already shown that 2′-PDE forms part of the soluble mitochondrial phase and therefore conclude that 2′-PDE is exclusively a mitochondrial matrix protein.
2′-PDE contains an EEP-type deadenylase domain and is an active 3′–5′ exoribonuclease
Sequence analyses show that 2′-PDE contains a so-called EEP nuclease domain and hence shares similarity with the cytoplasmic deadenylase families CCR4, Nocturnin and ANGEL (Supplementary Figure S5). To characterize the in vitro deadenylase activity of 2′-PDE, the enzyme was heterologously expressed and purified from E. coli. In doing so, the 2′-PDE615-His and 2′-PDEΔmTP600-His-encoding templates were firstly subcloned into the IPTG-inducible vector pTriEx-3 Neo, and the constructs transformed into E. coli BL21(DE3). Then, prior to purification, IPTG-induced lysates expressing 2′-PDE615-His and 2′-PDEΔmTP600-His, respectively, were tested for their ability to degrade a 2′–5′-linked adenosine tetramer core (ApApApA) to uncover a potential influence of removing the mTP on enzyme activity. Specifically, the lysates were found to degrade the 2′–5′ substrate with similar efficiency, suggesting only a negligible impact (if any) of the mTP on enzyme catalysis (Supplementary Figure S6A). A lysate from cells transformed with an empty-pTriEx-3 Neo vector was used as a control and found to be inactive, in accordance with the fact that 2′–5′ RNA is not a natural substrate of any endogenous E. coli enzymes. Immunoblotting was used to confirm that the two constructs expressed equally well in the BL21(DE3) strain (Supplementary Figure S6B).
Consequently, based on an unaltered activity response (compared to full-length 2′-PDE) the 2′-PDEΔmTP600-His was chosen and purified to homogeneity (Figure 5A), so as to simulate the mature mitochondrial form as far as possible. During the expression and purification of 2′-PDEΔmTP600-His, a stable degradation product of ∼45 kDa, referred to as 2′-PDEΔN344-His, was also isolated (Figure 5A). Both 2′-PDEΔmTP600-His and its truncated form were active in degradation of 2′–5′-linked substrates, but surprisingly, the truncated protein appeared to be more active than 2′-PDEΔmTP600-His (Figure 5B). Mapping of the N-terminus of 2′-PDEΔN344-His by N-terminal sequencing showed that it lacks the first 256 residues compared to 2′-PDEΔmTP600-His, and starts just upstream of the core 2′-PDE nuclease domain (Figure 5C). His-tag specific immunoblotting was likewise carried out confirming that in both 2′-PDEΔmTP600-His and 2′-PDEΔN344-His, the C-terminal His-tag is preserved (Figure 5D).
Figure 5.
Purification and validation of 2′-PDE expressed in E. coli BL21(DE3). (A) Coomassie blue-stained gel after SDS–PAGE showing 20 µg of purified 2′-PDEΔmTP600-His and 2′-PDEΔN344-His. Pop2p is a purified DEDD-type deadenylase from Schizosaccharomyces pombe used as a control in (B) (20). Arrows specify the position of the indicated proteins. The M lane contains marker proteins of different molecular masses. (B) 2′–5′ oligoadenylate degradation assay on the purified protein fractions from (A) using a 2′–5′ oligoadenylate tetramer, ApApApA, as substrate. The deadenylase Pop2p is only active on 3′–5′-linked RNA and thus constitutes a negative control for 2′–5′ oligoadenylate degradation. Reactions were performed in triplicate (mean/SEM) using 10 µg of protein. (C) The 2′-PDEΔN344-His truncation fragment and comparison to 2′-PDEΔmTP600-His. The five N-terminally sequenced residues of 2′-PDEΔN344-His are shown (brown box). (D) Immunoblotting of the 2′-PDE proteins from (A) using a His-tag specific antibody (Genscript). A HEK293 total cell lysate expressing the 2′-PDE615-His was employed as a positive control. Arrows denote individual proteins and the arrowhead designates the singly processed form of 2′-PDE615-His present in the HEK293 cell lysate. Of total protein, 10 µg were loaded into each lane.
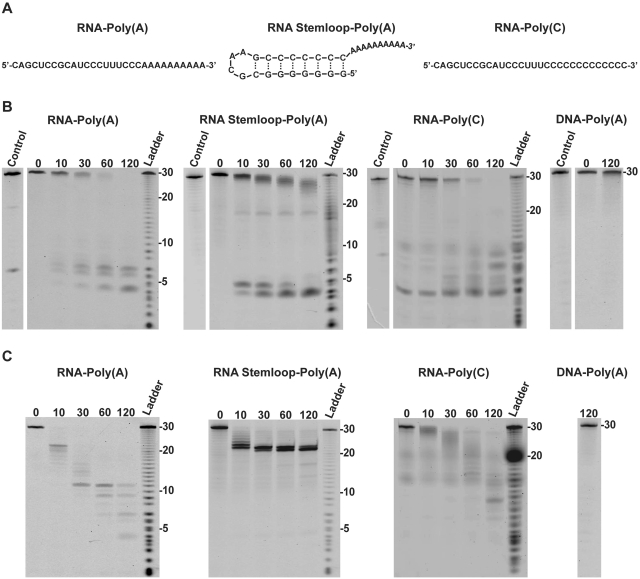
To examine a possible role of 2′-PDE in RNA deadenylation, an in vitro degradation assay was set up using defined 30-mer 3′–5′-linked RNA oligos fluorescently labeled at the 5′-end (Figure 6A). RNA–Poly(A) substrate contained a 20-residue generic unstructured RNA sequence followed by 10 adenosine residues, while RNA Stemloop–Poly(A) contained an 8-bp strong helical structure followed by 10 adenosine residues (20). The degradation of the substrates by 2′-PDE was monitored in time-course experiments carried out with either 2′-PDEΔmTP600-His or 2′-PDEΔN344-His using denaturing RNA gels (Figure 6B and C, respectively). When 2′-PDEΔmTP600-His is incubated with RNA–Poly(A), the substrate is removed in a slow and processive manner as seen by the immediate build-up of small RNA fragments of 5–8 nt in size, without the appearance of intermediate length RNAs. In contrast, the truncated protein, 2′-PDEΔN344-His, initially only removes the poly(A) tail from RNA–Poly(A), as seen by accumulation of completely deadenylated RNAs of 20 nt after ∼10 min (Figure 6C). However, upon longer incubation the RNA is degraded into small fragments of 4–11 nt in size. 2′-PDEΔmTP600-His also degrades RNA–Poly(C) with reasonable efficiency, demonstrating that 2′-PDE is not specific towards adenosine. The RNA–Poly(C) substrate is likewise broken down by 2′-PDEΔN344-His, although at a much slower rate compared to the RNA–Poly(A) substrate. These results suggest that 2′-PDE confers some degree of sequence preference towards adenosine, yet still possess the ability to degrade other sequences. This behavior is very similar to what is observed for the canonical, cytoplasmic deadenylation enzymes (21). The 2′-PDEΔN344-His is not only faster than 2′-PDEΔmTP600-His, but appears to work in a more distributive degradation mode, indicating an inherent functional significance of the N-terminal domain. RNAs containing a strong secondary structure are also processed differently by the two 2′-PDE variants. While 2′-PDEΔmTP600-His is able to remove a strong secondary structure element down to fragments of 4–5 nt, the short variant, 2′-PDEΔN344-His, arrests 2 nt before the stem. This suggests that the N-terminus of 2′-PDE contains elements that affect both processivity and secondary structure unwinding of the main exonuclease domain, most likely through an RNA-binding domain. Unfortunately, bioinformatics did not seem to support this notion, suggesting that if present, this RNA-binding domain of 2′-PDE may substantially deviate from known RNA-binding regions. None of the 2′-PDE variants were able to degrade a 2′-deoxy substrate, DNA–Poly(A), which is equivalent to RNA–Poly(A) but on the DNA form. Furthermore, a BL21(DE3)-specific protein fraction obtained from cells transformed with an empty-pTriEx-3 Neo vector and purified according to the same parameters as used for the 2′-PDEs was inactive on all RNA substrates (data not shown), strongly suggesting that any contaminating traces of co-purified nuclease activity is unlikely to contribute to the results seen in Figure 6.
Figure 6.
The 3′–5′ exoribonuclease activity of human 2′-PDE. (A) Sequence and predicted secondary structure of the RNA substrates used. (B and C) Time-course degradation assays where purified 2′-PDEΔmTP600-His (B) or 2′-PDEΔN344-His (C) were incubated with defined RNAs (or DNA where specified) at 30°C for the indicated amount of minutes. The DNA–Poly(A) oligomer was similar to the RNA–Poly(A) but on the DNA form. Ladder is a single-nucleotide ladder produced by limited alkaline hydrolysis of the substrates. Control lanes specify reactions in which the RNAs (or DNA) were incubated for 120 min at 30°C in the absence of enzyme.
In conclusion, 2′-PDE exhibits a general 3′–5′ riboexonucleolytic activity with a subtle preference for adenosine like that observed for the canonical deadenylases.
2′-PDE degrades 3′–5′-linked RNA with a preference for adenosine
To address the apparent preference for adenosine further, two 3′–5′-linked substrates (ApApApA, oligo-A and CpCpCpC, oligo-C) were used to determine Michaelis–Menten Km and Vmax values for 2′-PDE. Both 2′-PDEΔmTP600-His and 2′-PDEΔN344-His were able to degrade these short substrates completely down to a nucleoside (adenosine or cytidine, respectively) and a 5′-monophosphate nucleotide (AMP or CMP, respectively) as judged by the chromatographic analyses of products and determination of peak ratios (∼1:3) (Supplementary Figure S7). For both 2′-PDE forms, the Km on oligo-A is approximately half of that measured for oligo-C with a Vmax 1.4–1.7-fold higher for the oligo-A substrate (Table 1). Thus, 2′-PDE both has a higher affinity and activity for polyadenosine 3′–5′-linked RNA and therefore closely matches the substrate specificity of cytosolic deadenylases, defined as 3′–5′ exoribonucleases that prefer RNAs composed of adenosines (21).
Table 1.
Kinetic parameters
| Substrate | 2′-PDEΔmTP600-His |
2′-PDEΔN344-His |
||
|---|---|---|---|---|
| Vmax(fmol AMP*/s*/ pmol) | Km (µM) | Vmax(fmol AMP*/sec*/ pmol) | Km (µM) | |
| oligo-A, ApApApA | 11.4 ± 0.5 | 74 ± 13 | 18.7 ± 0.8 | 72 ± 14 |
| oligo-C, CpCpCpC | 8.2 ± 0.4 | 126 ± 26 | 10.8 ± 0.6 | 142 ± 31 |
2′-PDEΔmTP600-His and 2′-PDEΔN344-His were incubated with varying substrate concentrations of oligo-A, ApApApA or oligo-C, CpCpCpC, substrates at 37°C for 20 min followed by determination of the Michaelis–Menten Km and Vmax values based on the MonoQ elution profiles obtained as in Supplementary Figure S7. The Km and Vmax parameters were derived by non-linear regression using the GraphPad Prism Software. All reactions were performed in triplicate and is shown as mean±SEM.
A negative correlation exists between 2′-PDE and mitochondrial mRNA expression levels
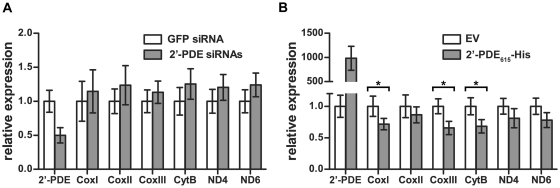
To elucidate if 2′-PDE is functionally involved in mitochondrial RNA turnover, either as an RNA deadenylase or alternatively as a general RNA decay enzyme, HeLa cells were transfected with a mix of four 2′-PDE siRNAs to downregulate the natural protein. 72 hours post-transfection, total cellular RNA was then obtained and qRT-PCR performed to evaluate an effect of 2′-PDE knockdown on chosen mitochondrial mRNAs, i.e. the cytochrome c oxidase subunit I (CoxI), CoxII, CoxIII, cytochrome b (CytB), NADH dehydrogenase subunit 4 (ND4) and ND6. Specifically, down-regulation of 2′-PDE by ∼50% was found not to evoke any statistical differences in mitochondrial mRNA expression levels, compared with cells transfected with a GFP control siRNA (Figure 7A). Despite hereof, the results seemed to indicate a slight negative correlation between 2′-PDE and mitochondrial mRNAs, suggesting that 2′-PDE may ultimately act to destabilize these RNAs.
Figure 7.
2′-PDE destabilizes mitochondrial mRNAs. qRT-PCR analyses of mitochondrial mRNAs following (A) siRNA knockdown, or (B) plasmid-mediated overexpression, of 2′-PDE in HeLa cells. In (A), cells were transfected with a pool of 2′-PDE siRNAs and incubated for 72 h prior to performing qRT-PCR analysis. Cells transfected with a GFP-specific siRNA were used as a negative control. In (B), 2′-PDE615-His transfectants were analyzed by qRT-PCR 36 h post-transfection. Cells treated with empty vector (EV) pcDNA3 were employed as a negative control. (A and B) Relative expression was calculated by normalization to RPL13A expression levels followed by gene-specific normalization, setting the controls to 1. Data are averages of triplicate qRT-PCR reactions, and the error bars denote standard deviations. Both experiments were repeated three times, each time with similar results. Asterisks denote those mRNAs exhibiting differential expression.
In order to test and validate this hypothesis, the full-length 2′-PDE615-His construct (in pcDNA3) was transfected into HeLa cells and the protein overexpressed for a period of 36 h. Any given effect of 2′-PDE on mitochondrial mRNAs was then evaluated by qRT-PCR as above. Overexpression of the 2′-PDE was found to markedly destabilize mitochondrial mRNAs, compared with cells transfected with control empty vector pcDNA3 (Figure 7B). Specifically, three of the six mRNAs were significantly down-regulated, counting the CoxI (28%↓), CoxIII (34%↓) and CytB (32%↓) mRNAs. The remaining mRNAs were only insignificantly reduced. Collectively, these data therefore put forward the 2′-PDE as a likely catabolic player involved in mitochondrial RNA turnover.
DISCUSSION
Human 2′-PDE is thought to control the cellular level of 2-5As through specific degradation, thereby acting as a negative regulator of the IFN-induced 2-5A system (9). In this article, we present strong evidence that the enzyme contains an N-terminal mitochondrial signal peptide and possesses a sub-cellular localization to the mitochondrial matrix. At present, two matrix-localized peptidases, MPP and mitochondrial intermediate peptidase (MIP), are known to participate in maturation of mitochondrial proteins (53). A small number of nuclear-encoded proteins are matured through sequential cleavage by MPP and MIP (54). The appearance of an intermediate-sized 2′-PDE band in HeLa cells could suggest that 2′-PDE undergoes such two-step maturation. Sequential maturation of precursor proteins occur through recognition of the R10 motif, xRx|(F/L/I)xx(S/T/G)xxxx↓, where the vertical line and arrow indicate the cleavage sites for MPP and MIP, respectively (40). Thus MPP leaves an intermediate with an N-terminal octapeptide extension which is subsequently removed by MIP (54,55). However, two findings indicate that the final step in maturation of 2′-PDE is not catalyzed by MIP. First, an R2 motif, specific for MPP and not MIP rather than the R10 motif was found in 2′-PDE. Second, the size difference between the ‘intermediate’ and ‘mature’ forms of ∼7 kDa is not compatible with the removal of an octapeptide (∼1 kDa). It is therefore possible that the second cleavage event in maturation of 2′-PDE results from the activity of a yet unidentified peptidase (56,57). Three similar forms of 2′-PDE have previously been observed in human cells, but in this case the phenomenon was ascribed to the usage of alternate translation start codons triggered by ‘weak’ Kozak signals (9). Specifically, the Kozak consensus for initiation of translation in vertebrates is: (G/A)CCATGG (58). In this sense, a ‘strong’ Kozak signal contains a purine (either G or A) at position −3 and a G nucleotide at position +4 relative to the adenine of the ATG initiator positioned at +1 (there is no number 0 position). Further, an ‘adequate’ consensus contains only one of these sites, while a ‘weak’ consensus has neither. Indeed the genome sequence around the ATG start codon in 2′-PDE, TTCATGT, represents a ‘weak’ Kozak signal, thus rendering this ATG triplet prone to ribosome skipping. However, the presence of a ‘strong’ Kozak sequence at the second ATG triplet, GCGATGG, encoding Met35 makes ribosome skipping at this position highly unlikely. Furthermore, even though an ATG codon at Met35 is in accordance with the observed size difference between the ‘precursor’ and the ‘intermediate’ form (2–4 kDa), the next ATG triplet found at Met63 is not compatible with the ∼7 kDa that separates the ‘intermediate’ and ‘mature’ band. Consequently, we argue that the appearance of three 2′-PDE forms results from protein processing, and not the usage of alternate translation start codons.
The observed localization of 2′-PDE to the mitochondrial matrix immediately seemed puzzling in light of its known role within the predominant nuclear- and cytosolic-based 2-5A system, suggesting an additional more mitochondria-specific activity of this enzyme (24–29). Based on both a functional and structural similarity of 2′-PDE to RNA deadenylases, this might very well be in mitochondrial RNA turnover (9,15–17). The current finding of an inverse relationship between 2′-PDE and mitochondrial mRNA expression levels, supports this hypothesis and demonstrates that 2′-PDE may act to degrade these RNAs. Whether this is the result of a principal role in RNA deadenylation or in general RNA decay, however, remains to be elucidated.
In human mitochondria, both RNA deadenylation and general RNA decay remains an unresolved issue (59,60). Previously, mammalian polynucleotide phosphorylase (PNPase) was suggested to be responsible for RNA deadenylation in mitochondria (61). However, the discovery that PNPase is localized to the mitochondrial intermembrane space and that knockdown does not affect RNA levels inside the matrix compartment, recently questioned this hypothesis (59). The data reported here defining 2′-PDE as a general 3′–5′ exoribonuclease with some preference for adenosine, is compatible to what is seen for most deadenylases, and therefore consistent with a role in RNA deadenylation (21,22,30–33). For instance, in Saccharomyces cerevisiae, the bulk of cytoplasmic deadenylation is maintained by the Ccr4 subunit of the Ccr4-Not complex (62), a large complex composed of seven core proteins including two deadenylases (Ccr4 and Pop2p) (63,64). In vitro, both Ccr4 and Pop2p nucleases show a preference for poly(A)-containing substrates, yet are able to degrade other sequences as well (21,22,33). Like many RNA-degrading enzymes, it is possible that 2′-PDE associates with other proteins in vivo inside the mitochondria that more precisely defines its substrates, but this is at present pure speculation.
In contrast to the exclusive stabilizing effect imposed by poly(A) tails added to cytoplasmic mRNAs, the poly(A) tail of human mitochondrial mRNAs is known to provide both stability and instability to the RNA, the outcome being dictated by the exact site of poly(A) addition (61,65,66). Specifically, human mitochondrial mRNAs can be poly(A) modified by (i) internal polyadenylation subsequent to endonucleolytic RNA cleavage, or (ii) by ‘normal’ polyadenylation taking place at the mature 3′ termini. In the former case, transcripts are marked to be degraded, whereas in the latter, they are stabilized. Suppose 2′-PDE carries out mitochondrial RNA deadenylation, which poly(A) tails, i.e. the internal added tails or the mature 3′ terminal tails, then constitute the main target for 2′-PDE. Though this of course remains speculative, the capacity of 2′-PDE to destabilize and down-regulate mitochondrial mRNAs would be consistent with a role in processing of the mature 3′-terminal tails, normally known to stabilize their RNA targets. On the contrary, 2′-PDE could also, or exclusively, be processing the internal added tails, and by this means cause RNA destabilization.
As stated, the 2′-PDE needs not be delimited to a molecular role in RNA deadenylation, but might alternatively be generally involved in the degradation of entire mitochondrial RNAs. For instance, in yeast where mitochondrial RNAs, remarkably, are not polyadenylated (67), RNA decay is maintained by a large enzyme machinery termed the ‘mitochondrial degradosome’ containing RNA helicase (Suv3p) and 3′–5′ exoribonuclease (Dss1p) activities (68,69). Based on similar properties recognized here for the 2′-PDE, i.e. a general 3′–5′ exoribonuclease activity combined with the ability to unwind secondary structures, human mitochondrial RNAs may indeed be processed by this enzyme.
As for its role in the 2-5A system, we speculate that the 2-5A core molecules and not their 5′-phosphorylated variants, are the natural substrates for 2′-PDE. First, viral infection prompts the accumulation of 2-5A core molecules—not 2-5As—to levels of micromolar magnitudes suggesting that a 5′-phosphatase initiates 2-5A catabolism (11,70). Second, the dephosphorylated and hence less charged 2-5A core molecules are more likely to passively traverse the hydrophobic barrier formed by the mitochondrial inner membrane (71,72). Alternatively, if not translocated by diffusion, a membrane-positioned 2-5A core solute-carrier, akin to the ADP/ATP antiporter, may exist and transport 2-5A core molecules into the mitochondrial matrix. Finally, the micromolar Km values estimated for 2′-PDE on the phosphorylated 2′–5′ substrates (15) are much higher compared to the nanomolar 2-5A levels sufficient for activating RNase L (12), indicating that a 5′-phosphatase indeed performs the initial cleavage in the overall 2-5A catabolism.
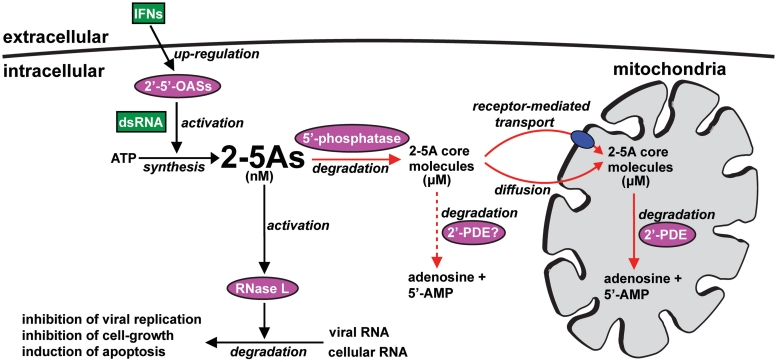
Based on these findings, we present a model (Figure 8) that besides accounting for the complete 2-5A catabolism includes a mechanism of how the cellular 2-5A level is regulated following viral exposure. In this scheme, RNase L and a 5′-phosphatase are both activated by 2-5A levels in the nanomolar range to (i) relieve cells of viral infection and (ii) to protect against the cytotoxic events resulting from build-up of 2-5As. Accordingly, when raised to nanomolar levels, 2-5As not only stimulate RNase L, but are simultaneously converted into the less cell-toxic 2-5A core molecules. The resulting ‘cores’ then concentrate to micromolar levels and are transported across the mitochondrial inner membrane into the matrix where they function as substrates for 2′-PDE. Notice that despite here having defined 2′-PDE as a mitochondrial protein, the possible occurrence of any non-mitochondrial forms hereof, e.g. arising by alternative splicing creating isoforms devoid of an N-terminal mTP, can at present not be excluded. Specifically, as the properties of such variants would be expected to be very similar to that of the mitochondrial protein, the 2-5A core molecules should also constitute their primary substrates. The hypothetical presence of non-mitochondrial 2′-PDEs were included in Figure 8, to illustrate yet a factor potentially involved in 2-5A catabolism.
Figure 8.
The 2-5A system—regulation and catabolism. Normally, the cellular concentration of 2-5As is kept below the nanomolar range (10,73), maintaining the pathway in an inactive state. Then, when increased to nanomolar levels by the IFN-induced 2′–5′-OASs, 2-5As activate RNase L and a 5′-phosphatase, which results in the degradation of viral RNA and in the conversion of the cytotoxic 2-5As into dephosphorylated 2-5A core molecules. Accumulation of 2-5A core molecules to micromolar concentrations subsequently result in transport across the mitochondrial inner membrane and into the matrix by a yet unidentified mechanism. Within the mitochondrial matrix, 2′-PDE completes 2-5A catabolism by degradation of the incoming 2-5A core molecules. The 2-5A catabolic pathway is specified by red arrows, and the possible involvement of yet to be discovered non-mitochondrial 2′-PDEs (2′-PDE?) is illustrated by a red dotted arrow.
In conclusion, we have shown that 2′-PDE, a down-regulator of the cytoplasmic 2-5A system, comprises a matrix-localized mitochondrial protein very likely also to be involved in organellar RNA turnover. Whether this role is in general RNA decay or in RNA deadenylation remains to be determined.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Danish Council for Independent Research (to P.M.M. and J.J.); Carlsberg Foundation (to J.J.); a PhD fellowship from Aarhus University (to J.B.P., K.H.K. and A.L.V.); Novo Nordisk Foundation (to D.E.B.); Lundbeck Foundation (to D.E.B.); a PhD fellowship from the Danish National Research Foundation, Centre for mRNP biogenesis and metabolism (to K.R.A.); Australian Research Council (to F.D., G.H.T., P.F. and N.H.). Funding for open access charge: The Danish Council for Independent Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Professor Michael Teitell for providing us with optimized primer sequences specific to each mitochondrial gene and the RPL13A gene, analyzed by qRT-PCR. We thank Anna Malgorzata Jurkiewicz for help and advice on confocal microscopy and Amelia Johnston, Ilya Levichkin and Nishant Aggarwal for guidance and useful discussions. A special thank is also given to Michael Toft Overgaard that helped verifying the identity of the synthesized 2′–5′ oligoadenylate tetramer core by MS.
REFERENCES
- 1.Samuel CE. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takaoka A, Yanai H. Interferon signalling network in innate defence. Cell. Microbiol. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- 3.Player MR, Torrence PF. The 2-5A system: modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacol. Therapeut. 1998;78:55–113. doi: 10.1016/S0163-7258(97)00167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lengyel P. Tumor-suppressor genes: news about the interferon connection. Proc. Natl Acad. Sci. USA. 1993;90:5893–5895. doi: 10.1073/pnas.90.13.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Justesen J, Hartmann R, Kjeldgaard NO. Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell Mol. Life. Sci. 2000;57:1593–1612. doi: 10.1007/PL00000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll SS, Chen E, Viscount T, Geib J, Sardana MK, Gehman J, Kuo LC. Cleavage of oligoribonucleotides by the 2′,5′-oligoadenylate- dependent ribonuclease L. J. Bio. Chem. 1996;271:4988–4992. doi: 10.1074/jbc.271.9.4988. [DOI] [PubMed] [Google Scholar]
- 7.Wreschner DH, McCauley JW, Skehel JJ, Kerr IM. Interferon action–sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature. 1981;289:414–417. doi: 10.1038/289414a0. [DOI] [PubMed] [Google Scholar]
- 8.Kerr IM, Brown RE, Hovanessian AG. Nature of inhibitor of cell-free protein synthesis formed in response to interferon and double-stranded RNA. Nature. 1977;268:540–542. doi: 10.1038/268540a0. [DOI] [PubMed] [Google Scholar]
- 9.Kubota K, Nakahara K, Ohtsuka T, Yoshida S, Kawaguchi J, Fujita Y, Ozeki Y, Hara A, Yoshimura C, Furukawa H, et al. Identification of 2′-phosphodiesterase, which plays a role in the 2-5A system regulated by interferon. J. Bio. Chem. 2004;279:37832–37841. doi: 10.1074/jbc.M400089200. [DOI] [PubMed] [Google Scholar]
- 10.Hearl WG, Johnston MI. Accumulation of 2′,5′-oligoadenylates in encephalomyocarditis virus-infected mice. J. Virol. 1987;61:1586–1592. doi: 10.1128/jvi.61.5.1586-1592.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hersh CL, Brown RE, Roberts WK, Swyryd EA, Kerr IM, Stark GR. Simian virus 40-infected, interferon-treated cells contain 2′,5′-oligoadenylates which do not activate cleavage of RNA. J. Biol. Chem. 1984;259:1731–1737. [PubMed] [Google Scholar]
- 12.Dong B, Xu L, Zhou A, Hassel BA, Lee X, Torrence PF, Silverman RH. Intrinsic molecular activities of the interferon-induced 2-5A-dependent RNase. J. Bio. Chem. 1994;269:14153–14158. [PubMed] [Google Scholar]
- 13.Doetsch PW, Suhadolnik RJ, Sawada Y, Mosca JD, Flick MB, Reichenbach NL, Dang AQ, Wu JM, Charubala R, Pfleiderer W, et al. Core (2′-5′)oligoadenylate and the cordycepin analog: inhibitors of Epstein–Barr virus-induced transformation of human lymphocytes in the absence of interferon. Proc. Natl Acad. Sci. USA. 1981;78:6699–6703. doi: 10.1073/pnas.78.11.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimchi A, Shure H, Revel M. Anti-mitogenic function of interferon-induced (2′-5′)oligo(adenylate) and growth-related variations in enzymes that synthesize and degrade this oligonucleotide. Euro. J. Biochem/FEBS. 1981;114:5–10. doi: 10.1111/j.1432-1033.1981.tb06163.x. [DOI] [PubMed] [Google Scholar]
- 15.Johnston MI, Hearl WG. Purification and characterization of a 2′-phosphodiesterase from bovine spleen. J. Biol. Chem. 1987;262:8377–8382. [PubMed] [Google Scholar]
- 16.Schmidt A, Chernajovsky Y, Shulman L, Federman P, Berissi H, Revel M. An interferon-induced phosphodiesterase degrading (2′-5′) oligoisoadenylate and the C-C-A terminus of tRNA. Proc. Natl Acad. Sci. USA. 1979;76:4788–4792. doi: 10.1073/pnas.76.10.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- 18.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat. Rev. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 19.Bernad A, Blanco L, Lazaro JM, Martin G, Salas M. A conserved 3′–5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59:219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- 20.Jonstrup AT, Andersen KR, Van LB, Brodersen DE. The 1.4-A crystal structure of the S. pombe Pop2p deadenylase subunit unveils the configuration of an active enzyme. Nucleic Acids Res. 2007;35:3153–3164. doi: 10.1093/nar/gkm178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen KR, Jonstrup AT, Van LB, Brodersen DE. The activity and selectivity of fission yeast Pop2p are affected by a high affinity for Zn2+ and Mn2+ in the active site. RNA. 2009;15:850–861. doi: 10.1261/rna.1489409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Chiang YC, Denis CL. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mol CD, Kuo CF, Thayer MM, Cunningham RP, Tainer JA. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature. 1995;374:381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- 24.Bayard BA, Gabrion JB. 2′,5′-Oligoadenylate-dependent RNAse located in nuclei: biochemical characterization and subcellular distribution of the nuclease in human and murine cells. Biochem. J. 1993;296(Pt 1):155–160. doi: 10.1042/bj2960155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Besse S, Rebouillat D, Marie I, Puvion-Dutilleul F, Hovanessian AG. Ultrastructural localization of interferon-inducible double-stranded RNA-activated enzymes in human cells. Exp. Cell Res. 1998;239:379–392. doi: 10.1006/excr.1997.3908. [DOI] [PubMed] [Google Scholar]
- 26.Chebath J, Benech P, Hovanessian A, Galabru J, Revel M. Four different forms of interferon-induced 2′,5′-oligo(A) synthetase identified by immunoblotting in human cells. J. Biol. Chem. 1987;262:3852–3857. [PubMed] [Google Scholar]
- 27.Floyd-Smith G, Yoshie O, Lengyel P. Interferon action. Covalent linkage of (2′-5′)pppApApA(32P)pCp to (2′-5′)(A)n-dependent ribonucleases in cell extracts by ultraviolet irradiation. J. Biol. Chem. 1982;257:8584–8587. [PubMed] [Google Scholar]
- 28.Marie I, Svab J, Robert N, Galabru J, Hovanessian AG. Differential expression and distinct structure of 69- and 100-kDa forms of 2-5A synthetase in human cells treated with interferon. J. Biol. Chem. 1990;265:18601–18607. [PubMed] [Google Scholar]
- 29.Yang K, Samanta H, Dougherty J, Jayaram B, Broeze R, Lengyel P. Interferons, double-stranded RNA, and RNA degradation. Isolation and characterization of homogeneous human (2′-5′)(a)n synthetase. J. Biol. Chem. 1981;256:9324–9328. [PubMed] [Google Scholar]
- 30.Astrom J, Astrom A, Virtanen A. Properties of a HeLa cell 3′ exonuclease specific for degrading poly(A) tails of mammalian mRNA. J. Biol. Chem. 1992;267:18154–18159. [PubMed] [Google Scholar]
- 31.Baggs JE, Green CB. Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr. Biol. 2003;13:189–198. doi: 10.1016/s0960-9822(03)00014-9. [DOI] [PubMed] [Google Scholar]
- 32.Korner CG, Wahle E. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J. Biol. Chem. 1997;272:10448–10456. doi: 10.1074/jbc.272.16.10448. [DOI] [PubMed] [Google Scholar]
- 33.Thore S, Mauxion F, Seraphin B, Suck D. X-ray structure and activity of the yeast Pop2 protein: a nuclease subunit of the mRNA deadenylase complex. EMBO Rep. 2003;4:1150–1155. doi: 10.1038/sj.embor.7400020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joseph SB, Bradley MN, Castrillo A, Bruhn KW, Mak PA, Pei L, Hogenesch J, O’Connell RM, Cheng G, Saez E, et al. LXR-dependent gene expression is important for macrophage survival and the innate immune response. Cell. 2004;119:299–309. doi: 10.1016/j.cell.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 36.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hurt EC, Pesold-Hurt B, Schatz G. The cleavable prepiece of an imported mitochondrial protein is sufficient to direct cytosolic dihydrofolate reductase into the mitochondrial matrix. FEBS Lett. 1984;178:306–310. doi: 10.1016/0014-5793(84)80622-5. [DOI] [PubMed] [Google Scholar]
- 38.Hay R, Bohni P, Gasser S. How mitochondria import proteins. Biochim Biophys. Acta. 1984;779:65–87. doi: 10.1016/0304-4157(84)90004-2. [DOI] [PubMed] [Google Scholar]
- 39.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. European J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 40.Gavel Y, von Heijne G. Cleavage-site motifs in mitochondrial targeting peptides. Protein Engineer. 1990;4:33–37. doi: 10.1093/protein/4.1.33. [DOI] [PubMed] [Google Scholar]
- 41.Roise D, Horvath SJ, Tomich JM, Richards JH, Schatz G. A chemically synthesized pre-sequence of an imported mitochondrial protein can form an amphiphilic helix and perturb natural and artificial phospholipid bilayers. EMBO J. 1986;5:1327–1334. doi: 10.1002/j.1460-2075.1986.tb04363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brix J, Dietmeier K, Pfanner N. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J. Biol. Chem. 1997;272:20730–20735. doi: 10.1074/jbc.272.33.20730. [DOI] [PubMed] [Google Scholar]
- 43.von Heijne G, Steppuhn J, Herrmann RG. Domain structure of mitochondrial and chloroplast targeting peptides. Eur. J. Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- 44.Peri S, Steen H, Pandey A. GPMAW–a software tool for analyzing proteins and peptides. Trends Biochem. Sci. 2001;26:687–689. doi: 10.1016/s0968-0004(01)01954-5. [DOI] [PubMed] [Google Scholar]
- 45.Cechetto JD, Soltys BJ, Gupta RS. Localization of mitochondrial 60-kD heat shock chaperonin protein (Hsp60) in pituitary growth hormone secretory granules and pancreatic zymogen granules. J. Histochem. Cytochem. 2000;48:45–56. doi: 10.1177/002215540004800105. [DOI] [PubMed] [Google Scholar]
- 46.Berger EG. Ectopic localizations of Golgi glycosyltransferases. Glycobiology. 2002;12:29R–36R. doi: 10.1093/glycob/12.2.29r. [DOI] [PubMed] [Google Scholar]
- 47.Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 49.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 50.Ryan MT, Voos W, Pfanner N. Assaying protein import into mitochondria. Methods Cell Biol. 2001;65:189–215. doi: 10.1016/s0091-679x(01)65012-x. [DOI] [PubMed] [Google Scholar]
- 51.Fujiki Y, Fowler S, Shio H, Hubbard AL, Lazarow PB. Polypeptide and phospholipid composition of the membrane of rat liver peroxisomes: comparison with endoplasmic reticulum and mitochondrial membranes. J. Cell Biol. 1982;93:103–110. doi: 10.1083/jcb.93.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin H, Eckerskorn C, Gartner F, Rassow J, Lottspeich F, Pfanner N. The yeast mitochondrial intermembrane space: purification and analysis of two distinct fractions. Analyt. Biochem. 1998;265:123–128. doi: 10.1006/abio.1998.2863. [DOI] [PubMed] [Google Scholar]
- 53.Schneider A, Behrens M, Scherer P, Pratje E, Michaelis G, Schatz G. Inner membrane protease I, an enzyme mediating intramitochondrial protein sorting in yeast. EMBO J. 1991;10:247–254. doi: 10.1002/j.1460-2075.1991.tb07944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalousek F, Hendrick JP, Rosenberg LE. Two mitochondrial matrix proteases act sequentially in the processing of mammalian matrix enzymes. Proc. Natl Acad. Sci. USA. 1988;85:7536–7540. doi: 10.1073/pnas.85.20.7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sztul ES, Chu TW, Strauss AW, Rosenberg LE. Import of the malate dehydrogenase precursor by mitochondria. Cleavage within leader peptide by matrix protease leads to formation of intermediate-sized form. J. Biol. Chem. 1988;263:12085–12091. [PubMed] [Google Scholar]
- 56.Lithgow T, Ristevski S, Hoj P, Hoogenraad N. High-level expression of a mitochondrial enzyme, ornithine transcarbamylase from rat liver, in a baculovirus expression system. DNA Cell Biol. 1991;10:443–449. doi: 10.1089/dna.1991.10.443. [DOI] [PubMed] [Google Scholar]
- 57.Kalousek F, Isaya G, Rosenberg LE. Rat liver mitochondrial intermediate peptidase (MIP): purification and initial characterization. EMBO J. 1992;11:2803–2809. doi: 10.1002/j.1460-2075.1992.tb05347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen HW, Rainey RN, Balatoni CE, Dawson DW, Troke JJ, Wasiak S, Hong JS, McBride HM, Koehler CM, Teitell MA, et al. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol. Cell. Biol. 2006;26:8475–8487. doi: 10.1128/MCB.01002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Minczuk M, Piwowarski J, Papworth MA, Awiszus K, Schalinski S, Dziembowski A, Dmochowska A, Bartnik E, Tokatlidis K, Stepien PP, et al. Localisation of the human hSuv3p helicase in the mitochondrial matrix and its preferential unwinding of dsDNA. Nucleic Acids Res. 2002;30:5074–5086. doi: 10.1093/nar/gkf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J. Biol. Chem. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- 62.Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bai Y, Salvadore C, Chiang YC, Collart MA, Liu HY, Denis CL. The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol. Cell. Biol. 1999;19:6642–6651. doi: 10.1128/mcb.19.10.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen J, Rappsilber J, Chiang YC, Russell P, Mann M, Denis CL. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J. Mol. Biol. 2001;314:683–694. doi: 10.1006/jmbi.2001.5162. [DOI] [PubMed] [Google Scholar]
- 65.Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation and degradation of human mitochondrial RNA: the prokaryotic past leaves its mark. Mol. Cell. Biol. 2005;25:6427–6435. doi: 10.1128/MCB.25.15.6427-6435.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Temperley RJ, Seneca SH, Tonska K, Bartnik E, Bindoff LA, Lightowlers RN, Chrzanowska-Lightowlers ZM. Investigation of a pathogenic mtDNA microdeletion reveals a translation-dependent deadenylation decay pathway in human mitochondria. Human Mol. Genet. 2003;12:2341–2348. doi: 10.1093/hmg/ddg238. [DOI] [PubMed] [Google Scholar]
- 67.Gagliardi D, Stepien PP, Temperley RJ, Lightowlers RN, Chrzanowska-Lightowlers ZM. Messenger RNA stability in mitochondria: different means to an end. Trends Genet. 2004;20:260–267. doi: 10.1016/j.tig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 68.Malecki M, Jedrzejczak R, Stepien PP, Golik P. In vitro reconstitution and characterization of the yeast mitochondrial degradosome complex unravels tight functional interdependence. J. Mol. Biol. 2007;372:23–36. doi: 10.1016/j.jmb.2007.06.074. [DOI] [PubMed] [Google Scholar]
- 69.Py B, Higgins CF, Krisch HM, Carpousis AJ. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 70.Rice AP, Roberts WK, Kerr IM. 2-5A accumulates to high levels in interferon-treated, vaccinia virus-infected cells in the absence of any inhibition of virus replication. J. Virol. 1984;50:220–228. doi: 10.1128/jvi.50.1.220-228.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hovanessian AG, Wood JN. Anticellular and antiviral effects of pppA(2′p5′A)n. Virology. 1980;101:81–90. doi: 10.1016/0042-6822(80)90485-7. [DOI] [PubMed] [Google Scholar]
- 72.Kimchi A, Shure H, Revel M. Regulation of lymphocyte mitogenesis by (2′–5′) oligo-isoadenylate. Nature. 1979;282:849–851. doi: 10.1038/282849a0. [DOI] [PubMed] [Google Scholar]
- 73.Brown RE, Kerr IM. (p)pp(A2′p)nA is rare in normal mouse tissues while (A2′p)nA but not (p)pp(A2′p)nA appears to be present in E coli. Prog. Clin. Biol. Res. 1985;202:3–10. [PubMed] [Google Scholar]
- 74.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.