Abstract
Quinolones inhibit bacterial type II DNA topoisomerases (e.g. DNA gyrase) and are among the most important antibiotics in current use. However, their efficacy is now being threatened by various plasmid-mediated resistance determinants. Of these, the pentapeptide repeat-containing (PRP) Qnr proteins are believed to act as DNA mimics and are particularly prevalent in Gram-negative bacteria. Predicted Qnr-like proteins are also present in numerous environmental bacteria. Here, we demonstrate that one such, Aeromonas hydrophila AhQnr, is soluble, stable, and relieves quinolone inhibition of Escherichia coli DNA gyrase, thus providing an appropriate model system for Gram-negative Qnr proteins. The AhQnr crystal structure, the first for any Gram-negative Qnr, reveals two prominent loops (1 and 2) that project from the PRP structure. Deletion mutagenesis demonstrates that both contribute to protection of E. coli DNA gyrase from quinolones. Sequence comparisons indicate that these are likely to be present across the full range of Gram-negative Qnr proteins. On this basis we present a model for the AhQnr:DNA gyrase interaction where loop1 interacts with the gyrase A ‘tower’ and loop2 with the gyrase B TOPRIM domains. We propose this to be a general mechanism directing the interactions of Qnr proteins with DNA gyrase in Gram-negative bacteria.
INTRODUCTION
Quinolones, particularly fluoroquinolones, are among the most widely used antibiotics on account of their broad spectrum of activity that encompasses many pathogenic Gram-negative (Enterobacteriaceae) and Gram-positive (staphylococci and streptococci) bacteria (1,2). They are particularly effective against urinary tract and sexually acquired infections, but more recently have found application in the treatment of gastrointestinal, skin and soft tissue, bone and joint and some respiratory infections. Newer fluoroquinolones (ofloxacin, gatifloxacin, moxifloxacin) also show promise as candidates for inclusion in multidrug anti-tuberculosis therapy (3). Quinolones target the bacterial type IIA DNA topoisomerases DNA gyrase and DNA topoisomerase IV. These heterotetrameric enzymes manipulate DNA topology by introduction of transient double-stranded breaks in bound DNA (G-segment) through which a second DNA fragment (T-segment) may be passed (4). Binding, by intercalation (5), of quinolone antibiotics to the complex of enzyme and the cut G-segment stabilizes this so-called cleavage complex, leading to accumulation, and eventual release, of double-stranded DNA breaks that are ultimately lethal to the cell (6).
As quinolones are fully-synthetic drugs, transferable resistance was long considered to be unlikely in the absence of a pool of environmental resistance genes emanating from producer organisms. Until recently, quinolone resistance was known to arise by mutation of specific regions of type IIA DNA topoisomerases [the quinolone resistance determinant region (QRDR) (7)] and/or by the acquisition or upregulation of efflux pump genes (8). However, the situation has changed markedly with the discovery of a variety of plasmid-borne resistance determinants [reviewed in ref. (9)]. These include the DNA topoisomerase protection protein Qnr (10,11), the fluoroquinolone acetyltransferase Aac(6′)-Ib-cr (12) and transferable fluoroquinolone efflux pumps such as QepA (13) and OqxAB (14,15). Of these, the mechanism by which expression of qnr confers clinical fluoroquinolone resistance is the least understood.
Qnr was first identified in the early 1990s as a plasmid-borne gene in a clinical strain of Klebsiella pneumoniae (11). Subsequent investigations have established that qnr genes have a worldwide distribution in a range of (mainly Gram-negative opportunist) bacterial pathogens, particularly Enterobacteriaceae (e.g. Klebsiella pneumoniae, Escherichia coli) (16). New family members continue to be described (17,18). Sequence comparison of plasmids isolated from clinical Gram-negative strains differentiates five distinct qnr subfamilies: qnrA (A1–A7), qnrB (B1–B25), qnrS (S1–S4) (19) and most recently qnrC (17) and qnrD (18). While high levels of amino acid sequence conservation (∼90% similarity) are observed between members of the same subfamily, the five different subfamilies are less closely related (60% similarity or less). Furthermore, qnr-like open reading frames (orfs) are present upon the chromosomes of numerous other bacteria, including both Gram-positive organisms such as enterococci (20) and mycobacteria (21) and a range of environmental Gram-negative organisms [e.g. Shewanella (22), Stenotrophomonas (23,24) and Vibrio (25,26) spp.]. The relationship between these qnr-like orfs and the qnr genes found in clinical isolates varies—qnrA is essentially identical to orfs found in strains of Shewanella algae (22), whereas chromosomal qnr-like genes from other organisms may be more distantly related to qnrs of clinical origin [Supplementary Figures S1 and S2; (23)].
The sequences of Qnr and related proteins are largely composed of degenerate pentapeptide repeats. Crystal structures, now available for Qnr-like proteins from Mycobacterium tuberculosis [MfpA; (27)] and Enterococcus faecalis [EfsQnr; (28,29)], as well as for a number of other pentapeptide repeat proteins [PRPs; (30–33)] show that these sequences fold to a distinctive right-handed β-helix. In the case of MfpA and EfsQnr, the dimensions and surface charge distribution of this helix approximate to those of B-form DNA (27). Accordingly, it has been suggested that Qnr and related proteins act as DNA mimics that compete with DNA for binding to type IIA DNA topoisomerases. Consistent with this observation, MfpA binds directly to the DNA gyrase holoenzyme (27), while QnrA binds to both the DNA gyrase and DNA topoisomerase IV holoenzymes and to their constituent subunits (34,35). However, different pentapeptide repeat proteins affect DNA topoisomerase activity in different ways. MfpA is a potent inhibitor of DNA gyrase in the absence of antibiotics (27,36), whereas for Qnr proteins (QnrB4) this effect is only weakly observed (36). QnrA1 (34,37), QnrB1 (38) and QnrB4 (36) all protect DNA gyrase from inhibition by fluoroquinolones, whereas no such effect has been demonstrated for MfpA (36). QnrA1 also shows equivalent protection of DNA topoisomerase IV (35). Very recently, EfsQnr was shown to act both as a DNA gyrase inhibitor and to partially protect the enzyme from inhibition by ciprofloxacin (29).
Many aspects of the mechanism of action of Qnr proteins are thus unclear. It remains to be established: (i) why inhibition of DNA gyrase activity varies between different pentapeptide repeat proteins and (ii) how Qnr proteins counteract quinolone inhibition of type IIA DNA topoisomerases without overly affecting the normal function of these essential cellular enzymes. Furthermore, to date no Qnr or Qnr-related protein from a Gram-negative organism has been structurally characterized, an important consideration as the majority of known Qnr proteins from both clinical and environmental sources are of Gram-negative origin. Our initial investigations, using K. pneumoniae QnrB2, were hampered by the poor solubility of the recombinant protein, while we also experienced difficulty in expressing recombinant QnrS1. Accordingly, we here use a bioinformatics approach to identify AhQnr, a Qnr-like protein encoded on the chromosome of the Gram-negative aquatic bacterium [and occasional opportunist pathogen, (39)] Aeromonas hydrophila, as a Qnr family member of predicted high solubility, and demonstrate that AhQnr is readily over-expressed and purified from recombinant E. coli. AhQnr protects DNA gyrase from quinolone inhibition in a similar fashion to previously characterized Qnr proteins, and is thus a suitable model system in which to undertake biophysical and structural studies of Qnr proteins. The crystal structure of AhQnr reported here is, however, distinguished from those of previously reported family members from Gram-positive bacteria (MfpA and EfsQnr) by possession of two distinctive loops projecting from the overall β-helix structure. Deletion of the larger of these abolishes the ability of AhQnr to protect DNA gyrase from inhibition by the fluoroquinolone ciprofloxacin. Sequence comparisons indicate that both of these loops are conserved in Qnr proteins from other Gram-negative species of both environmental and pathogenic origin, with the larger one being highly conserved. Our results suggest a specific mechanism directing interactions of Qnr proteins from Gram-negative bacteria with DNA gyrase.
MATERIALS AND METHODS
AhQnr cloning, expression and purification
Detailed procedures are given in Supplementary Data. The qnr-like open reading frame AhQnr (GenBank accession no. ABK38882.1, from the chromosome of Aeromonas hydrophila ATCC 7966; generous gift of Dr Antonio Corriea, University of Aveiro, Portugal) was cloned into the T7 expression vector pET-26b (Novagen) by standard procedures. Recombinant protein was purified from E. coli strain BL21 (DE3) or B834 (DE3) (selenomethionine-substituted material) by anion exchange, heparin affinity and size exclusion chromatography.
Circular dichroism spectroscopy
Circular dichroism (CD) spectra were measured on a JASCO J-815 spectropolarimeter fitted with a Peltier temperature controller. AhQnr solutions were dialyzed into buffer D (20 mM sodium phosphate pH 8.0, 10% glycerol) and prepared in 1 mm path-length cuvettes. Spectra were recorded at 5°C using a 1 nm interval, 1 nm bandwidth and 2 s response time.
Analytical ultracentrifugation
AhQnr in buffer D was diluted to concentrations of 10.6, 7.0 and 5.3 μM. Samples were centrifuged at 10 000, 12 000 and 14 000 rpm at 10°C in 12 mm path-length cells in a Beckman XL-I analytical ultracentrifuge until sedimentation had reached equilibrium, and absorbance at 280 nm was recorded as a function of centrifugal radius. Data were fitted globally using UltraScan [http://www.ultrascan.uthscsa.edu/; (40)] with the partial specific volume (0.7205 ml/g) and the solvent density (1.0311 g/ml) calculated, based on the sequence of AhQnr and buffer composition, using SEDNTERP (41). Fits were evaluated on the basis of random variation in the residuals, where the residuals represent the difference between each data point and the corresponding theoretical point on the fitted curve. For data plotting absorbance readings were converted to protein concentration using an extinction coefficient of 22 970 M−1 cm−1 calculated from the AhQnr amino acid sequence using the algorithm of Gill and von Hippel (42).
DNA gyrase supercoiling assays
Assays were carried out using a commercial E. coli DNA gyrase supercoiling assay kit (Inspiralis, Norwich, UK) in a total reaction volume of 30 μl. Relaxed plasmid pBR322 (500 ng) was incubated with DNA gyrase in the supplied buffer (35 mM Tris–HCl pH 7.5, 24 mM KCl, 2 mM DTT, 1.8 mM spermidine, 1 mM ATP, 6.5% glycerol, 0.1 mg/ml BSA) with appropriate amounts of ciprofloxacin and AhQnr in buffer D (AhQnr) or buffer H (25 mM Tris-HCl pH 7.5, 2 mM DTT, 5% glycerol; AhQnr-6His and mutants) for 1 h at 37°C. The entire reaction was loaded on a 1% agarose gel in 1 × TAE buffer for electrophoresis (2 V/cm, 16 h). Gels were stained in 1 × TAE buffer supplemented with 1 μg/ml ethidium bromide and visualized under UV illumination.
Crystallization and structure determination
Full methods for crystallization and structure determination are given in Supplementary Data. Briefly, AhQnr was crystallized by hanging-drop vapour diffusion from 0.1 M sodium cacodylate pH 6.8, 1.5 M sodium acetate, 20 mM DTT, yielding crystals of orthorhombic symmetry that diffracted to 2.2 Å resolution at beamline 10.1 of the SRS, Daresbury, UK. As this single crystal could not be reproduced we re-screened for additional crystal forms using protein modified by reductive methylation of surface lysine residues (43). Crystals diffracting to 2.0 Å resolution at beamline I03 of the Diamond Light Source grew from 0.1 M bis–tris propane pH 7.5, 0.2 M potassium thiocyanate, 16–20% PEG 3350, 8–16% glycerol, 40 mM DTT, 20 mM spermine–HCl. These were reproduced for selenomethionine (SeMet)-substituted protein and a 9-fold redundant data set to 3.0 Å resolution collected at the Se absorption edge (maximal f″) at Diamond beamline I02. The structure was solved by a combination of single-wavelength anomalous dispersion (SAD) and molecular replacement as described in Supplementary Data, with subsequent phase extension and refinement against the native data set. The refined structure was then used as a search model to solve the structure of the orthorhombic crystal form by molecular replacement. Data collection and refinement statistics are given in Table 1.
Table 1.
Data collection and refinement statistics
| Native | SeMet-SAD | Orthorhombic | |
|---|---|---|---|
| Data collection | |||
| X-ray source | DIAMOND I03 | DIAMOND I02 | SRS 10.1 |
| Wavelength (Å) | 0.97630 | 0.97920 | 1.11700 |
| Space group | P21 | P21 | P21212 |
| Cell dimensions | |||
| a, b, c (Å) | 52.23, 57.71, 85.12 | 52.45, 57.67, 86.22 | 88.73, 116.2, 55.10 |
| α, β, γ (°) | 90, 106.96, 90 | 90, 107.25, 90 | 90, 90, 90 |
| Resolution (Å) | 49.96–2.00 | 49.86–3.00 | 43.43–2.20 |
| (2.11–2.00) | (3.16–3.00) | (2.32–2.20) | |
| Rmerge | 0.101 (0.562) | 0.052 (0.086) | 0.132 (0.424) |
| No. reflections (total) | 237 955 (34 930) | 178 653 (26 231) | 127 598 (17 522) |
| No. reflections (unique) | 32 765 (4744) | 10 020 (1442) | 28 777 (4050) |
| I/σI | 12.8 (3.5) | 47 (29.4) | 8 (3.1) |
| Completeness (%) | 99.9 (99.9) | 99.8 (100) | 98.1 (98.6) |
| Redundancy | 7.3 (7.4) | 9.1 (9.2) | 4.4 (4.3) |
| Refinement | |||
| Resolution (Å) | 2.0 | 2.2 | |
| No. reflections | 32 807 | 29 576 | |
| Rwork/Rfree | 17.87/23.28 | 21.08/26.99 | |
| No. atoms | |||
| Protein | 3422 | 3409 | |
| Ligand/ion | 0 | 91 | |
| Water | 302 | 249 | |
| B-factors | |||
| Protein | 34.785 | 19.689 | |
| Ligand/ion | 38.934 | ||
| Water | 37.651 | 26.411 | |
| R.m.s.d. | |||
| Bond lengths (Å) | 0.0095 | 0.0118 | |
| Bond angles (°) | 1.2112 | 1.3641 |
Figures in parentheses relate to data collected in the highest resolution shell. Note that for the SeMet SAD data sets Bijvoet pairs were not merged.
Data deposition
Co-ordinates and structure factors for the structures of AhQnr in the monoclinic and orthorhombic crystal forms have been deposited with the Protein Data Bank (www.rcsb.org/pdb) with accession codes 3PSS and 3PSZ for release upon publication. The co-ordinates of the modeled complex of AhQnr and the E. coli GyrA 59 kDa and GyrB TOPRIM fragments are available for download from http://www.bristol.ac.uk/cellmolmed/staff/spencer.html.
Production and purification of AhQnr deletion mutants
Full details are given in Supplementary Data. AhQnr deletion mutants were generated in a variant of the recombinant protein, bearing a C-terminal hexahistidine tag and expressed in the pET-26b vector, using the Quikchange II directed mutagenesis kit (Agilent). Mutant proteins were expressed as above (AhQnr-6His and Δloop2) or in E. coli ArcticExpress (DE3) (Agilent; Δloop1 and Δdouble) and purified by nickel-affinity and size exclusion chromatography.
Modelling of the AhQnr:DNA gyrase complex
A partial model of E. coli DNA gyrase was constructed by combining the crystal structure of the E. coli GyrA 59 kDa fragment [PDB accession 1AB4; (44)] and an homology model of the E. coli TOPRIM domain. The latter was generated by fitting the E. coli TOPRIM sequence (GyrB residues 406–453, 622–638 and 728–799; i.e. removing the insertion region) onto the equivalent portion of Streptococcus pneumoniae DNA topisomerase IV [(5); PDB accession 3K9F chain C] using Swiss-PdbViewer (45) and SwissModel [(46); http://swissmodel.expasy.org/]. The relative orientation of the E. coli GyrA and TOPRIM fragments in the complex was obtained by superposition onto their counterparts in S. pneumoniae topoisomerase IV using PyMOL. AhQnr (monoclinic crystal form) was then manually docked into the G-segment binding groove of the resulting structure. Of the available type IIA DNA topoisomerase crystal structures, that of S. pneumoniae topoisomerase IV (5) was chosen as the most appropriate template as this both includes DNA (ensuring that the individual protein domains are appropriately oriented to accommodate the AhQnr structure) and is assembled from individual subunits rather than a single fused polypeptide.
The modelled complex was then edited by removal of the methyl groups from methylated lysines in the AhQnr structure, automatic addition of missing heavy atoms and manual rotation, along the Cα-Cβ bonds, of the side chains of clashing residues, using Swiss-PdbViewer. The resulting structure was then prepared for minimization using the tLEaP module of the AMBER 10 software package (47) by neutralizing with Na+ ions and solvating in a truncated octahedron of TIP3P water molecules (48) with a minimal thickness of 5 Å around the proteins. The resulting system of 137 856 atoms was minimized using the SANDER module of AMBER 10 and the 03 force field described by Duan et al. (49) for 1000 steps (500 steepest decent and 500 conjugate gradient). The AMBER calculations were carried out at the Center for Macromolecular Modelling and Materials Design at California State Polytechnic University, Pomona, CA, USA.
RESULTS AND DISCUSSION
AhQnr, the Qnr family member from Aeromonas hydrophila ATCC 7966
Our initial investigations of Qnr proteins of clinical origin, were hampered by the poor solubility and tendency to aggregate of the recombinant protein (QnrB2) or difficulties with expression (QnrS1). Accordingly, we sought to identify a soluble Qnr homologue from a Gram-negative organism that was more amenable to structural and biophysical characterization. Bioinformatics methods [Supplementary Figure S3; (50)] identified an orf from Aeromonas hydrophila ATCC 7966 (AhQnr) as a Gram-negative Qnr family member of likely high solubility, while sequence alignment and phylogenetic analysis (Supplementary Figures S1 and S2) confirmed this protein to be a member of the Qnr superfamily that is distinct from orfs found in Gram-positive organisms.
AhQnr cloned in the pET-26b T7 expression vector was readily over-expressed in E. coli BL21 and related strains and purified to homogeneity by ion-exchange, heparin affinity and size exclusion chromatography. Far-UV CD spectroscopy (Figure 1A) showed the purified protein to adopt the predicted β-conformation and yields a spectrum resembling that previously reported for the pentapeptide repeat protein Rfr32 (30). The results of analytical ultracentrifugation (AUC) experiments (Figure 1B), performed at a range of spin speeds and protein concentrations, were well described by a dimer-only model (MW 46 200 Da) as evidenced by the random deviation of the residual plot (Figure 1B; inset), and in excellent agreement with the molecular weight of 48 473 Da calculated from the amino acid sequence. We found no evidence of dissociation to monomers across the entire concentration range of our experiments (minimum concentration of 4.5 µM as estimated from the y-axis intercept on Figure 1B). Importantly, basal levels of AhQnr expression (i.e. in the absence of inducer) conferred an increase in minimal inhibitory concentration (MIC) for various quinolone antibiotics of 2- to 4-fold on host E. coli strains, consistent with levels of protection previously reported when other Qnr family members from environmental bacteria are over-expressed in E. coli [Supplementary Table S2; (23,51,52)] and indicative of an ability to protect E. coli type IIA DNA topoisomerases from quinolone action. In vitro assays of the supercoiling activity of E. coli DNA gyrase confirmed this. In line with previous reports of the activity of other Qnr proteins (34,36–38), but distinct from the activity of the Gram-positive Qnr homologue MfpA (36), micromolar concentrations of AhQnr relieve supercoiling inhibition by the fluoroquinolone ciprofloxacin (Figure 1C).
Figure 1.
AhQnr is a β-structured dimeric protein that protects DNA gyrase from inhibition by fluoroquinlones. (A) Far-UV CD spectrum of purified recombinant AhQnr (21.1 µM). (B) Analytical ultracentrifugation (14 000 rpm) of AhQnr (5.3 µM). Open circles are raw data, solid line shows best fit to a dimer-only model of MW 46 200 kDa. Inset shows deviation of experimental data from fit (residuals). (C) AhQnr rescues DNA gyrase (2 U) from inhibition by the fluoroquinolone ciprofloxacin (6 µM). Assay of ATP-dependent supercoiling activity of E. coli DNA gyrase. Lanes: N, negative control (relaxed pBR322 only; 500 ng); P, positive control (relaxed pBR322 plus DNA gyrase); 0, relaxed DNA plus DNA gyrase plus ciprofloxacin (6 µM); 0.175–1.4; as zero plus AhQnr at given concentrations (µM). Positions of migration for relaxed forms and supercoiled pBR322 DNA are labeled as R and S, respectively.
Structure of AhQnr
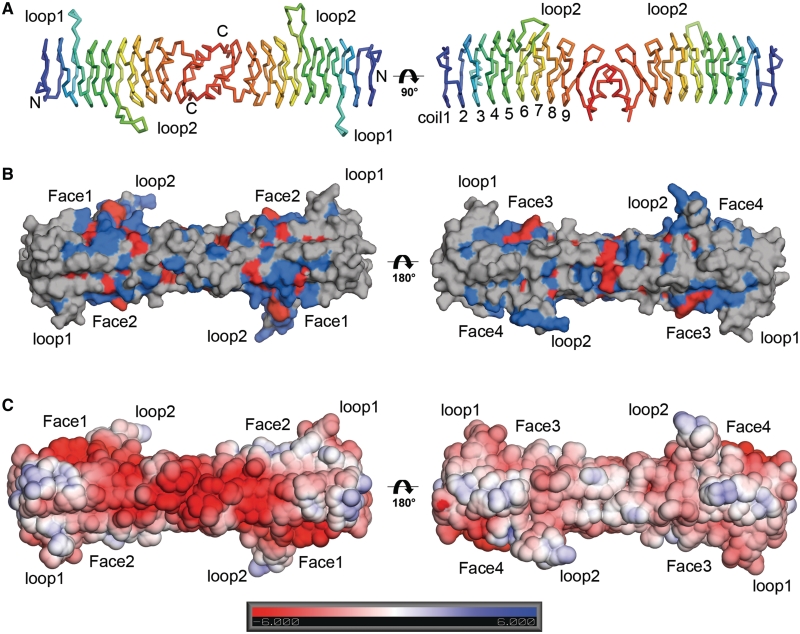
In an effort to establish a structural basis for the distinctive activities of AhQnr, and of other Qnr family members from Gram-negative bacteria, we determined structures of AhQnr in two crystal forms to a maximal resolution of 2.0 Å. MolProbity (53) analysis of the final refined structures reveals both to be of excellent stereochemical quality. To our knowledge, these are the first crystal structures for any Qnr family member from a Gram-negative bacterium. AhQnr (Figure 2) folds into the right-handed β-helix structure characteristic of PRPs (54), in which each face of the quadrilateral β-helix is formed by 5 amino acids, and 20 amino acids make up one complete coil (Supplementary Figure S4). Nine complete coils are present in the AhQnr structure (Figure 2A). The AhQnr dimer is formed by interactions between the C-termini of each monomer involving a β-strand (residues 195–199)–α-helix–β-strand (residues 211–215) dimerization motif. PISA (55) analysis of the dimer interface reveals a buried surface area of 1019.2 Å2 per monomer and a total of 13 hydrogen bonds involved in interdomain interactions. Notably, strand-exchange between the final strand of the β-helix structure (residues 195–199) and the C-terminal β-strand (residues 211–215) of the opposing subunit makes a prominent contribution to the dimer interface. Although the AhQnr dimer forms an essentially straight rod-like structure, the helical axis of each individual monomer bends towards its C-terminal end. Two structural features contribute to this distortion. First, between residues 167 and 171 the sequence of β-strands is interrupted by a short α-helix, opening up the β-helix structure between coils 7 and 8, and 8 and 9. Second, at this point the side chain of Trp-168 penetrates the interior of the β-helix, further displacing coil 9 away from coil 8.
Figure 2.
Crystal structure of AhQnr. (A) Ribbon diagram of AhQnr dimer (monoclinic crystal form). Individual monomers are colour-ramped from N- (blue) to C- (red) termini. Individual coils are numbered 1–9 from N- to C-terminus (right hand panel; A chain). (B) Rendered surface of AhQnr structure showing pattern of sequence conservation across Qnr family members from Gram-negative bacteria derived from the sequence alignment shown in Supplementary Figure S1. Faces are labelled as defined from the pentapeptide repeat sequence as shown in Supplementary Figure S4. Residues coloured red are completely conserved, those coloured blue show over 80% similarity. (C) Rendered surface of AhQnr structure showing electrostatic surface potential as calculated using APBS (60). This figure, and Figure 3 were generated using PyMOL (www.pymol.org).
Significantly, the AhQnr β-helix structure is also interrupted by two loop insertions: a smaller loop (loop1; residues 49–55) in the middle of coil 3, and a larger one (loop2; residues 105–115) between coils 5 and 6. While the conformation of loop1 is essentially fixed, that of loop2 differs in the two (independently refined) subunits of the AhQnr dimer and between the structures determined in the two crystal forms (Supplementary Figure S4). In both the monoclinic and orthorhombic structures, loop2 is also involved in crystallographic contacts between adjacent dimers in the crystal. Taken together, these results indicate that loop2 is likely to be conformationally flexible in solution.
To look for additional functional insights, we analysed the pattern of amino acid conservation and the distribution of surface charge in the AhQnr crystal structure. Mapping conserved amino acids [identified from an alignment of 61 Qnr family members from Gram negative bacteria (Supplementary Figure S1)] onto the surface of the AhQnr structure (Figure 2B) shows that, while sequence conservation is more evenly distributed than charge across all faces of the protein, face 1 contains a cluster of conserved amino acids in the central portion of the β-helix structure. Furthermore, loop2 is largely composed of conserved residues, including positions (Ile-107, Phe-113 and Cys-114) that are invariant across all 61 proteins in the alignment, whereas the composition of loop1 is more variable. Electrostatic surface calculations indicate that AhQnr is a strongly negatively charged protein, but that this charge is not uniformly distributed. Specifically, negative charge is localized in a distinctive ‘stripe’ across one surface of the protein (formed by faces 1 and 2; Figure 2C) whereas the opposite side is comparatively less strongly charged.
Comparison of Qnr family structures
Comparison of the AhQnr structure with those of Qnr family members from Gram-positive bacteria: M. tuberculosis MfpA [PDB accession 2BM4; (27)] and E. faecalis EfsQnr [PDB accession 2W7Z; (28,29)], reveals several features that distinguish AhQnr from these relatives (Figure 3). AhQnr contains nine complete PRP coils compared to MfpA (eight) or EfsQnr (eight complete coils and an incomplete ninth). The structures differ at their N-termini: the first (N-terminal) coil of the AhQnr structure is interrupted by a short α-helix (residues 13–18), whereas the MfpA N-terminus is part of the PRP structure. The EfsQnr N-terminus packs onto the exterior of the structure before crossing the centre of the quadrilateral β-helix and forming an incomplete first coil. The degree and nature of distortion along the β-helix axis also differs between the three structures. The AhQnr β-helix bends between coils 7 and 9 (see above); whereas in MfpA Pro-81 introduces a distortion between coils 4 and 5. The first seven coils of EfsQnr form a straight rod that is disrupted by insertion of the side chain of Trp-149 into the β-helix interior, bending the helical axis at this point and moving coil 8 away from coil 7. Trp-149 does not however occupy an equivalent position to AhQnr Trp-168 (see above). The structures also differ in the relative orientations of the two monomers in the overall dimeric structure. While the AhQnr dimer forms a straight rod, both the MfpA and EfsQnr structures bend about the dimer interface, inducing a degree of curvature to the overall structure that is most pronounced for EfsQnr. Comparison of the electrostatic surface potentials of the three proteins (Figure 3D) shows that, while all are negatively charged, AhQnr both carries the strongest charge and has the most localized charge distribution.
Figure 3.
Structures of Qnr family members. (A) AhQnr (chain A, blue; chain B, green); (B) M. tuberculosis MfpA [(27); PDB accession 2BM4; chain A, cyan; chain B, magenta]; (C) E. faecalis EfsQnr [(28); PDB accession 2W7Z; chain A, orange; chain B, red]. Residues introducing bends into the PRP structure (AhQnr Trp-168; MfpA Pro-81; EfsQnr Trp-149) are shown as sticks. (D) Electrostatic surface potentials (as calculated using APBS) mapped onto rendered surfaces for AhQnr (left), MfpA (centre) and EfsQnr (right).
Mutation of AhQnr loops
The most striking difference between the AhQnr structure and those of MfpA and EfsQr remains the presence of the two insertion sequences, loops 1 and 2 (above), in the AhQnr PRP β-helix structure. While similar loops are present in some other PRPs (31–32), this feature has not been previously observed in structures of Qnr family members determined to date [i.e. MfpA (27) and EfsQnr (28,29)]. Further, the high degree of sequence conservation in the region of loop2 leads us to hypothesize that this loop in particular is likely to represent a common feature of Qnr family members from Gram-negative bacteria. To investigate whether AhQnr loops 1 and 2 are of functional importance, we therefore constructed deletion mutants in which either (Δloop1, deletion of residues 49–54; Δloop2, deletion of residues 105–114) or both (Δdouble) loops were removed, and tested their ability to protect the supercoiling activity of DNA gyrase from ciprofloxacin inhibition. In order to facilitate purification of poorly expressed mutant proteins these were constructed in a variant of recombinant AhQnr (AhQnr-6His) bearing a C-terminal hexahistidine tag. DNA gyrase supercoiling protection assays confirmed that, in line with previous reports for other recombinant Qnr proteins (35–37), incorporation of this tag had no effect upon AhQnr activity (compare Figures 1C and 4A). Importantly, CD spectroscopy indicates that all three mutants retained wild-type like secondary structure (Supplementary Figure S5A) and size exclusion chromatography confirms that all form dimers in solution (Supplementary Figures S5B and S5C).
Figure 4.
Deletion of surface loops abrogates DNA gyrase protection from ciprofloxacin by AhQnr. ATP-dependent supercoiling activity of DNA gyrase (2 U) in the presence of ciprofloxacin (6 µM), AhQnr and mutants. (A) AhQnr-6His, (B) Δloop1, (C) Δloop2, (D): Δdouble. Lane Q: pBR322 DNA plus AhQnr only (4 µM). Other lanes are labelled as above (Figure 1C).
The results of supercoiling protection assays are shown in Figure 4. For the wild-type His-tagged protein (AhQnr-6His), inhibition of DNA gyrase supercoiling activity by ciprofloxacin (6 µM) is significantly relieved at submicromolar AhQnr concentrations (Figure 4A). In the case of the Δloop1 mutant, micro-, rather than submicro-molar, AhQnr concentrations are required to achieve equivalent levels of protection, but protection activity is nonetheless clearly evident (Figure 4B). In contrast, for both the Δloop2 (Figure 4C) and Δdouble (Figure 4D) mutants, no protective activity is observed in this AhQnr concentration range (up to 5 µM AhQnr). These data demonstrate that, while both loops 1 and 2 participate in protection of DNA gyrase activity from inhibition by quinolones, loop2 makes the major contribution to this process. None of the mutants showed detectable inhibition of supercoiling activity in the absence of ciprofloxacin (Supplementary Figure S6) at AhQnr concentrations of up to 40 µM. Although few studies to date have investigated the effects of introduced mutations on the activity of Qnr proteins (and none to our knowledge on protection of DNA gyrase from quinolones in an in vitro supercoiling assay), these results are in accord with a previous investigation of QnrA1, QnrB1 and QnrS1, where quinolone MICs were reduced by mutations in the highly conserved FCSA motif within loop2 (AhQnr numbering from Phe-113) (56). Taken together, these results suggest that the conserved loop2 may play a common role in the protection of DNA topoisomerases from quinolone action by Qnr proteins of Gram-negative bacteria.
A model for the interaction of AhQnr with DNA gyrase
To further investigate the possible roles of loops 1 and 2 in the interaction of AhQnr with DNA gyrase, we constructed a model of the AhQnr:DNA gyrase interaction using the monoclinic AhQnr structure and the E. coli gyrase A 59 kDa and gyrase B TOPRIM domain fragments (respectively, 82 and 84% identical in sequence to their counterparts in the A. hydrophila enzyme) as described. The results are shown in Figure 5. Similar to previously published observations with MfpA (27), AhQnr may be readily placed into the gyrase structure such that the negative charge on faces 1 and 2 provides electrostatic complementarity with the positively charged ‘saddle’ of GyrA. However, this orientation also enables both loops 1 and 2 to make additional contacts with the GyrAB complex on opposite sides of the central DNA-binding cleft. First, loop1 is positioned to contact the GyrB TOPRIM domain on the exterior face of the molecule (i.e. outside and perpendicular to the DNA-binding groove) via the α-helices (residues 771–781 and 785–794, E. coli numbering) at its C-terminus. Second, loop2 contacts the surface of the GyrA ‘tower’ domain, formed by the α-helices 272–284 and 313–323 (E. coli numbering), that extends above the central groove. Notably, the spacing of the two loops along the long axis of the AhQnr dimer is such that loops 1 and 2 of each monomer are positioned to make equivalent contacts with the GyrAB complex, with the spacing of loop1 matching that of the exterior surfaces of the two GyrB TOPRIM domains. This orientation also positions the majority of conserved residues on the AhQnr surface (Figure 2B above) in direct contact with gyrase (Figure 5, right-hand panel). Significantly, the surfaces of the GyrA tower and GyrB TOPRIM domains, that our model suggests may be involved in interactions with AhQnr, are also highly conserved across DNA gyrases from Gram-negative bacteria. Together, these observations indicate that the mode of binding that we postulate here may be generally applicable to interactions between DNA gyrase enzymes and Qnr proteins in Gram-negative bacteria.
Figure 5.
Model for interaction of AhQnr with DNA Gyrase. Figure shows minimized model of the complex of AhQnr with DNA gyrase A (59 kDa fragment) plus gyrase B TOPRIM domain. GyrA is coloured blue except for winged helix (residues 56–136, cyan) and tower (residues 236–327, green) domains. GyrB TOPRIM domain is coloured magenta. AhQnr is coloured according to sequence conservation of Qnr family members from Gram-negative bacteria as described (Supplementary Figure S1). Left hand and centre panels: views of complex in two orientations at 90° separation. Right-hand panel: view as centre with one GyrA and one GyrB fragments removed showing involvement of conserved AhQnr residues in proposed interaction.
Concluding remarks
The data reported here represent the first structural characterization for a Qnr protein from a Gram-negative bacterium, and the most comprehensive characterization to date of a Qnr protein from an environmental organism, as opposed to a clinical isolate. We show that, consistent with previous results from other aquatic organisms such as Vibrio, Shewanella and Stenotrophomonas spp. (22–26), A. hydrophila contains a chromosomal pentapeptide repeat protein (AhQnr) that in its recombinant form both increases the quinolone MIC of host E. coli and protects E. coli DNA gyrase from quinolone inhibition in in vitro assays of supercoiling activity. These data show AhQnr to be a novel member of the Qnr protein family and indicate that, as has previously been the case for organisms such as Shewanella spp. (22), Aeromonas spp. represent an additional potential source of new Qnr resistance determinants that may in the future be exploited by more clinically important organisms. Thus our study provides further evidence that environmental, particularly aquatic, bacteria contain functional resistance genes with the potential to impact on the effectiveness of clinically important antibiotics (57,58).
The AhQnr crystal structure reveals a number of features that appear common to the various Qnr proteins of Gram-negative origin. While AhQnr has the expected pentapeptide repeat fold and forms dimers in solution and in the crystalline state, it is distinguished from known structures of other Qnr family members by virtue of the two extended loops, loop1 and loop2, that project from the pentapeptide repeat β-helix structure. Our mutagenesis experiments show that, while both loops make significant contributions to the quinolone-protection activity of AhQnr, the larger of the two, loop2, is essential to this function. Multiple sequence alignments indicate that both loops are likely to be present, and that the sequence of loop2 is well conserved, in Qnr family members from other Gram-negative bacteria. Docking experiments suggest that both loops are well positioned to interact with both the GyrA and GyrB subunits of DNA gyrase. We propose this to be a structural and functional motif, important to the DNA gyrase:Qnr interaction, that is common to Qnr proteins from Gram-negative bacteria of both environmental and clinical origin.
One of the most intriguing aspects of Qnr biochemistry is the difference between the reported activities of characterized family members from Gram-positive (M. tuberculosis MfpA) and Gram-negative (E. cloacae QnrB4) organisms. Specifically, MfpA is a sub-micromolar DNA gyrase inhibitor that does not affect the interaction with quinolones, whereas QnrB4 is a weak DNA gyrase inhibitor that protects the enzyme from quinolone inhibition (36). Our data show that in this respect AhQnr is similar to QnrB4, suggesting that the combination of weak DNA gyrase inhibition with quinolone protection activity may be common to Qnr proteins of Gram-negative origin. While we cannot provide a definitive explanation for the differing properties of the various Qnr family members, our model of the AhQnr:DNA gyrase interaction has features pertinent to this issue. In comparison with MfpA, AhQnr (and by implication other Qnr proteins from Gram-negative bacteria) possesses additional structural features, loops1 and 2, that make more extensive interactions with DNA gyrase that in part involve regions outside the central DNA-binding groove. In the case of loop1, these interactions involve the GyrB (TOPRIM domain), rather than the GyrA, subunit of DNA gyrase. Available crystal structures for the DNA gyrase DNA binding and cleavage core (59) indicate that the GyrB TOPRIM domain can adopt multiple orientations in the context of the GyrAB heterotetramer. Thus, binding of Qnr proteins may be sensitive to DNA gyrase conformation; i.e. a specific orientation of the GyrA and GyrB subunits relative to one another may be required for high-affinity Qnr binding. The implication is that the appropriate conformation may be stabilized in quinolone-containing complexes, enabling Qnr proteins to disrupt the quinolone:DNA gyrase interaction, but is less favoured in their absence, resulting in only weak inhibition of DNA gyrase activity by Qnr proteins when quinolones are absent. We propose that the interactions, involving loops 1 and 2, that we have here identified represent a possible mechanism by which this discrimination between DNA gyrase conformations may be achieved.
Note added in proof
While this manuscript was in revision, a crystal structure of K. pneumoniae QnrB1 was deposited in the PDB (accession 2XTW). The structure confirms the presence of loops 1 and 2 in a Qnr protein of clinical origin.
ACCESSION NUMBERS
PDB Accession nos 3PSS, 3PSZ.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
British Society for Antimicrobial Chemotherapy (GA741 to J.S.); UK Overseas Research Students Awards Scheme (to X.X.); University of Bristol, award of a postgraduate scholarship (to X.X.); Human Frontiers Science Program (RGP0031/2007 to E.H.C.B. and D.N.W.); Research Corporation for Science Advancement, Cottrell College Science Award (ID: 10493 to P.O.); UK Biotechnology and Biological Sciences Research Council [grant nos 719/B15474 and 719/REI20571 to UK Northwest Structural Genomics Centre (NWSGC) MAD10 beamline at SRS (UK)]; North West Development Agency project award (N0002170). Funding for open access charge: British Society for Antimicrobial Chemotherapy; UK ORSAS; the University of Bristol; the Human Frontiers Science Program.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff of the synchrotron beamlines used in this study for invaluable assistance with X-ray data collection; Dr Antonio Corriea for the gift of the Aeromonas hydrophila strain, Dr Andrea Hadfield for crystallographic discussions; and Dr Paul Race and Dr Darryl Hill and Prof. Anthony Maxwell, for their critical reading of the article.
REFERENCES
- 1.Hooper DC. Clinical applications of quinolones. Biochim. Biophys. Acta. 1998;1400:45–61. doi: 10.1016/s0167-4781(98)00127-4. [DOI] [PubMed] [Google Scholar]
- 2.Hooper DC. In: Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. 6 edn. Mandell GL, Douglas RG, Bennett JE, Dolin R, editors. New York: Elsevier/Churchill Livingstone; 2005. pp. 451–467. [Google Scholar]
- 3.Rustomjee R, Lienhardt C, Kanyok T, Davies GR, Levin J, Mthiyane T, Reddy C, Sturm AW, Sirgel FA, Allen J, et al. A Phase II study of the sterilising activities of ofloxacin, gatifloxacin and moxifloxacin in pulmonary tuberculosis. Int. J. Tuberc. Lung Dis. 2008;12:128–138. [PubMed] [Google Scholar]
- 4.Maxwell A, Costenaro L, Mitelheiser S, Bates AD. Coupling ATP hydrolysis to DNA strand passage in type IIA DNA topoisomerases. Biochem. Soc. Trans. 2005;33:1460–1464. doi: 10.1042/BST0331460. [DOI] [PubMed] [Google Scholar]
- 5.Laponogov I, Sohi MK, Veselkov DA, Pan X-S, Sawhney R, Thompson AW, McAuley KE, Fisher LM, Sanderson MR. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 2009;16:667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 6.Drlica K, Malik M, Kerns RJ, Zhao X. Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 2008;52:385–392. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J. Antimicrob. Chemother. 2003;51:1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 8.Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 2006;19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 2009;22:664–689. doi: 10.1128/CMR.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordmann P, Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 2005;56:463–469. doi: 10.1093/jac/dki245. [DOI] [PubMed] [Google Scholar]
- 11.Martínez-Martínez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid. Lancet. 1998;351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 12.Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, Bush K, Hooper DC. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 2006;12:83–88. doi: 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- 13.Yamane K, Wachino J-i, Suzuki S, Kimura K, Shibata N, Kato H, Shibayama K, Konda T, Arakawa Y. New plasmid-mediated fluoroquinolone Efflux Pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 2007;51:3354–3360. doi: 10.1128/AAC.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim HB, Wang M, Park CH, Kim EC, Jacoby GA, Hooper DC. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob. Agents Chemother. 2009;53:3582–3584. doi: 10.1128/AAC.01574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen LH, Johannesen E, Burmolle M, Sorensen AH, Sorensen SJ. Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli. Antimicrob. Agents Chemother. 2004;48:3332–3337. doi: 10.1128/AAC.48.9.3332-3337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6:629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 17.Wang M, Guo Q, Xu X, Wang X, Ye X, Wu S, Hooper DC, Wang M. New plasmid-mediated quinolone resistance gene, qnrC, found in a clinical isolate of Proteus mirabilis. Antimicrob. Agents Chemother. 2009;53:1892–1897. doi: 10.1128/AAC.01400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavaco LM, Hasman H, Xia S, Aarestrup FM. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob. Agents Chemother. 2009;53:603–608. doi: 10.1128/AAC.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacoby G, Cattoir V, Hooper D, Martinez-Martinez L, Nordmann P, Pascual A, Poirel L, Wang M. qnr Gene nomenclature. Antimicrob. Agents Chemother. 2008;52:2297–2299. doi: 10.1128/AAC.00147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodríguez-Martínez JM, Velasco C, Briales A, Garcia I, Conejo MC, Pascual A. Qnr-like pentapeptide repeat proteins in gram-positive bacteria. J. Antimicrob. Chemother. 2008;61:1240–1243. doi: 10.1093/jac/dkn115. [DOI] [PubMed] [Google Scholar]
- 21.Montero C, Mateu G, Rodriguez R, Takiff H. Intrinsic resistance of Mycobacterium smegmatis to fluoroquinolones may be influenced by new pentapeptide protein MfpA. Antimicrob. Agents Chemother. 2001;45:3387–3392. doi: 10.1128/AAC.45.12.3387-3392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel L, Rodríguez-Martínez JM, Mammeri H, Liard A, Nordmann P. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob. Agents Chemother. 2005;49:3523–3525. doi: 10.1128/AAC.49.8.3523-3525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sánchez MB, Hernández A, Rodríguez-Martínez JM, Martínez-Martínez L, Martínez JL. Predictive analysis of transmissible quinolone resistance indicates Stenotrophomonas maltophilia as a potential source of a novel family of Qnr determinants. BMC Microbiol. 2008;8:148. doi: 10.1186/1471-2180-8-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu K, Kikuchi K, Sasaki T, Takahashi N, Ohtsuka M, Ono Y, Hiramatsu K. Smqnr, a new chromosome-carried quinolone resistance gene in stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2008;52:3823–3825. doi: 10.1128/AAC.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cattoir V, Poirel L, Mazel D, Soussy CJ, Nordmann P. Vibrio splendidus as the source of plasmid-mediated QnrS-like quinolone resistance determinants. Antimicrob. Agents Chemother. 2007;51:2650–2651. doi: 10.1128/AAC.00070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poirel L, Liard A, Rodriguez-Martinez J-M, Nordmann P. Vibrionaceae as a possible source of Qnr-like quinolone resistance determinants. J. Antimicrob. Chemother. 2005;56:1118–1121. doi: 10.1093/jac/dki371. [DOI] [PubMed] [Google Scholar]
- 27.Hegde SS, Vetting MW, Roderick SL, Mitchenall LA, Maxwell A, Takiff HE, Blanchard JS. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science. 2005;308:1480–1483. doi: 10.1126/science.1110699. [DOI] [PubMed] [Google Scholar]
- 28.Vetting MW, Hegde SS, Blanchard JS. Crystallization of a pentapeptide-repeat protein by reductive cyclic pentylation of free amines with glutaraldehyde. Acta Crystallogr. D Biol. Crystallogr. 2009;65:462–469. doi: 10.1107/S0907444909008324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegde SS, Vetting MW, Mitchenall LA, Maxwell A, Blanchard JS. Structural and biochemical analysis of the Pentapeptide Repeat Protein, EfsQnr, a potent DNA gyrase inhibitor. Antimicrob. Agents Chemother. 2011 doi: 10.1128/AAC.01158-10. doi:10.1128/AAC.01158-10, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buchko GW, Ni S, Robinson H, Welsh EA, Pakrasi HB, Kennedy MA. Characterization of two potentially universal turn motifs that shape the repeated five-residues fold–crystal structure of a lumenal pentapeptide repeat protein from Cyanothece 51142. Protein Sci. 2006;15:2579–2595. doi: 10.1110/ps.062407506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buchko GW, Robinson H, Pakrasi HB, Kennedy MA. Insights into the structural variation between pentapeptide repeat proteins–crystal structure of Rfr23 from Cyanothece 51142. J. Struct. Biol. 2008;162:184–192. doi: 10.1016/j.jsb.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Ni S, Sheldrick GM, Benning MM, Kennedy MA. The 2A resolution crystal structure of HetL, a pentapeptide repeat protein involved in regulation of heterocyst differentiation in the cyanobacterium Nostoc sp. strain PCC 7120. J. Struct. Biol. 2009;165:47–52. doi: 10.1016/j.jsb.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 33.Vetting MW, Hegde SS, Hazleton KZ, Blanchard JS. Structural characterization of the fusion of two pentapeptide repeat proteins, Np275 and Np276, from Nostoc punctiforme: resurrection of an ancestral protein. Protein Sci. 2007;16:755–760. doi: 10.1110/ps.062637707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tran JH, Jacoby GA, Hooper DC. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 2005;49:118–125. doi: 10.1128/AAC.49.1.118-125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran JH, Jacoby GA, Hooper DC. Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob. Agents Chemother. 2005;49:3050–3052. doi: 10.1128/AAC.49.7.3050-3052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merens A, Matrat S, Aubry A, Lascols C, Jarlier V, Soussy C-J, Cavallo J-D, Cambau E. The pentapeptide repeat proteins MfpAMt and QnrB4 exhibit opposite effects on DNA gyrase catalytic reactions and on the ternary gyrase-DNA-quinolone complex. J. Bacteriol. 2009;191:1587–1594. doi: 10.1128/JB.01205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran JH, Jacoby GA. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl Acad. Sci. USA. 2002;99:5638–5642. doi: 10.1073/pnas.082092899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, Hooper DC. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 2006;50:1178–1182. doi: 10.1128/AAC.50.4.1178-1182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janda JM, Abbott SL. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010;23:35–73. doi: 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Demeler B. In: Modern Analytical Ultracentrifugation: Techniques and Methods. Scott DJ, Harding SE, Rowe AJ, editors. London: Royal Society of Chemistry (UK); 2005. pp. 210–229. [Google Scholar]
- 41.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding SE, Rowe AJ, Horton JC, editors. Cambridge, UK: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 42.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 43.Walter TS, Meier C, Assenberg R, Au K-F, Ren J, Verma A, Nettleship JE, Owens RJ, Stuart DI, Grimes JM. Lysine methylation as a routine rescue strategy for protein crystallization. Structure. 2006;14:1617–1622. doi: 10.1016/j.str.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morais Cabral JH, Jackson AP, Smith CV, Shikotra N, Maxwell A, Liddington RC. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 45.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 46.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 47.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, et al. San Francisco: AMBER10. University of California; 2008. [Google Scholar]
- 48.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 49.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 50.Wilkinson DL, Harrison RG. Predicting the solubility of recombinant proteins in Escherichia coli. Biotechnology. 1991;9:443–448. doi: 10.1038/nbt0591-443. [DOI] [PubMed] [Google Scholar]
- 51.Gordon NC, Wareham DW. Novel variants of the Smqnr family of quinolone resistance genes in clinical isolates of Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2010;65:483–489. doi: 10.1093/jac/dkp476. [DOI] [PubMed] [Google Scholar]
- 52.Saga T, Kaku M, Onodera Y, Yamachika S, Sato K, Takase H. Vibrio parahaemolyticus chromosomal qnr homologue VPA0095: demonstration by transformation with a mutated gene of its potential to reduce quinolone susceptibility in Escherichia coli. Antimicrob. Agents Chemother. 2005;49:2144–2145. doi: 10.1128/AAC.49.5.2144-2145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vetting MW, Hegde SS, Fajardo JE, Fiser A, Roderick SL, Takiff HE, Blanchard JS. Pentapeptide repeat proteins. Biochemistry. 2006;45:1–10. doi: 10.1021/bi052130w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 56.Rodríguez-Martínez JM, Briales A, Velasco C, Conejo MC, Martínez-Martínez L, Pascual A. Mutational analysis of quinolone resistance in the plasmid-encoded pentapeptide repeat proteins QnrA, QnrB and QnrS. J. Antimicrob. Chemother. 2009;63:1128–1134. doi: 10.1093/jac/dkp111. [DOI] [PubMed] [Google Scholar]
- 57.Canton R. Antibiotic resistance genes from the environment: a perspective through newly identified antibiotic resistance mechanisms in the clinical setting. Clin. Microbiol. Infect. 2009;15(Suppl. 1):20–25. doi: 10.1111/j.1469-0691.2008.02679.x. [DOI] [PubMed] [Google Scholar]
- 58.Martinez JL. Antibiotics and antibiotic resistance genes in natural environments. Science. 2008;321:365–367. doi: 10.1126/science.1159483. [DOI] [PubMed] [Google Scholar]
- 59.Schoeffler AJ, May AP, Berger JM. A domain insertion in Escherichia coli GyrB adopts a novel fold that plays a critical role in gyrase function. Nucleic Acids Res. 2010;38:7830–7844. doi: 10.1093/nar/gkq665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.