Abstract
A TG microsatellite in the 3′-untranslated region (UTR) of FGF9 mRNA has previously been shown to modulate FGF9 expression. In the present study, we investigate the possible interacting protein that binds to FGF9 3′-UTR UG-repeat and study the mechanism underlying this protein–RNA interaction. We first applied RNA pull-down assays and LC-MS analysis to identify proteins associated with this repetitive sequence. Among the identified proteins, FUBP3 specifically bound to the synthetic (UG)15 oligoribonucleotide as shown by supershift in RNA-EMSA experiments. The endogenous FGF9 protein was upregulated in response to transient overexpression and downregulated after knockdown of FUBP3 in HEK293 cells. As the relative levels of FGF9 mRNA were similar in these two conditions, and the depletion of FUBP3 had no effect on the turn-over rate of FGF9 mRNA, these data suggested that FUBP3 regulates FGF9 expression at the post-transcriptional level. Further examination using ribosome complex pull-down assay showed overexpression of FUBP3 promotes FGF9 expression. In contrast, polyribosome-associated FGF9 mRNA decreased significantly in FUBP3-knockdown HEK293 cells. Finally, reporter assay suggested a synergistic effect of the (UG)-motif with FUBP3 to fine-tune the expression of FGF9. Altogether, results from this study showed the novel RNA-binding property of FUBP3 and the interaction between FUBP3 and FGF9 3′-UTR UG-repeat promoting FGF9 mRNA translation.
INTRODUCTION
Microsatellites (MSs) are simple sequence repetitive motifs that are ubiquitous and frequently polymorphic in mammalian genomes. More than 30 000 MSs have been identified so far (1) and they comprise ∼3% of the human genome (2). Many studies have demonstrated that MSs in the non-coding region function in gene regulation, presumably by forming specific DNA structures-like Z-DNA (3) or H-DNA (4). In addition, tissue- and cell type-specific regulation of polymorphic MS motifs on target gene expression has been reported for human SNCA (5), TH (6), and HMGA2 (7); this suggests a common mechanism for this type of regulation. Furthermore, these findings indicate that the different effects might be controlled by the interacting proteins that are expressed in various cells and tissues and bind to these regulatory elements (7).
Increasing numbers of MSs have been found in the UTRs of protein-coding genes and have quantitative effects on gene expression through post-transcriptional regulation (8). For example, CA dinucleotide repeats in the 3′-UTR of the mouse CD154 gene regulates CD154 expression by poly(A) tail shortening (9). CA repeats in the 3′-UTR of human Bcl-2 mRNA also contribute to constitutive mRNA decay (10). Furthermore, a (T)8 MS motif embedded in the 3′-UTR of human CEACAM1 gene forms a hairpin structure and contributes to mRNA stability (11). Evidence from other species also showed that RNA-binding proteins regulate mRNA stability or translation through the binding to 3′-UTR UG repeats. For example, one of the circadian controlled translational regulators (CCTRs) in green algae, CHLAMY 1, negatively regulates the activity of nitrogen metabolism components by binding to UG repeats residing in the 3′-UTR of those genes (12). Results from these studies revealed that MS repeats participate in different mechanisms to regulate gene expression and the specificity of the effect depends on the interacting protein.
Fibroblast growth factor 9 (FGF9) is a member of the secreted polypeptide family (13) and involved in many important processes, including developments of lung (14) and bone (15), and steroidogenesis in postnatal Leydig cells (16). In addition, Fgf9 knockout mice demonstrate male-to-female sex reversal, revealing a novel role for Fgf9 in testicular embryogenesis and sex determination (14). The expression of Fgf9 mRNA is ubiquitous at the early stage of embryonic development (17), but restricted to a few organs, including brain, kidney (18) and endometrium (19). Furthermore, abnormal expressions of FGF9 has been implicated in pathogenic conditions like glioma (20), prostate cancer (21), ovarian endometrioid adenocarcinoma (22), colorectal carcinoma (23) and endometriosis (19,24). These studies demonstrated that the expression of FGF9 must be tightly controlled. Unlike studies have shown that FGF9 is transcriptionally induced by many factors like prostaglandin E2 (PGE2) (25) and estrogen (24), the mechanisms involved in its post-transcriptional regulation remain largely unknown and are just starting to be illustrated. A previous study in our laboratory found that FGF9 has a TG MS in its 3′-UTR. The TG MS is polymorphic in Han Chinese population [i.e. (TG)13–16] and exhibits function to control FGF9 mRNA stability (26). Although the study provided the first evidence that the expression of FGF9 can be controlled post-transcriptionally, the factors involved and the underlying mechanism of this regulation remain unidentified and merit further investigation. Here, we report our findings on the identification of a putative RNA binding protein, FUBP3, which interacts with FGF9 3′-UTR UG repeats and positively controls FGF9 expression through increasing translation of FGF9 mRNA.
MATERIALS AND METHODS
Cell lines and plasmids
Human embryonic kidney 293 cells (HEK293) were routinely maintained in Eagle’s Minimum Essential Medium (MEM) supplemented with 10% horse serum and 1 mM sodium pyruvate under 5% CO2 at 37°C. In addition, the pluripotent human testicular embryonic carcinoma cell line, NTERA-2c1.D1 (NT2D1), which has the ability to be differentiated to neuronal cells was also used in this study. The NT2D1 cells were routinely maintained in Dulbecco’s Modified Eagle Medium (DMEM) with 4 mM l-glutamine, 4.5 g/l glucose, 1.0 mM sodium pyruvate and 10% heat-inactivated fetal bovine serum under 5% CO2 at 37°C.
The cloning vectors of pcDNA3.1 myc/His A+ (Invitrogen), pGL3-P (Promega, Madison, WI, USA), and pRL-TK (Promega) were used in the construction of various recombinant clones. Full-length CDS of human FUBP3 (nucleotides 106–1830 of NM_003934) was amplified by PCR using primers for FUBP3 (Forward: 5′-GTA ATG GCG GAG CTG GTG-3′ and Reverse: 5′-GTC CTA CTG CTC CTG GCT GT-3′) and cloned into yT&A vector (Yeastern Biotech, Taipei, Taiwan). The yT&A-FUBP3 clone was digested with HindIII and XbaI and subcloned into pcDNA3.1 myc/His A+ to obtain the pcDNA-FUBP3. Various pGL3-(TG)n plasmids were constructed by digesting constructs (26) with AvrII and XbaI enzymes to obtain partial FGF9 3′-UTR sequences containing different TG repeats (nucleotides 1605–2161 of NM_002010), and subcloned into pGL3-promoter. Plasmids for in vitro transcription assays were constructed by PCR amplification of FGF9 (NM_002010, nucleotides 1605–2161) and GAPDH (NM_002046, nucleotides 628–1079) fragments using primers for FGF9 (Forward: 5′-GCT TGG ATG GGA ATA TGC TG-3′, Reverse: 5′-AAA ATG CCT ATC TGG CCT GT-3′) and GAPDH (Forward: 5′-ACC ACA GTC CAT GCC ATC AC-3′; Reverse: 5′-TCC ACC ACC CTG TTG CTG TA-3′), followed by cloning into yT&A vector (Yeastern Biotech).
In vitro transcription
To prepare riboprobes for the in vitro binding assay, FGF9 and GAPDH plasmids were linearized by BamHI or XbaI restriction enzymes. One microgram of plasmid DNA was used as template for synthesizing T7-containing PCR products at their 5′- (for sense transcript) or 3′- (for antisense transcript) ends, then transcribed in vitro using biotin-11-CTP (Roche) as one-tenth of total CTP.
RNA secondary structure prediction
Sequences from FGF9 3′-UTR (nucleotides 1465–2077 of NM_002010) were subjected to secondary structure prediction using the minimum free energy method implemented in the RNAfold web server [http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi; (27)]. The graphic output was used for visual comparison at the region of interest.
UV-cross-linking
To confirm that the sequence of FGF9 3′-UTR TG repeats indeed does form RNA–protein complexes, biotinylated (UG)15 oligoribonucleotide were synthesized (Integrated DNA Technologies, Inc., Coralville, USA) and used for further analysis. UV-cross-linking experiments were carried out according to the procedure described earlier (28). Briefly, the RNA–protein binding reaction was performed in 10 μl containing 1× binding buffer (10 mM HEPES, 3 mM MgCl2, 5% glycerol and 1 mM DTT), 100 mM KCl, 0.5 μg/μl yeast tRNA, 20 U RNasin (Promega), 5 μg of protein extract, various amounts of unlabeled RNA probe and 20 fmol of biotinylated (UG)15 probe. After 20-min incubation at room temperature, 50 μg of heparin was added and irradiated for 30 min on ice with a UV-cross-linker (Stratagene). Samples were separated by 8% SDS–PAGE under denaturing conditions and blotted to PVDF membranes using a Mini Trans-Blot cell (Bio-Rad). RNA–protein complexes were detected with a LightShift Chemiluminescent EMSA Kit (Thermo scientific) according to the manufacturer’s recommendations.
RNA-electrophoretic mobility shift assay
RNA-electrophoretic mobility shift assay (RNA-EMSA) was conducted in 10 μl containing 1× EMSA binding buffer (10 mM Tris, 50 mM KCl, 1 mM DTT), 2.5% glycerol, 0.05% NP-40, 10 U RNasin (Promega), 0.5 μg/μl yeast tRNA, 1–2 μg of protein extract, unlabeled probe, 10 fmol biotinylated (UG)15 probe and nuclease-free water. For supershift assay, 1 μl of anti-FUBP3 antibody (sc-1110, Santa Cruz) or an equal amount of goat normal IgG was added to the reaction and incubated at room temperature for 20 min. This was followed by adding 50 μg of heparin into the reaction mix and incubated for 10 min. RNA–protein complexes were detected and analyzed using the LightShift Chemiluminescent EMSA Kit (Thermo scientific) according to the suggested protocol.
Identification of proteins interacting with FGF9 3′-UTR TG repeats
To identify proteins that specifically bind to FGF9 3′-UTR UG repeats, biotinylated (UG)15 oligoribonucleotide was used in pull-down assays. Cytoplasmic extract was prepared with NE-PER reagents (Thermo Scientific) in the presence of 1× complete protease inhibitors cocktail (Roche) and quantified by MicroBCA Protein Assay Kit (Thermo scientific). Approximately 200 fmol of biotinylated (UG)15 oligoribonucleotides were immobilized to 1 μg of streptavidin-conjugated Dynabeads (Invitrogen) according to the manufacturer’s recommendations. Cell lysates containing 100–200 μg of proteins were incubated with 400 μg RNA probe-coated Dynabeads in a total volume of 500 μl. The experiment was conducted as in the UV-cross-linking assay without the cross-linking step. The reaction with no biotinylated oligoribonucleotides was conducted in parallel to serve as a control. After extensive washing with 1× binding buffer, the precipitated proteins were separated on 4–12% NuPAGE in denaturing conditions (Invitrogen) and visualized by silver staining (29). The differentially stained bands were excised and submitted for protein identification using the LC/MS/MS service provided by the Proteomics Research Core at National Cheng Kung University. The searches for matched peptides were done by the Mascot algorithm (www.matrixscience.com).
Reverse transcription and conventional RT-PCR
One microgram of total RNA was isolated and reverse transcribed to cDNA by High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems). cDNA was diluted 2- and 5-fold for conventional and quantitative RT-PCR. Conventional RT-PCR was carried out with 1x PCR buffer, 3.5 mM MgCl2, 0.2 mM of each dNTP Mix Solution, 0.2 μM of each primer for FGF9 (Forward: 5′-GCG AGG GTG CAG TCT TAC TT-3′; Reverse: 5′-CAT AGC GGA CTT TGC CAT TT-3′) or GAPDH (Forward: 5′-ACC ACA GTC CAT GCC ATC AC-3′; Reverse: 5′-TCC ACC ACC CTG TTG CTG TA-3′.), 0.05 U/μl GoTaq (Promega), 2 μl diluted cDNA and water to a final volume of 10 μl. The PCR program was 1 min at 94°C followed by 25 or 33 cycles of 30 s at 94°C, 30 s at 55°C and 30 s at 72°C. The reaction was terminated by 72°C for 1 min.
RNA Immunoprecipitation
To confirm RNA–protein interactions, cell lysates were specifically prepared from 1 to 5 × 106 HEK293 cells and used in RNA immunoprecipitation (RNA-IP) assays according to the procedure described by Tenenbaum et al. (30). Briefly, 100 μl of ∼50% protein G-sepharose beads (GE Healthcare Bio-Sciences) was pre-coated with 4 μg of normal goat IgG, or goat anti-FUBP3 at 4°C overnight. The antibody-coated beads were washed five times with ice-cold NT2 buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1 mM MgCl2 and 0.05% NP-40) and resuspended in 850 μl of NT2 buffer. Two hundred units of RNase OUT (Invitrogen), 1 mM DTT, 20 mM EDTA and 100 μl of cell lysate were added to the binding reaction. After brief centrifugation and removal of a 100-μl aliquot to represent total cellular mRNA, the messenger ribonucleoprotein (mRNP) was incubated with antibody-coated beads for 2 h at room temperature. Immunoprecipitated mRNP complexes were washed six times with ice-cold NT2 buffer and resuspended in 100 μl of NT2 buffer containing 0.1% SDS and 30 μg of proteinase K. The RNA was released by incubation for 30 min at 55°C and isolated by RNeasy mini kit (QIAGEN, Hilden, Germany). Isolated RNA was further treated with a DNA-free kit (Ambion) to eliminate any DNA contamination, and reverse transcribed to cDNA (Applied Biosystems) for further applications.
Analysis of proteins bound to biotinylated RNA
Plasmids used for in vitro transcription were linearized by BamHI or XbaI restriction enzymes and 1 μg of plasmid DNA was used as template for the synthesis of biotinylated transcripts in vitro using biotin-11-CTP (Roche) as one-tenth of total CTP (31). To pull-down proteins that bind to RNA probes (32), 100 μg of cytoplasmic extract was incubated with 1 μg of biotinylated RNA or equimolar biotinylated (UG)15 oligoribonucleotides for 1 h at room temperature and subjected to western blotting.
Transient overexpression and shRNA knockdown
In FUBP3 overexpression experiments, ∼2 × 105 HEK293 cells/ml were inoculated in 24- or 96-well culture plates 1 day before transfection. This was followed by transfection of cells with various constructs using FuGENE HD Transfection Reagent (Roche) according to the manufacturer’s instructions. For endogenous FGF9 analysis, cells grown on 24-well plates were transfected by pcDNA3.1 or pcDNA-FUBP3 and incubated for another day. For reporter gene assay, pcDNA3.1 or pcDNA-FUBP3 was co-transfected in a mass ratio of 1:5 to pGL3-P or pGL3-(TG)n reporter constructs and assayed after 1 day by Dual-Reporter Assay (Promega) according to the manufacturer’s recommendations with minor modifications. Five microliters of cell lysate was mixed with 25 μl of LAR II reagent to measure the activity of firefly luciferase, followed by administration of 25 μl Stop&Glo reagent to estimate Renilla luciferase activity for normalization. pRL-TK was used as internal control in a mass ratio of 1:50 to the reporter plasmid. Total proteins and RNAs were isolated by RIPA lysis buffer (50 mM Tris–Cl, pH 7.5, 1% NP-40, 0.5% sodium deoxycholate, 0.05% SDS, 1 mM EDTA, 150 mM NaCl, 1x complete) and an RNeasy mini kit (QIAGEN).
As for FUBP3 knockdown experiments, pseudotyped lentiviruses against GFP (TRCN000072194) and FUBP3 (TRCN0000020576) were generated according to the protocols suggested by the National RNAi Core Facility (http://rnai.genmed.sinica.edu.tw/). Stable knockdown pools were obtained by transducing HEK293 cells with virus suspension (1:10 volume) and selecting in the presence of 2 μg/ml puromycin for 2–3 weeks. To perform the knockdown experiments, parental HEK293 cells were substituted with stable knockdown HEK293 cells (GFP or FUBP3) and similar procedures were used when FUBP3 overexpression was applied.
Enzyme-linked immunosorbent assay
One hundred micrograms of cell lysates from FUBP3-overexpressing or -knockdown HEK293 cells were used to determine the amount of FGF9 protein using enzyme-linked immunosorbent assay (ELISA) according to manufacturer’s protocol (R&D Systems, Oxfordshire, UK). Briefly, diluted capture antibody was added into 96-well plate and incubated at room temperature overnight. The antibody was removed from each well by washing the plate with wash buffer. Non-specific binding was blocked by adding Reagent Diluent to each well and incubated at room temperature for 1 h. After washing with wash buffer, cell lysates and standards were added to each well and incubated by shaking on an orbital shaker for 2 h at room temperature. After washing the plate, diluted detection antibody was added and incubated for 2 h at room temperature. After incubating with streptavidin-HRP and substrate solution each for 20 min, stop solution was added to terminate the color reaction. Optical density (OD) was determined by reading absorbance at 450 nm in an enzyme immunoassay plate reader. The amount of FGF9 protein was calculated by plotting the OD value of each sample into the standard curve.
Endogenous FGF9 turn-over
Approximately 2 × 105 stable knockdown HEK293 cells/ml (GFP or FUBP3) were inoculated in 24-well cell culture plates for 24 h, followed by adding 5 μg/ml of actinomycin D to stop the transcription from the FGF9 promoter. Total RNA was isolated at 0, 15, 30, 60 and 120 min thereafter. Relative FGF9 level was normalized to GAPDH at time zero by quantitative RT-PCR.
Ribosome complex pulldown
To investigate translational activity, ribosome complex pulldown assay was performed according to the procedure described in Yeh et al. (33). Briefly, cycloheximide (CHX) was added to HEK293 cells to a final concentration of 100 µg/ml. Five min later, cells were harvested in phosphate-buffered saline (PBS) and pelleted and suspended in lysis buffer (10 mM HEPES, pH 7.9, 100 mM KCl, 5 mM MgCl2, 1% Triton-X 100, 0.5% sodium deoxycholate, 10 U/ml RNaseOUT, 100 µg/ml CHX and 1X protease inhibitor cocktail). Cells were kept on ice for 10 min, and lysates were centrifuged at 8000 rpm for 10 min, and the supernatant was saved as cytoplasmic lysate. About 2 µg of ribosomal protein S6 antibody (Santa Cruz Biotechnology Inc., CA, USA) was added to 200 µg cytoplasmic lysate and incubated at 4°C overnight. Protein A/G agarose was added to the mixture to pull down the ribosome complex. The mRNAs bound with ribosome complex were extracted with TRI-reagent and analyzed the FGF9 mRNA expression level by quantitative RT-PCR.
Sucrose gradient centrifugation and polysomal profiles
Prior to lysis, cells were pre-treated with CHX (100 µg/ml) for 5 min and collected in PBS containing 100 µg/ml CHX. All subsequent steps were performed at 4°C. Cells were lysed on ice for 10 min in a RSB-150 buffer containing 10 mM Tris–HCl, pH 7.4, 3 mM MgCl2 and 150 mM NaCl, 100 µg/ml CHX, 40 µg/ml digitonin (Calbiochem, San Diego, CA, USA), 20 U/ml RNasin (Promega) and a protease inhibitor cocktail (Thermo Scientific Inc., Bremen, Germany). After incubation on ice, cells were disrupted by passage five to six times through a 26-gage needle. The cell lysates were collected by centrifugation in a microcentrifuge (5424R, Eppendorf AG, Hamburg, Germany) at 6000 rpm for 2 min, and clarified by further centrifugation at 11 000 rpm for 15 min. The samples were loaded on a linear gradient of 15–40% sucrose and centrifuged at 38 000 rpm at 4°C for 3 h in a Beckman SW41 rotor. After centrifugation, the polysome profile was monitored at 254 nm using a fractionation system (ISCO, Lincoln, NE, USA). Sucrose gradients were split into 11 subfractions each, starting with 1 (top) to 11 (bottom). Total RNA was isolated using the phenol–chloroform extraction followed by ethanol precipitation. The abundance of FGF9 and GAPDH mRNAs of individual fraction was measured by quantitative RT-PCR. The translational efficiency was calculated in the ratio of polyribosome-associated FGF9 or GAPDH mRNAs (7–11 fractions) to total mRNA (all fractions).
Quantitative RT-PCR
Quantitative RT-PCR of FGF9 and GAPDH was performed with TaqMan assays (Applied Biosystems, Foster City, CA, USA). The levels of FGF9 mRNA expression in HEK293 cells with and without FUBP3 overexpression were measured (i.e. the 2−ΔΔCt method) and normalized to the expression level of GAPDH. All measurements were performed in triplicate and the experiments were repeated at least twice.
Western blotting analysis, chemicals and antibodies
All chemicals were from Sigma (Sigma-Aldrich Co., Steinheim, Germany). The cocktail of complete protease inhibitor was provided by Roche (Roche Applied Science, Indianapolis, USA). Cell culture media and supplements were from Invitrogen (Invitrogen Inc., Carlsbad, CA, USA). Primary antibodies used were FUBP3 (sc-11103, Santa Cruz Biotechnology), GAPDH (sc-32233, Santa Cruz Biotechnology), AUF1 (07-260, Upstate), and goat normal IgG (sc-2028, Santa Cruz Biotechnology). HRP-conjugated secondary antibodies were supplied by Jackson ImmunoResearch Laboratories, Inc.
Protein blots were blocked with 3% non-fat dry milk (NFDM) in TBS-T buffer at room temperature for 1 h. Primary antibodies were diluted in blocking buffer or immunological enhancer solution (CanGetSignal, Solution 1, TOYOBO Co., Ltd., Japan) and incubated overnight at 4°C. After extensive washing with TBS-T, blots were incubated with HRP-conjugated secondary antibodies in blocking buffer or CanGetSignal Solution 2 (TOYOBO) and detected using chemiluminescent substrate (Millipore, Immobilon) for X-ray film exposure.
Statistical analysis
All experimental data were analyzed using GraphPad Prism 3.0 (GraphPad Software, Inc., San Diego, CA, USA; www.graphpad.com) and presented as mean ± SEM. Results were further analyzed using t-test or one-way ANOVA, and post-test was processed using Newman–Kuels post test. The significance level for all statistical tests was 0.05.
RESULTS
Prediction of FGF9 3′-UTR RNA structures
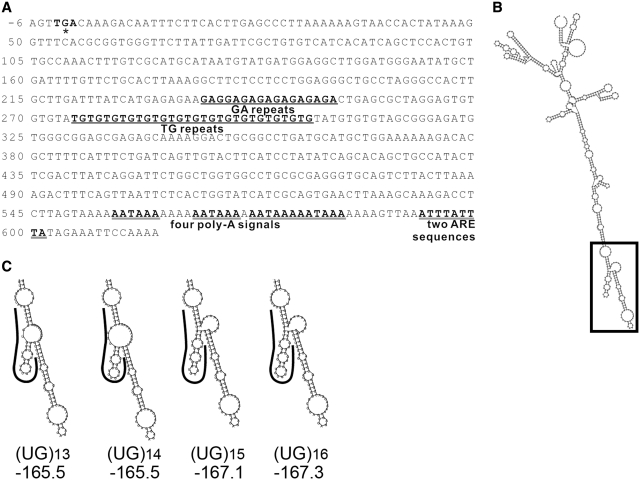
Previously we found the TG MS in FGF9 3′-UTR had four polymorphic alleles [c.*275TG(13_16)] in Han Chinese population and different lengths of the TG repeat affected luciferase gene expression (26). To investigate whether any sequence motifs are embedded in the TG repeats of FGF9 3′-UTR, the 613 nt downstream from the coding region of human FGF9 (Figure 1A) were submitted for secondary structure prediction by RNAfold (27). The 3′-UTR sequence of human FGF9 is highly structured as indicated by the presence of many stems and loops (Figure 1B). Interestingly, different repeat numbers in the UG MS influence the local folding of RNA, and the varied structures contain different numbers of stem–loops with different minimal free energy (MFE) (Figure 1C). Among these, (UG)13 and (UG)14 are similar in their structures and folding MFE. Likewise, (UG)15 and (UG)16 are similar in their structures and folding MFE. We therefore hypothesized that the predicted secondary structures of human FGF9 3′-UTR sequences, especially around the UG-repeat region can be recognized by specific RNA-binding proteins and the interaction between FGF9 RNA and protein may affect the FGF9 expression.
Figure 1.
FGF9 3′-UTR RNA structure prediction. (A) Sequence analysis of human FGF9 3′-UTR. The sequence corresponding to the 3′-end of the gene is depicted. The TGA nucleotide sequence marked in bold type corresponds to the translation stop codon. The (GA) and (TG) MS motifs are marked in bold and underlines, and the AU-rich element (ARE) sequence and poly-A signals are marked with double underlines. (B) Representative RNA structure of full-length FGF9 3′-UTR sequence (NM_002010; 613 nt). (C) Enlargement of boxed area in B. Curved lines mark the locations of indicated UG repeats. MFE of predicted structure under each indicated UG repeat in units of kcal/mol.
Specific proteins bind to (UG)15 oligoribonucleotide
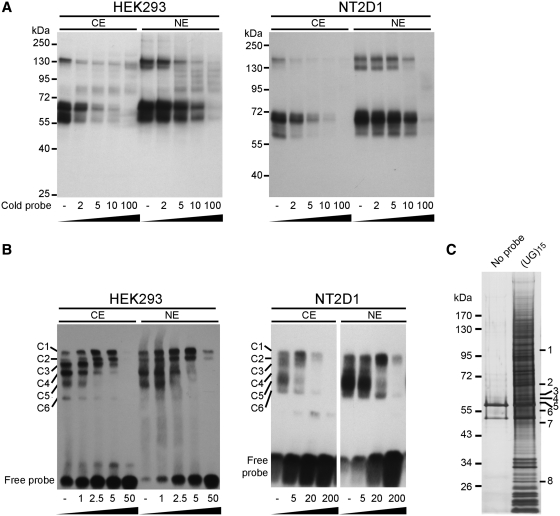
To determine whether specific proteins bind to (UG)n repeat motifs to form RNA–protein complexes, synthetic (UG)15 oligoribonucleotide, the dominant allele of the UG MSs in human FGF9 3′-UTR (26), was incubated with cytoplasmic or nuclear extracts prepared from HEK293 and NT2D1 cells. Two major protein groups ranging from 55 to 70 kDa and ∼130 kDa were detected by UV-cross-linking assays (Figure 2A). The specificity of the interaction between UG repeats and protein extracts was confirmed by competition assays showing that the binding affinity of these proteins was reduced with increased doses of unlabeled probes. Moreover, the RNA–protein interaction was stronger in nuclear than cytoplasmic extract. Similar results were obtained using the RNA-EMSA approach. At least six RNA–protein complexes (C1–C6) were formed in both cytoplasmic and nuclear extracts (Figure 2B). Together, these data showed that the structural motif of the (UG)15 MS attracted many proteins to form RNA–protein complexes.
Figure 2.
Riboprotein complex formation by UG repeats of FGF9 3′-UTR. (A) UV-cross-linking was conducted using biotinylated (UG)15 oligoribonucleotide and protein extracts (cytoplasmic extract, CE; nuclear extract, NE) prepared from HEK293 (left) and NT2D1 (right) cells. Excess molar ratio of unlabeled cold probe was added as indicated in the competition assay. (B) The same labeled probe, cold probe and protein extracts as in (A), were used in RNA-EMSA and showed similar results in HEK293 (left) and NT2D1 (right) cells. C1–C6: complexes 1–6. (C) (UG)15 binding proteins identified by LC–MS. Proteins pulled down in the absence (left lane) or the presence of riboprobe (UG)15 (right lane) were assayed by silver stain. Differentially expressed bands (marked by 1–8) were excised for protein identification by LC–MS.
To identify the specific (UG)n–binding proteins, biotin-labeled (UG)15 oligoribonucleotide was used as a bait to pull down interacting proteins from HEK293 cells. Compared with the control, silver stain revealed several differentially pulled-down proteins (Figure 2C). Following LC–MS–MS analysis, many proteins were identified as potential (UG)15-binding proteins (Figure 2C and Table 1). The identified proteins, their identification scores, and sequence coverage are provided in Table 1. Among these, the molecular weights of far upstream element (FUSE) binding protein 3 (FUBP3), Ras-GTPase-activating protein SH3-domain-binding protein (G3BP1), non-POU domain containing octamer-binding (NONO), vimentin (VIM), and DNA polymerase delta interacting protein 3 (POLDIP3) isoform 1, ranged between 55 and 70 kDa and represented the most promising candidates for further investigation.
Table 1.
Proteins identified by LC/MS/MS
| No.a | Gene name (gene symbol) | Matched Gene ID | Mass (kDa) | Score | Coverage (%) |
|---|---|---|---|---|---|
| 1 | Splicing factor proline/glutamine-rich (polypyrimidine tract binding protein associated), isoform CRA_a (SFPQ) | gi|119627826 | 105 | 506 | 22 |
| 2 | FUSE binding protein 3(FUBP3) | gi|100816392 | 67 | 123 | 17 |
| 3 | Ras-GTPase-activating protein SH3-domain-binding protein (G3BP1) | gi|5031703 | 65 | 180 | 22 |
| 4 | Non-POU domain containing, octamer-binding (NONO) | gi|34932414 | 62 | 633 | 20 |
| 5 | Vimentin (VIM) | gi|340219 | 58 | 108 | 28 |
| 6 | DNA polymerase delta interacting protein 3 isoform 1(POLDIP3) | gi|29837655 | 56 | 234 | 24 |
| 7 | TAR DNA binding protein (TARDBP) | gi|6678271 | 47 | 235 | 16 |
| 8 | Cleavage and polyadenylation specific factor 5 (NUDT21) | gi|5901926 | 30 | 126 | 14 |
aNo: number. The number follows the order indicated in Figure 2C.
FUBP3 binds to FGF9 3′ (UG)15
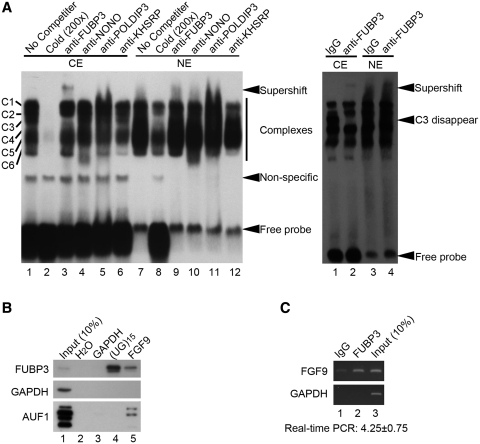
We carried out RNA-EMSA with specific antibodies to confirm the binding of identified proteins to (UG)15 sequence. The presence of many RNA–protein complexes in protein extracts incubated with (UG)15 oligoribonucleotide only was in agreement with the results shown in Figure 2B (Figure 3A). Among four proteins examined, a distinct complex formed by the (UG)15 probe interacted with anti-FUBP3 antibody in HEK293 cells (Figure 3A, lane 3 in left panel). Furthermore, a weak but detectable supershift was also observed with anti-FUBP3 antibody in the NT2D1 cells (Figure 3A, lane 2 in right panel), but not with mouse IgG (Lanes 1 and 3 in the right panel); complete disappearance of the C3 band was accompanied by a band shift (Figure 3A). It is of note that the interaction between FUBP3 and the (UG)15 probe was specific to the cytoplasmic fraction (Lanes 1–6 and 1–2 in left and right panel, respectively) but not the nuclear fraction (Lanes 7–12 and 3–4 in left and right panel, respectively).
Figure 3.
Identification of FUBP3 as UG-repeat binding protein. (A) The RNA–protein complex was detected using specific antibodies as indicated in RNA-EMSA in the protein extracts from NT2D1 (left) and HEK293 (right) cells. CE, cytoplasmic extract; NE, nuclear extract. IgG was used as a control. (B) Pull-down assays were performed using biotinylated (UG)15 riboprobe and in vitro-transcribed FGF9 3′-UTR containing (UG)15 repeats in the cytoplasmic extract from HEK293 cells. Specific antibodies to FUBP3, GAPDH and AUF1 were used to detect the pulled-down proteins. H2O and in vitro-transcribed GAPDH probe were used as negative controls for pull-down assays. (C) Immunoprecipitated RNA–protein complexes using anti-FUBP3 antibody showed the presence of endogenous FGF9, but not GAPDH mRNA in HEK293 cells (lane 2). Goat normal IgG was used as a control for the immunoprecipitation reaction. The enrichment of FGF9 mRNA precipitated by anti-FUBP3 antibody is shown relative to goat normal IgG. Data were normalized to GAPDH.
To confirm FUBP3 binding to the (UG)15 of FGF9 mRNA, we first applied the original membrane used in UV-cross-linking experiment for western blot analysis. Using FUBP3-specific antibody, our data showed a signal locates on the top of 55–70 kDa cluster that indicates FUBP3 (Supplementary Figure S1). Further, biotin-labeled (UG)15 oligoribonucleotide or in vitro transcribed FGF9 3′-UTR RNA were used to pull down interacting proteins from HEK293 cytoplasmic extract, while in vitro-transcribed GAPDH was used as negative control. It was clearly demonstrated that both biotin-labeled (UG)15 and FGF9 3′-UTR RNA, but not GAPDH, interacted with FUBP3 protein (Figure 3B, top panel). In addition, AU-rich element (ARE) binding factor 1 (AUF1), that is known to control FGF9 gene expression by interacting with the FGF9 3′-UTR ARE (34), was present in the protein pools pulled down by FGF9 3′-UTR RNA but not (UG)15 oligoribonucleotide (Figure 3B, bottom panel), thus suggesting that the protein–RNA complexes and their interactions are specific. We then investigated the ability of FUBP3 to bind to the FGF9 (UG)15 in vivo. HEK293 cell lysates were immunoprecipitated with anti-FUBP3 antibody and the presence of endogenous FGF9 mRNA in the immunoprecipitates was determined by quantitative RT-PCR. A distinct PCR product was detected in the immunoprecipitates for FGF9, but not for GAPDH, and a 4.25-fold enrichment of FGF9 level in the immunoprecipitates for FUBP3 relative to those for IgG was obtained (Figure 3C). Collectively, these data demonstrated that FUBP3 specifically binds to FGF9 3′-UTR (UG)15 and the interaction may participate in the regulation of FGF9 gene expression.
FUBP3 positively regulates FGF9 expression through an increase in translational efficiency
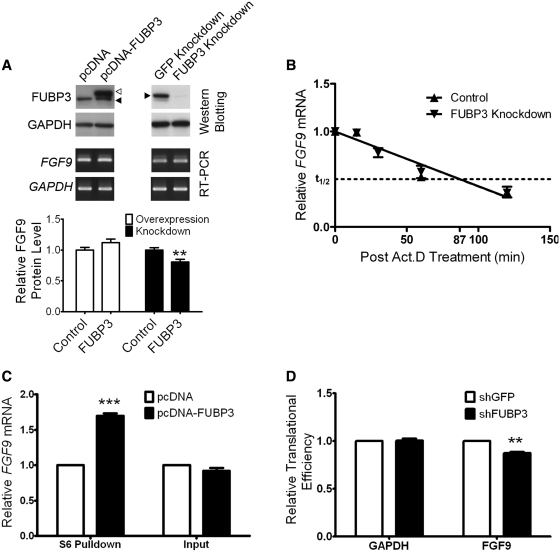
To address how FUBP3 interacts with FGF9 3′-UTR (UG)15 and participates in the regulation of FGF9 gene expression, we first determined the level of endogenous FGF9 in response to alteration of the cellular level of FUBP3 in HEK293 cells. Either full-length FUBP3 cDNA or FUBP3-specific shRNA constructs were transfected into HEK293 cells. The overexpression or knockdown of FUBP3 expression was confirmed by western blot using anti-FUBP3 antibodies (Figure 4A, top panel) and the levels of FGF9 mRNA and protein were measured. While the levels of FGF9 mRNA were similar in response to FUBP3 overexpression (1.16 ± 0.01) and knockdown (1.19 ± 0.04) (Figure 4A, middle panel), FGF9 protein increased to 1.12-fold (P = 0.056) under FUBP3 overexpressing condition and significantly declined to 0.81-fold (P < 0.01) in FUBP3-knockdown cells (Figure 4A, bottom panel). These data showed that the FUBP3 had no effect on FGF9 transcription and suggested that FUBP3 regulates FGF9 expression at the post-transcriptional level by stabilizing mRNA or increasing translational efficiency.
Figure 4.
FUBP3 increased the expression of endogenous FGF9 and reporter proteins containing FGF9 3′-UTR. (A) Representative western blots and RT-PCR results from transient FUBP3 overexpression and shRNA knockdown. Endogenous FUBP3 (filled triangle) and overexpressed FUBP3 (open triangle) were shown in the top panel. Relative mRNA level (RT-PCR; middle panel) and protein amount (ELISA; bottom panel) were shown under each treatments. (B) RNA stability assay showed that knockdown of FUBP3 had no effect on FGF9 turn-over rate. (C) FUBP3 overexpression promotes FGF9 protein synthesis in HEK293 cells. The ribosome complexes were pulled down by 40S ribosomal protein S6 antibody from HEK293 cells overexpressing recombinant FUBP3. The mRNAs bound with ribosome complexes were extracted and analyzed by quantitative RT-PCR. FGF9 mRNA expression level was detected and normalized with the mRNA expression level of GAPDH. The total RNA was also extracted from HEK293 cell with or without FUBP3-overexpression (Right), and the FGF9 mRNA level was detected as an experimental comparison to the results showed in (Left). (D) Relative translational efficiency representation of FGF9 and GAPDH in the polysomal fractions. Ribosome-associated transcripts were measured using quantitative RT-PCR and translational efficiency of FGF9 and GAPDH under FUBP3-knockdown were normalized with the translational efficiency in GFP-knockdown cells. **P < 0.01; ***P < 0.001.
To define the physiological significance of the interaction between FUBP3 and UG repeats in the regulation of FGF9 expression, we determined the decay of FGF9 mRNA either with or without FUBP3 using a cell-based decay system. FUBP3-specific shRNA was transfected into HEK293 cells to knockdown the FUBP3 expression and the effect on mRNA stability in cells with and without FUBP3 knockdown were measured and compared. After 0, 15, 30, 60 and 120 min after adding actinomycin D (Act D), RNA was extracted and the relative amounts of FGF9 to GAPDH mRNA at different time points were measured using quantitative RT-PCR. The two regression lines almost overlapped and the estimated half-life (t1/2) for cells transfected and untransfected with FUBP3 shRNA were the same (P = 0.58; Figure 4B). As the stability of FGF9 mRNA did not change upon FUBP3 depletion, these results ruled out the possibility that FUBP3 regulates FGF9 expression through mRNA stabilization and suggested that FUBP3 contributes to FGF9 expression through an increase in translational efficiency.
To test whether FUBP3 translationally controls endogenous FGF9 protein expression, we first performed ribosome complex pulldown assay (33) under the condition of overexpressing FUBP3. RNA was isolated from the immunoprecipated pellet as well as the initial lysate, and amplified using quantitative RT-PCR. We found that overexpression of FUBP3 protein increased the ribosome-bound FGF9 mRNA by ∼1.7-fold (P = 0.001; Figure 4C, left) while there was no difference of FGF9 mRNA level in the initial lysates (Figure 4C, right). Next, we conducted sucrose gradient analysis of ribosome-bounded FGF9 mRNA to examine the FGF9 translational efficiency in FUBP3-knockdown HEK293 cells. The polysome profiling revealed that translational efficiency of FGF9 mRNA drops 13% (P < 0.002; Figure 4D, right) while there is no change in the translational efficiency of GAPDH gene (Figure 4D, left). Taken together, these results support our hypothesis that FUBP3 positively regulates the FGF9 protein expression by promoting translational activity.
Synergistic effect of FUBP3 with FGF9 (UG)n repeats
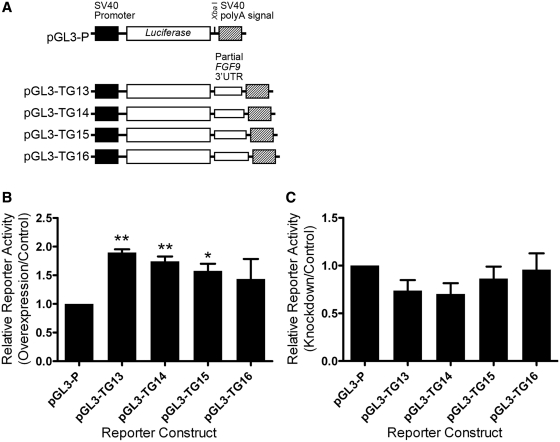
Four alleles with different length of TG repeats in FGF9 3′-UTR were found in Han Chinese (26). To determine whether various numbers of UG repeats influence the interaction with FUBP3 and thus have different effects, we used constructs containing different numbers of TG repeats (Figure 5A) in reporter gene assays under FUBP3 overexpression or knockdown in HEK293 cells. Overexpression of FUBP3 significantly promoted reporter gene activity (Figure 5B). In addition, reporter constructs containing (TG)13 (P < 0.01) and (TG)14 (P < 0.01) repeats had a more prominent effect than that containing (TG)16 repeats in responding to FUBP3 overexpression. On the other hand, the structural effect of (TG) repeat in FUBP3 knockdown cells was not statistically distinguishable (Figure 5C). These data mirror the results presented in the structure prediction (Figure 1B) and suggest that the number of TG repeats in FGF9 3′-UTR and levels of FUBP3 protein collaboratively fine tune the expression of FGF9.
Figure 5.
Synergistic effect of FUBP3 with FGF9 (UG)n repeats. (A) Schematic diagram of reporter constructs containing partial FGF9 3′-UTR with various TG repeats. Reporter activities of various constructs from HEK293 cells overexpressing (B) or knockdown (C) FUBP3. All values were normalized to Renilla luciferase activity produced from a cotransfected control plasmid. Error bars represent standard deviations from three independent transfections. Statistical analyses were conducted by one-way ANOVA. *P < 0.05; **P < 0.01.
DISCUSSION
In the present study, we found that FUBP3 is able to interact with FGF9 3′-UTR (UG) repeats to control FGF9 expression in both HEK293 and NT2D1 cells. Furthermore, FUBP3 overexpression or knockdown resulted in an increased or reduced FGF9 expression. We also demonstrated, while depletion of FUBP3 had no effect on FGF9 mRNA stability, the altered FGF9 protein expressions under FUBP3 overexpression and knockdown are in association with concordant changes of ribosome-bound FGF9 mRNAs. These results show that FUBP3 regulates FGF9 expression through a mechanism involved in translational efficiency.
FGF9 is a potent mitogen, angiogenic, osteogenic and morphogenic factor besides its role in lung (review see (35) and gonad developments [review see (36)]. Aberrant expression of FGF9 usually results in dramatic effects in physiological and pathological processes. Thus, FGF9 expression needs to be tightly controlled to properly regulate various physiological processes. In this study, we found that knockdown of FUBP3 reduced FGF9 protein by 20%, a change that is not enormously huge but should be biologically relevant. It is quite common under physiological conditions that small fluctuation of ligands (such as neurotransmitters or ions) usually leads to a series of chain reaction that ultimately results in huge physiological response. Behr et al. (37) demonstrated in a mouse model that only 50% decrease in fgf9 (i.e. fgf9+/−) severely impair bone repair and vessel formation. Considering that FGF9 modulates various signaling pathways including VEGF (38) and sonic hedgehog (39), and the knockdown is not 100% efficient in the current study, the actual magnitude of FUBP3-regulated FGF9 protein expression might be greater and effect of FGF9 will be augmented in vivo.
Human FUBP3 belongs to a regulatory gene family consisting of three members, FUBP1, KH-type splicing regulatory protein (KHSRP, also known as FUBP2), and FUBP3 (40). Although the three members share highly conserved sequence and structure (40), their functions are quite diverse. FUBP1 is a single-strand DNA-binding protein which activates c-myc proto-oncogene expression by binding to the non-coding strand cis FUSE, encompassing positions −1554 to −1526 of c-myc (41). KHSRP interacts with ARE-containing mRNAs and is a key mediator of mRNA decay (42). Independent of the role in RNA degradation, KHSRP also regulates the maturation of a subset of miRNAs (43). Although FUBP3 is the strongest activator among this family (44), it has no effect on the transcription of FGF9 as shown in this study and a survey of genes regulated by siFBPs (44). Our data indicated that FUBP3 executed its effect at the post-transcriptional rather than at the pre-transcriptional level. Unlike the other two members, there was no evidence indicating an association of FUBP3 with RNA (42,45,46). The subcellular localization of FUBP3 in both cytoplasm and nucleus also distinguished it from the other two, that were restricted to the nucleus (44), implying its distinct role in gene regulation. However, we here report the first evidence that FUBP3 binds to FGF9 mRNA and the specific protein–RNA interaction translationally controls FGF9 gene expression.
Earlier, we provided evidence that the TG repeat located on the FGF9 3′-UTR is a functional element that controls FGF9 mRNA stability and regulates FGF9 expression (26). It is worth noting that the factor we identified in this study, FUBP3, has no destabilizing effect on FGF9 mRNA. Instead, it positively regulates FGF9 expression through a mechanism related to translational efficiency. Thus though FUBP3 binds to FGF9 UG repeats and contributes to the regulation of FGF9 expression, it is not the factor we originally found in our earlier study. Indeed, data from RNA-EMSA demonstrated that at least six proteins can form complexes with (UG)15 riboprobe in the cytoplasmic fraction, while five protein–RNA complexes were found in the nuclear fraction. These studies demonstrated that other factors are involved in the regulation of FGF9 mRNA stability. Nevertheless, the FUBP3-(UG)15 complex only exists in the cytoplasmic fraction (i.e. C3 complex), suggesting the effect of FUBP3 is involved in the later process of gene expression where the interaction mainly occurs in the cytoplasmic compartment.
Recent studies demonstrated that mRNAs that encode proteins with similar functions are coordinately regulated by the same RBP to form a post-transcriptional network called an RNA regulon (47). On the other hand, many RBPs may coordinately bind to the same transcript to regulate gene expression. Beside the TG repeats, we also found a functional AU-rich element (ARE) in the 3′-UTR of the FGF9 gene. The interaction between FGF9 ARE and AU-rich element binding protein 1 (AUF1) contributes to the destabilization of FGF9 mRNA and leads to a reduction of FGF9 expression (34). Together with the findings from this study, these results demonstrated the presence of two cis-elements that communicate with different cellular factors and trigger different mechanisms to regulate FGF9 expression post-transcriptionally.
Our team has been engaging to study the function and regulation of FGF9 for a long time. We are the first group to report FGF9 function is involved in steroidogenesis (16), and associated with human male-to-female sex reversal (26) and endometriosis (19). In addition, we identified PGE2 promotes FGF9 mRNA expression (25) and AUF1 downregulate FGF9 mRNA stability (34). These studies make us believed the expression of FGF9 is under many layers of controls to keep the FGF9 homeostasis in a cell. Our goal is to establish the regulatory network of FGF9 expression and illustrate the consequence of deregulated FGF9 expression. The current study demonstrates that translational regulation is another level that cells will use to regulate FGF9 protein production, an important step in the central dogma control of gene expression. Furthermore, results from this study strongly support the existence of a complex regulatory network of FGF9 gene expression to fine-tune FGF9 homeostasis. Finally, our study reports the novel RNA-binding property of FUBP3 and its role in regulation of translational efficiency. Thus it raises the possibility that some diseases induced by FGF9 over-expression might be a consequence of FUBP3 dysregulation. Further studies are necessary to elucidate this proposed possibility.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Science Council, Taiwan (NSC 98-3112-B-006-002 to H.S.S.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Iain Bruce for the excellent help in English editing and National RNAi Core Facility, Taiwan for various RNAi constructs.
REFERENCES
- 1.Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Rothenburg S, Koch-Nolte F, Rich A, Haag F. A polymorphic dinucleotide repeat in the rat nucleolin gene forms Z-DNA and inhibits promoter activity. Proc. Natl Acad. Sci. USA. 2001;98:8985–8990. doi: 10.1073/pnas.121176998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fabregat I, Koch KS, Aoki T, Atkinson AE, Dang H, Amosova O, Fresco JR, Schildkraut CL, Leffert HL. Functional pleiotropy of an intramolecular triplex-forming fragment from the 3′-UTR of the rat Pigr gene. Physiol. Genomics. 2001;5:53–65. doi: 10.1152/physiolgenomics.2001.5.2.53. [DOI] [PubMed] [Google Scholar]
- 5.Chiba-Falek O, Nussbaum RL. Effect of allelic variation at the NACP-Rep1 repeat upstream of the alpha-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum. Mol. Genet. 2001;10:3101–3109. doi: 10.1093/hmg/10.26.3101. [DOI] [PubMed] [Google Scholar]
- 6.Albanese V, Biguet NF, Kiefer H, Bayard E, Mallet J, Meloni R. Quantitative effects on gene silencing by allelic variation at a tetranucleotide microsatellite. Hum. Mol. Genet. 2001;10:1785–1792. doi: 10.1093/hmg/10.17.1785. [DOI] [PubMed] [Google Scholar]
- 7.Borrmann L, Seebeck B, Rogalla P, Bullerdiek J. Human HMGA2 promoter is coregulated by a polymorphic dinucleotide (TC)-repeat. Oncogene. 2003;22:756–760. doi: 10.1038/sj.onc.1206073. [DOI] [PubMed] [Google Scholar]
- 8.Wren JD, Forgacs E, Fondon JW, III, Pertsemlidis A, Cheng SY, Gallardo T, Williams RS, Shohet RV, Minna JD, Garner HR. Repeat polymorphisms within gene regions: phenotypic and evolutionary implications. Am. J. Hum. Genet. 2000;67:345–356. doi: 10.1086/303013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamilton BJ, Wang XW, Collins J, Bloch D, Bergeron A, Henry B, Terry BM, Zan M, Mouland AJ, Rigby WF. Separate cis-trans pathways post-transcriptionally regulate murine CD154 (CD40 ligand) expression: a novel function for CA repeats in the 3′-untranslated region. J. Biol. Chem. 2008;283:25606–25616. doi: 10.1074/jbc.M802492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JH, Jeon MH, Seo YJ, Lee YJ, Ko JH, Tsujimoto Y. CA repeats in the 3′-untranslated region of bcl-2 mRNA mediate constitutive decay of bcl-2 mRNA. J. Biol. Chem. 2004;279:42758–42764. doi: 10.1074/jbc.M407357200. [DOI] [PubMed] [Google Scholar]
- 11.Ruggiero T, Olivero M, Follenzi A, Naldini L, Calogero R, Di Renzo MF. Deletion in a (T)8 microsatellite abrogates expression regulation by 3′-UTR. Nucleic Acids Res. 2003;31:6561–6569. doi: 10.1093/nar/gkg858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mittag M. The function of circadian RNA-binding proteins and their cis-acting elements in microalgae. Chronobiol Int. 2003;20:529–541. doi: 10.1081/cbi-120022411. [DOI] [PubMed] [Google Scholar]
- 13.Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-3-reviews3005. REVIEWS3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colvin JS, Green RP, Schmahl J, Capel B, Ornitz DM. Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell. 2001;104:875–889. doi: 10.1016/s0092-8674(01)00284-7. [DOI] [PubMed] [Google Scholar]
- 15.Robinson D, Hasharoni A, Evron Z, Segal M, Nevo Z. Synovial chondromatosis: the possible role of FGF 9 and FGF receptor 3 in its pathology. Int. J. Exp. Pathol. 2000;81:183–189. doi: 10.1046/j.1365-2613.2000.00154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin YM, Tsai CC, Chung CL, Chen PR, Sun HS, Tsai SJ, Huang BM. Fibroblast growth factor 9 stimulates steroidogenesis in postnatal Leydig cells. Int. J. Androl. 2010;33:545–553. doi: 10.1111/j.1365-2605.2009.00966.x. [DOI] [PubMed] [Google Scholar]
- 17.Colvin JS, Feldman B, Nadeau JH, Goldfarb M, Ornitz DM. Genomic organization and embryonic expression of the mouse fibroblast growth factor 9 gene. Dev. Dyn. 1999;216:72–88. doi: 10.1002/(SICI)1097-0177(199909)216:1<72::AID-DVDY9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 18.Miyamoto M, Naruo K, Seko C, Matsumoto S, Kondo T, Kurokawa T. Molecular cloning of a novel cytokine cDNA encoding the ninth member of the fibroblast growth factor family, which has a unique secretion property. Mol. Cell Biol. 1993;13:4251–4259. doi: 10.1128/mcb.13.7.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai SJ, Wu MH, Chen HM, Chuang PC, Wing LY. Fibroblast growth factor-9 is an endometrial stromal growth factor. Endocrinology. 2002;143:2715–2721. doi: 10.1210/endo.143.7.8900. [DOI] [PubMed] [Google Scholar]
- 20.Miyagi N, Kato S, Terasaki M, Aoki T, Sugita Y, Yamaguchi M, Shigemori M, Morimatsu M. Fibroblast growth factor-9 (glia-activating factor) stimulates proliferation and production of glial fibrillary acidic protein in human gliomas either in the presence or in the absence of the endogenous growth factor expression. Oncol. Rep. 1999;6:87–92. [PubMed] [Google Scholar]
- 21.Giri D, Ropiquet F, Ittmann M. FGF9 is an autocrine and paracrine prostatic growth factor expressed by prostatic stromal cells. J. Cell Physiol. 1999;180:53–60. doi: 10.1002/(SICI)1097-4652(199907)180:1<53::AID-JCP6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 22.Hendrix ND, Wu R, Kuick R, Schwartz DR, Fearon ER, Cho KR. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 2006;66:1354–1362. doi: 10.1158/0008-5472.CAN-05-3694. [DOI] [PubMed] [Google Scholar]
- 23.Abdel-Rahman WM, Kalinina J, Shoman S, Eissa S, Ollikainen M, Elomaa O, Eliseenkova AV, Butzow R, Mohammadi M, Peltomaki P. Somatic FGF9 mutations in colorectal and endometrial carcinomas associated with membranous beta-catenin. Hum. Mutat. 2008;29:390–397. doi: 10.1002/humu.20653. [DOI] [PubMed] [Google Scholar]
- 24.Wing LY, Chuang PC, Wu MH, Chen HM, Tsai SJ. Expression and mitogenic effect of fibroblast growth factor-9 in human endometriotic implant is regulated by aberrant production of estrogen. J. Clin. Endocrinol. Metab. 2003;88:5547–5554. doi: 10.1210/jc.2003-030597. [DOI] [PubMed] [Google Scholar]
- 25.Chuang PC, Sun HS, Chen TM, Tsai SJ. Prostaglandin E2 induces fibroblast growth factor 9 via EP3-dependent protein kinase Cdelta and Elk-1 signaling. Mol. Cell Biol. 2006;26:8281–8292. doi: 10.1128/MCB.00941-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen TM, Kuo PL, Hsu CH, Tsai SJ, Chen MJ, Lin CW, Sun HS. Microsatellite in the 3′ untranslated region of human fibroblast growth factor 9 (FGF9) gene exhibits pleiotropic effect on modulating FGF9 protein expression. Hum. Mutat. 2007;28:98. doi: 10.1002/humu.9471. [DOI] [PubMed] [Google Scholar]
- 27.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin R. Protein synthesis: methods and protocols. Totowa, NJ: Humana Press; 1998. [Google Scholar]
- 29.Yan JX, Wait R, Berkelman T, Harry RA, Westbrook JA, Wheeler CH, Dunn MJ. A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometry. Electrophoresis. 2000;21:3666–3672. doi: 10.1002/1522-2683(200011)21:17<3666::AID-ELPS3666>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 30.Tenenbaum SA, Lager PJ, Carson CC, Keene JD. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- 31.Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000;19:2340–2350. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl Acad. Sci. USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeh CH, Hung LY, Hsu C, Le SY, Lee PT, Liao WL, Lin YT, Chang WC, Tseng JT. RNA-binding protein HuR interacts with thrombomodulin 5′ untranslated region and represses internal ribosome entry site-mediated translation under IL-1 beta treatment. Mol. Biol. Cell. 2008;19:3812–3822. doi: 10.1091/mbc.E07-09-0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen TM, Hsu CH, Tsai SJ, Sun HS. AUF1 p42 isoform selectively controls both steady-state and PGE2-induced FGF9 mRNA decay. Nucleic Acids Res. 2010;38:8061–8071. doi: 10.1093/nar/gkq717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warburton D, Perin L, Defilippo R, Bellusci S, Shi W, Driscoll B. Stem/progenitor cells in lung development, injury repair, and regeneration. Proc. Am. Thorac. Soc. 2008;5:703–706. doi: 10.1513/pats.200801-012AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piprek RP. Genetic mechanisms underlying male sex determination in mammals. J. Appl. Genet. 2009;50:347–360. doi: 10.1007/BF03195693. [DOI] [PubMed] [Google Scholar]
- 37.Behr B, Leucht P, Longaker MT, Quarto N. Fgf-9 is required for angiogenesis and osteogenesis in long bone repair. Proc. Natl Acad. Sci. USA. 2010;107:11853–11858. doi: 10.1073/pnas.1003317107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White AC, Lavine KJ, Ornitz DM. FGF9 and SHH regulate mesenchymal Vegfa expression and development of the pulmonary capillary network. Development. 2007;134:3743–3752. doi: 10.1242/dev.004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lavine KJ, White AC, Park C, Smith CS, Choi K, Long F, Hui CC, Ornitz DM. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev. 2006;20:1651–1666. doi: 10.1101/gad.1411406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis-Smyth T, Duncan RC, Zheng T, Michelotti G, Levens D. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J. Biol. Chem. 1996;271:31679–31687. doi: 10.1074/jbc.271.49.31679. [DOI] [PubMed] [Google Scholar]
- 41.Duncan R, Bazar L, Michelotti G, Tomonaga T, Krutzsch H, Avigan M, Levens D. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 1994;8:465–480. doi: 10.1101/gad.8.4.465. [DOI] [PubMed] [Google Scholar]
- 42.Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 43.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chung HJ, Liu JH, Dundr M, Nie ZQ, Sanford S, Levens D. FBPs are calibrated molecular tools to adjust gene expression. Mol. Cell. Biol. 2006;26:6584–6597. doi: 10.1128/MCB.00754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irwin N, Baekelandt V, Goritchenko L, Benowitz LI. Identification of two proteins that bind to a pyrimidine-rich sequence in the 3′-untranslated region of GAP-43 mRNA. Nucleic Acids Res. 1997;25:1281–1288. doi: 10.1093/nar/25.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rothe F, Gueydan C, Bellefroid E, Huez G, Kruys V. Identification of FUSE-binding proteins as interacting partners of TIA proteins. Biochem. Biophys. Res. Commun. 2006;343:57–68. doi: 10.1016/j.bbrc.2006.02.112. [DOI] [PubMed] [Google Scholar]
- 47.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.