Abstract
Unlike bacteria, many eukaryotes initiate DNA replication from genomic sites that lack apparent sequence conservation. These loci are identified and bound by the origin recognition complex (ORC), and subsequently activated by a cascade of events that includes recruitment of an additional factor, Cdc6. Archaeal organisms generally possess one or more Orc1/Cdc6 homologs, belonging to the Initiator clade of ATPases associated with various cellular activities (AAA+) superfamily; however, these proteins recognize specific sequences within replication origins. Atomic resolution studies have shown that archaeal Orc1 proteins contact double-stranded DNA through an N-terminal AAA+ domain and a C-terminal winged-helix domain (WHD), but use remarkably few base-specific contacts. To investigate the biochemical effects of these associations, we mutated the DNA-interacting elements of the Orc1-1 and Orc1-3 paralogs from the archaeon Sulfolobus solfataricus, and tested their effect on origin binding and deformation. We find that the AAA+ domain has an unpredicted role in controlling the sequence selectivity of DNA binding, despite an absence of base-specific contacts to this region. Our results show that both the WHD and ATPase region influence origin recognition by Orc1/Cdc6, and suggest that not only DNA sequence, but also local DNA structure help define archaeal initiator binding sites.
INTRODUCTION
The decision to initiate DNA replication is a critical event in the life of a cell. Not only must replicative enzymes be properly assembled to permit the faithful copying of parental chromosomes, the onset of replisome assembly and activity must be matched precisely to the cell cycle to avoid changes in ploidy. In organisms with more than one replication start site (or ‘origin’) per chromosome, the replication of each genomic region with respect to its neighbors must also be controlled to prevent local alterations in DNA copy number that can lead to genetic instabilities (1).
Cells use many disparate approaches to regulate replication initiation. One particularly important line of control derives from the use of dedicated, ATP-dependent ‘initiators’, factors that both recognize replication origins and assist with replisome formation (2). Although the assembly states, accessory proteins and regulation of initiators can differ between and within the three domains of life (bacteria, archaea and eukaryotes), bioinformatic studies have indicated that one or more initiator proteins is always a member of the ATPases associated with various cellular activities (AAA+) superfamily (3). All AAA+ initiators serve two roles in the context of origin firing, functioning both as factors that identify and mark replication start sites, and as molecular switches that use ATP to promote activities such as initiator assembly and helicase deposition (2,4,5). Structural efforts have shown that in prokaryotes, the AAA+ module is augmented with a C-terminal, origin-binding domain consisting of either a NarL/FixJ-class helix-turn-helix (HTH) fold (in bacteria), or a winged helix domain (WHD) (in archaea) (6,7). Sequence predictions suggest the latter configuration is likely to extend to certain eukaryotic initiator subunits as well (6,8).
Given the degree of molecular conservation among cellular initiators, it is notable that replication origins are quite distinct between the major cellular lineages. Prokaryotes generally rely on sequence repeats, such as DnaA boxes (in bacteria) or origin recognition boxes (ORBs, in archaea), to guide their respective initiators to replication start sites (9,10). By contrast, metazoan origins do not have discernable sequence conservation (11); instead, sequence and chromosome context are thought to play a dominant role in initiator binding and function (12). Yeast appear to occupy a middle ground, possessing both relatively well-defined ARS sequences (13), but also allowing for contextual cues, such as nucleosome positioning, to regulate initiator binding (14,15).
A combination of structural and biochemical investigations has begun to reveal molecular mechanisms for origin recognition and activation. A structure of the HTH origin-binding domain of DnaA (the bacterial initiator) bound to a consensus DnaA box showed that a number of highly-conserved, signature amino acids in the protein make several base-specific contacts within the origin binding site (16). Two views of the full-length archaeal initiator, Orc1, bound to a target ORB site similarly show that the WHD constitutes a primary DNA-binding element of these factors (17,18). However, these origin recognition complex (Orc1)/DNA complex structures unexpectedly showed that archaeal initiators make very few base-specific contacts to target duplex sequences (Figure 1). Moreover, these studies revealed that, in their ADP-bound form, initiator ATPase regions directly engage the double stranded DNA via a characteristic initiator specific motif (ISM) that extends from the core AAA+ fold. The joint action of the Orc1 AAA+ region and the WHD appears to significantly deform and underwind the DNA target.
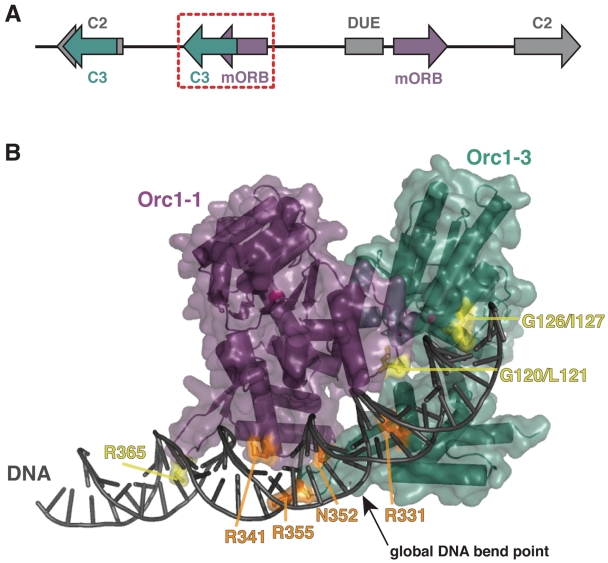
Figure 1.
Initiator-origin interactions. (A) Schematic of S. solfataricus oriC2. Orc1-1-bound mORB sites are shown in purple, Orc1-3-bound C3 sites are shown in teal, the Orc1-2-bound C2 sites are shown in gray. The AT-rich DUE element is depicted as a gray box while the overlapping mORBa/C3b sites are highlighted by a dashed-red box. (B) The four residues observed to make base-specific contacts in the Orc1-1/Orc1-3/origin DNA structure (PDB 2QBY and 17) are highlighted in orange on a cartoon of the complex. Residues that make non-specific interactions but which are proposed to be important to origin recognition, are colored yellow. Protein, purple and teal; DNA, gray; ADP, black sticks; magnesium ions, magenta spheres. Mutations of arginine and asparagine residues in the WHDs to alanines are designed to weaken DNA binding by disrupting hydrogen bond interactions, whereas mutations of glycine-leucine/isoleucine sequences within the AAA+ ISMs (to leucine–aspartate or aspartate–aspartate pairs) are intended to disturb DNA interactions through steric hinderance and charge repulsion with the DNA phosphate backbone.
Together, these findings raise a number of questions regarding the molecular mechanisms underlying origin identification and remodeling by archaeal Orc1 initiators. For example, the degree to which particular, conserved amino acids in the Orc1 WHD and AAA+ domains contribute to general DNA affinity versus ORB specificity is not known. How origin discrimination arises from a relatively small number of direct base contacts likewise is undefined. Other unresolved issues include whether the deformations seen in crystallographic models are static or plastic in their placement, or how different Orc1 regions collaborate in controlling DNA deformation. Finally, in light of the diversity of origin sequences throughout nature, and the clear evolutionary kinship between initiators, it is not evident whether origin recognition strategies used by prokaryotic initiators are extensible to their eukaryotic counterparts.
To begin to address these issues, we investigated the origin-binding properties of two initiator paralogs, Orc1-1 and Orc1-3, from the archaeon S. solfataricus. Biochemical data demonstrate that the WHD contributes to affinity and specificity, consistent with its relatively large interaction surface with origin DNA. Unexpectedly, we find that the AAA+ ISM element also influences specificity, along with the position of deformation sites within the binding locus, and possibly the binding stoichiometry of initiator-DNA complexes as well. Together, our observations implicate both origin DNA sequence and structure as Orc1 recognition determinants in archaeal DNA replication. Compared with prior studies of the yeast ORC, these findings further suggest that eukaryotic initiators may retain some vestige of these functional properties. Overall, this effort establishes archaea as a middle ground within a continuum of origin recognition strategies that ranges from hard-encoded, cis-acting DNA sequences to more context-based cues.
METHODS
Site-directed mutagenesis, expression and purification of Orc1 proteins
Point mutations were introduced into His6-Orc1-1 (residues 15-397) and His6-Orc1-3 (residues 14-394) wild-type constructs (17) using QuickChange mutagenesis (Stratagene), and sequenced at the UC Berkeley DNA Sequencing facility. All His6-Orc1 proteins were expressed and purified as ADP-bound proteins as previously described (17). All Orc1 mutants expressed at similar levels as the WT initiators, and eluted from a final size-exclusion column as sharp, symmetrical, monomer peaks, with retention times identical to WT Orc1-1 and Orc1-3 (Supplementary Figure S1).
DNA substrates for fluorescence anisotropy assays
Oligonucleotides of S. solfataricus origin DNA, including oligonucleotides with 5′-fluorescein tags, were synthesized by MWG Biotech and shipped as lyophilized pellets. Oligonucleotides were dissolved in annealing buffer (10 mM Tris pH 8, 1 mM MgCl2) and quantified by A260. For annealing, complementary oligonucleotide pairs were added together in equimolar amounts, heated to 95°C and slow-cooled to 4°C. Annealed oligonucleotide duplexes were then flash-frozen in liquid nitrogen and stored at −20°C.
Fluorescence anisotropy assays
Orc1-1 and Orc1-3 binding affinities for origin DNA sequences were determined by monitoring the change in fluorescence anisotropy of short, fluorescein-tagged dsDNA substrates over a range of initiator concentrations. Specific Orc1-1 binding was assayed using its cognate mORB sequence, 5′-GATTTTCAGATGAAACG. Specific Orc1-3 binding was assayed using the sequence for the C3 site it prefers, 5′-GAAACGTAGGAAATTTAC. Non-specific DNA binding of either protein was assayed using the randomized sequence, 5′-GCAATATCTATGAGATAC. For each of the DNA substrates, the complementary strand was fluorescein-labeled at the 5′-end. DNA oligonucleotides were first mixed with initiator at high salt concentration (Orc1 storage buffer; [50 mM HEPES pH 7.5, 750 mM sodium chloride, 10% (v/v) glycerol, 5 mM β-ME]), then rapidly diluted 10-fold to final assay conditions of 0–7 µM initiator, 17 nM DNA, 28 mM HEPES pH 7.5, 75 mM sodium chloride, 10% (v/v) glycerol, 3 mM DTT, 0.5 mM β-ME, 1 mM MgCl2 and 0.1 mg/ml BSA. Samples were incubated at 37°C for 30 min before transferring aliquots to 384-well, black, polystyrene plates (Molecular Devices). The plates were spun briefly to remove bubbles, incubated for 10 min at 37°C and read in a Victor3V (Perkin Elmer) multi-label plate reader at 37°C. Binding assays were performed in triplicate and the average anisotropy value plotted as a function of initiator concentration. Binding curves were fit to a simple, single-site binding equation using Kaleidagraph version 3.8 (Synergy Software):
where A is the anisotropy value, Amax is the anisotropy value at saturation, A0 is the baseline anisotropy value for oligo alone, [P]T is the total protein concentration and Kd is the apparent dissociation constant. A few binding curves were fit to a modified version of the binding equation that took into account an apparent second binding event (presumed to be due to weak protein–protein interactions):
where Kd1 and Kd2 refer to the strong and weak apparent dissociation constants, respectively. Only the higher affinity interaction is reported in Table 1.
Table 1.
Summary of Orc1 binding measurements
| Orc1 variant |
Kd, µM |
Percent change in specificity | |
|---|---|---|---|
| mORB 17-mer | Random 18-mer | ||
| Orc1-1 | |||
| WT | 0.39 | 4.7 | – |
| G120L/L121D (AAA+) | 2.2 | 7.0 | −74 |
| G120D/L121D (AAA+) | 0.60 | 3.8 | −48 |
| R341A (HTH) | 6.1 | 11.8 | −84 |
| R365A (Wing) | 4.6 | 17.3 | −69 |
| R341A/R365A (HTH/Wing) | 22.8 | 47.1 | −83 |
| C3 site 18-mer | Random 18-mer | ||
| Orc1-3 | |||
| WT | 0.027 | 7.7 | – |
| G126L/I127D (AAA+) | 0.070 | 18.4 | −5 |
| G126D/I127D (AAA+) | 0.088 | 98.9 | 305 |
| R331A (HTH) | 6.5 | 29.9 | −98 |
| N352A (Wing) | 2.4 | 15.0 | −97 |
| R355A (Wing) | 0.82 | 25.5 | −89 |
| R331A/R355A (HTH/Wing) | no binding | no binding | nd |
Radiolabeling of DNA substrates
T4 polynucleotide kinase (NEB) was used to radiolabel 100 ng of either top or bottom strand of an 70-mer oriC2 fragment (SIGMA Genosys) containing a mORB and a C3 site (17). Following annealing performed in the presence of a 2-fold molar excess of complementary oligonucleotide, the DNA probe was passed through a Sephadex G50 microspin column (GE Healthcare) and used for DNA footprinting.
Maxam-Gilbert AG sequencing
A mixture of 100 fmol of DNA substrate, 8 µg of sonicated salmon sperm DNA and 3 μl of 88% formic acid were incubated for 7 min at 37°C. Subsequently, 150 μl of a 10% piperidine solution were added and incubated at 90°C for 30 min. DNA precipitation was achieved by addition of 1.2 ml of butanol, followed by vortexing and spinning. The resulting pellet was resuspended in 150 μl of a 1% SDS solution, to which 1.2 ml of butanol was added, followed by vortexing and spinning. The pellet was dried and resuspended in 25 μl of TE buffer, and desalted by using a Sephadex G50 microspin column (GE Healthcare).
Copper phenanthroline footprinting
Binding reactions were performed in 20 µl volumes under the same conditions described above, except for the absence of glycerol, using a fixed concentration 3-fold greater than the Kd calculated by fluorescence anisotropy for each Orc1 mutant. This protocol was used to ensure near maximal occupancy of specific DNA binding sites, while minimizing non-specific binding to adjacent, non-cognate sequences. Following a 15-min incubation at 55°C, 1 µl of a freshly prepared mix of 1 mM 1,10 phenanthroline and 0.23 mM CuSO4 was added to the binding reaction followed by 1 µl of 58 mM 3 mercaptopropionic acid and incubation was continued for 3 min. Reactions were stopped by adding 1 µl of 28 mM Neocuproine (SIGMA) and an equal volume of urea-gel loading dye (8 M urea, 89 mM Tris, 28.5 mM Taurine, 0.5 mM EDTA, 0.5% bromophenol blue). Samples were subsequently heated at 95°C for 5 min prior to loading on a denaturing PAGE gel run in 1× TTE buffer (89 mM Tris, 28.5 mM Taurine, 0.5 mM EDTA) for two 2.5 h at 60 W constant power. Gels were scanned using a Fujifilm FLA-5000 phosphorimager and individual-lane profiles calculated using the Aida Image Analyzer software (Raytest). Band profiles were processed using Microsoft Excel, subtracting no-protein control lanes from each wild-type and mutant protein lane. Differences in digested-band intensities were represented on a heat map generated using Heatmap Builder (19).
RESULTS AND DISCUSSION
Orc1 affinity and specificity for origin DNA sequences
Sulfolobus solfataricus encodes three Orc1 paralogs, two of which (Orc1-1 and Orc1-3) can simultaneously bind to an overlapping region within the oriC2 origin. As illustrated in Figure 1A, Orc1-1 binds to the minimal ORB sequence, or mORB site, while Orc1-3 binds to a distinct origin sequence called the C3 site (10). To first understand the contribution of specific amino acids and domains in Orc1-1 and Orc1-3 toward origin recognition, we made mutations in both proteins that were designed to disrupt initiator-DNA interactions based on a crystal structure of the Orc1-1/Orc1-3/DNA ternary complex (Figure 1B) (17). Proteins were purified to homogeneity bound to ADP, the default state for many prokaryotic initiators (17,18,20). The affinities of these mutants for origin DNA were measured by fluorescence anisotropy using fluorescein-labeled, ‘single-site’ oligonucleotides (the 17 bp ‘mORB’ and 18bp ‘C3′ sites for Orc1-1 and Orc1-3, respectively); longer DNA sequences resulted in biphasic binding curves that complicated initial analyses, likely due to non-specific initiator interactions with regions flanking the minimal single sites. Representative binding curves are shown in Figure 2, and all binding data are summarized in Table 1 (see also Supplementary Figure S2).
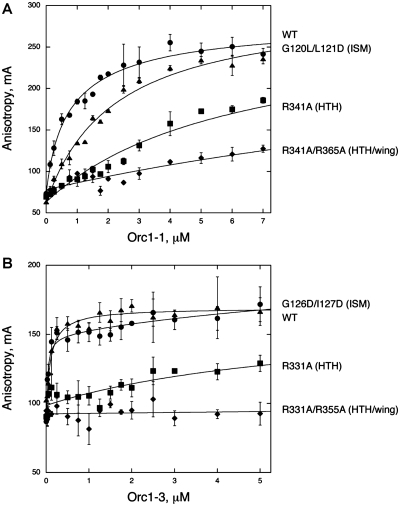
Figure 2.
Orc1 binding to origin DNA sequences. Fluorescence anisotropy values of short, fluorescein-labeled origin DNA sequences are plotted as a function of increasing (A) Orc1-1 and (B) Orc1-3 variant concentration. As fluorescence anisotropy increases with the decrease in tumbling rate of the labeled species, DNA bound to Orc1 protein will have higher anisotropy values than free DNA. Only a subset of Orc1 variants is shown for clarity. Data shown are averaged from triplicate experiments and fit to the single-site binding equation to determine affinity (Table 1), with the exception of WT Orc1-3 and G126L/I127D, which are fit to a two-site binding equation due to a weak second binding event (possibly the result of protein-protein interactions, not shown). Mutations within the WHD and AAA+ ISM have varying effects on the affinity of Orc1 protein for origin DNA.
Alanine substitutions of each of the four residues that make base-specific contacts in the complex structure (R341A for Orc1-1 and R331A, N352A and R355A for Orc1-3) considerably weaken Orc1 binding to origin sequences, decreasing affinity between 15- and 240-fold. R341 and R331 sit in the HTH motif of the WHD, whereas R355 resides in the wing with N352 (Figure 1B). The magnitude of the R341A and R331A defects are in line with previous mutagenesis studies of the WHDs of archaeal Orc1 (10,21–23). Although the Orc1-1 wing residue, R365 (equivalent to R355 in Orc1-3), does not make specific sidechain/base contacts in the complex, changing it to alanine reduces binding to a similar degree as the R341A mutation located within the HTH element of Orc1-1 (Supplementary Figure S2). As R365 engages the phosphate backbone of DNA, this result indicates that the interaction seen in the structure is functionally significant. Simultaneously mutating both wing and HTH arginine residues (R341A/R365A) synergistically weakens Orc1-1 binding ∼60-fold and the equivalent mutations completely abolish Orc1-3 binding (a Kd could not be determined for R331A/R355A). Interestingly, although Orc1-1 and Orc1-3 AAA+ ISM residues do not make specific base contacts with origin DNA, mutations in the ISM of either protein affect binding, albeit to a lesser degree than mutations in either WHD element (Figure 2, Table 1).
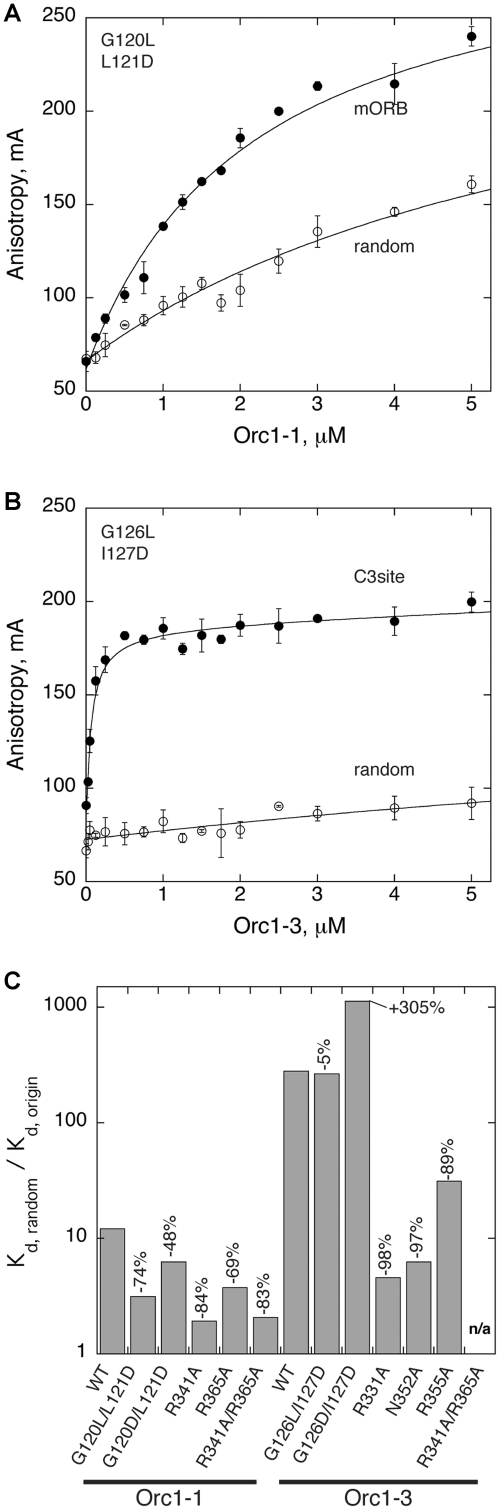
Next, we wished to determine whether our selected mutations weakened Orc1 binding only with respect to origin sequences, or disrupted DNA binding in general. Therefore, we measured the affinity of WT and mutant Orc1 proteins for a randomized oligonucleotide of similar length and base composition as the mORB and C3 site targets (Figure 3, Table 1). All Orc1-1 and Orc1-3 variants demonstrate weaker binding to the randomized oligo than to their cognate origin recognition sequence (Supplementary Figure S2). These findings are summarized in Figure 3C, where tight binding to origin sequences, paired with weak binding to a randomized sequence, results in a large specificity factor (the ratio Kd,random/Kd,origin). Larger disparities between the binding affinities for cognate and non-cognate sequences reflect greater differences in specificity, and thus reveal those initiator-origin DNA interactions that govern target site selectivity.
Figure 3.
Orc1 specificity for origin DNA sequences. Representative binding curves of (A) Orc1-1 and (B) Orc1-3 ISM mutants demonstrate tighter binding to origin sequences than to a randomized DNA sequence. As illustrated in the (C) summary plot of Kd ratios (Kd,random/Kd,origin), most Orc1 mutants decrease the specificity of Orc1 proteins for their cognate origin DNA recognition sequences (larger ratios reflect greater specificity). The measured percentage change in specificity is indicated above each bar.
Wild-type Orc1-1 and Orc1-3 both show high selectivity towards their respective origin recognition sites, with specificity differences for randomized targets versus origin DNA sequences of 12-fold for Orc1-1 and 280-fold for Orc1-3. Each WHD and AAA+ mutation decreases initiator specificity, except for the Orc1-3 ISM mutant (G126D/I127D), whose binding to the randomized sequence was sufficiently weak to actually increase the specificity difference as compared to WT Orc1-3. We were unable to determine the specificity of the R331A/R355A mutant because we could not measure its binding to either the C3 site or the randomized target. Taken together, these studies confirm that residues within the WHDs, which make base-specific interactions with origin sequences, not only provide substantial binding energy to initiator-DNA complexes, but also help establish Orc1 specificity. In particular, arginine residues within the HTH motif that contact guanine bases (R341 for Orc1-1 and R331 for Orc1-3) appear to make key contributions, with mutation of these residues showing the most deleterious effects on initiator binding and specificity compared to any other single point mutants. Combining these HTH mutations with arginine to alanine substitutions in the wing elements (R365A and R355A for Orc1-1 and Orc1-3, respectively), further reduces or even completely abrogates binding.
In general, the mutated Orc1-3 WHD residues appear to have a more dramatic effect on binding than equivalent mutations in Orc1-1. Furthermore, WT Orc1-3 binding to the C3 site is >20-fold more specific, as well as ∼10-fold tighter, than that of Orc1-1 to the mORB site. This finding is consistent with the observation that Orc1-3 makes more base-specific contacts to origin DNA than does Orc1-1 (Figure 1). By contrast, the effects of AAA+ mutations on initiator specificity are unexpected. These mutations, which were designed to disrupt what appeared to be non-specific interactions with origin DNA, mostly result in modest affinity decreases for Orc1-1 and Orc1-3 on both origin and non-specific DNA sequences. However, while comparable mutations in the ISM loops weaken binding to origin DNAs to a similar degree between the two paralogs (1.5- to 5-fold), these same substitutions exhibit quite variable effects on initiator specificity. For instance, whereas the Orc1-1 ISM mutant proteins both show lowered specificity of binding to cognate sites as compared to WT Orc1-1, the equivalent Orc1-3 mutations either have little effect, or even increased specificity for origin loci (Figure 3C).
These findings raise two interesting questions. First, how do ISM residues, which appear to make non-specific, van der Waals interactions with DNA sequences, nonetheless influence initiator specificity? Second, how do Orc1-1 and Orc1-3 establish specificity for origin sequences with minimal base-specific interactions—indeed, only a single sidechain-base contact in the case of Orc1-1? One explanation may be that origin recognition by archaeal initiators depends more on local features such as sequence context (e.g. nearest neighbor effects) and/or the shape and deformability of target elements, as opposed to direct base pair readout. An analysis of conserved prokaryotic and eukaryotic initiator-recognition motifs within replication origins has suggested that these DNA elements are readily deformable, and that initiators may utilize such duplex distortions for binding (24). Consistent with this prediction, the DNA in Orc1/origin complexes is deformed, being both bent (∼20–35° overall) and significantly underwound as compared to idealized B-form DNA (17,18). This observation suggests that ISM mutations may affect discrimination by perturbing more ideal AAA+–DNA interactions. Given that the AAA+ domains also constitute a point of contact between Orc1-1 and Orc1-3 (17), this effect may further help promote rearrangements within or between Orc1 protomers to influence binding.
DNA distortion in Orc1 recognition of origin sequences
To explore how specificity-determining interactions could be linked to DNA structure, we employed copper phenanthroline (Cu-OP) footprinting to monitor the impact of each initiator variant (both alone and in combination with its WT Orc1 paralog) upon binding to origin DNA (Figure 4). The nucleolytic activity of Cu-OP is sensitive to protein-induced changes in DNA conformation and provides a relatively high-resolution footprint (25). In these studies, each Orc1 variant was added to origin DNA at a concentration 3-fold greater than the Kd-value measured for its specific recognition sequence. This procedure was used to ensure essentially complete binding at the specific site and to minimize binding to non-cognate DNA regions—binding conditions that best enabled us to distinguish Orc1 effects on DNA distortion from simple affinity defects. Differences in Cu-OP modification (as compared to a footprinting control consisting of DNA alone) can be seen most readily when plotted as a ‘heat map’ along the top and bottom strands of the origin sequence for individual initiator variants and initiator pairs (Figure 5). The double mutants R331A/R355A and R341A/R365A were not tested owing to their weak binding.
Figure 4.
DNA distortion monitored by Cu-OP footprinting. Cu-OP footprints of the (A–B) top and (C–D) bottom strands of a 70 bp fragment of origin DNA illustrate areas of protection and hypersensitivity upon binding of either individual Orc1 variants (A and C) or Orc1 pairings (B and D). Maxam-Gilbert AG sequencing is shown in the far left lane of panels (A) and (C). The location of Orc1-1 (purple) and Orc1-3 (teal) binding sites are marked according to the initiator-DNA complex structure. The control lane corresponds to a mock footprinting experiment with no protein added.
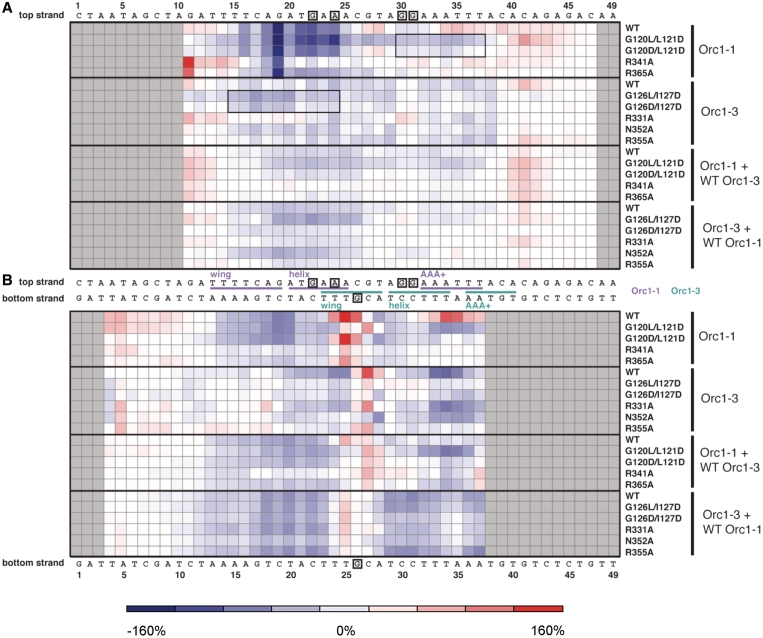
Figure 5.
Heat map of Orc1-induced Cu-OP modifications. Positions along the origin DNA sequence that show increased Cu-Op modification (DNA distortion) for each individual Orc1 variant and Orc1 pairing, as compared to DNA alone control, are colored red. Areas of decreased modification (DNA protection) are colored blue. The strength of the modification of the (A) top or (B) bottom strand is reflected in the intensity of the color plotted. A color key on the bottom indicates the correspondence between color and percentage-difference in band intensity. Gray squares indicate regions of the DNA sequence that were not resolved in the footprinting gel. Squares boxed by a thin black line indicate regions of extended protection arising from the mutation of ISM elements. Orc1-1 (purple) and Orc1-3 (teal) binding sites, as defined by the initiator-DNA complex structure, are highlighted on the origin sequence, with base-specific contacts shown boxed.
From this analysis, we find that WT Orc1-1 by itself strongly protects conserved mORB positions bound by the Orc1-1 WHD, while at the same time distorting origin DNA 3′ to the WHD and AAA+ binding sites (150 and 80% increases in band intensity for positions 25 and 34 of the bottom strand, respectively, compared to DNA alone controls). The latter effect likely results from a repositioning of the Orc1-1 ISM from a major groove contact to an adjacent minor groove contact when Orc1-1 binds to the origin DNA in the absence of Orc1-3 (a site occluded by the Orc1-3 WHD in the heterodimeric complex structure (17)); such a configuration is observed in the Orc1/ORB complex structure from the archeaon Aeropyrum pernix, in which the AAA+ ISM binds and deforms the minor groove adjacent to the HTH-major groove interaction, bending the DNA by ∼35° (18). The other area of strong, Orc1-1-induced Cu-OP modification corresponds to the global DNA bend point (position 25 on the bottom strand) located in the region where Orc1-1 and Orc1-3 binding sites overlap in the complex structure (Figures 1 and 5). WT Orc1-3 alone also causes increased distortion of this region (although shifted slightly compared to Orc1-1 to position 27, a 40% intensity increase), along with modest protection of its WHD and AAA+ binding sites.
Mutation of the single Orc1-1 base-specific contact (HTH mutant R341A) leads to a new hypersensitive region 5′ to the wing binding locus (25–50% intensity increases for positions 12–15, top strand), along with a marked decrease both in the overall protection afforded by the WHD and in modification of the global DNA bend site (Figure 5). The R365A wing mutation of Orc1-1 shows similar, albeit slightly more modest effects. Unexpectedly, both of these Orc1-1 mutations also dampen Cu-OP modification of the AAA+ binding site (positions 32-37, bottom strand). In contrast to the global effects arising from WHD alterations, mutations in the Orc1-1 AAA+ domain act more locally. For example, G120D/L121D nearly abolishes Cu-OP modification of the Orc1-1 AAA+ binding site, but retains a near wild-type pattern of protection around the WHD. The G120L/L121D mutant is similar to the double-aspartate ISM mutant in its WHD/origin interactions, but actually shows enhanced protection across the region normally contacted by the Orc1-1 ISM (40–60% intensity decreases at positions 32-37, bottom strand). Moreover, protection in this mutant extends 5′ from the wild-type AAA+ binding site, covering the region that would be contacted by the Orc1-3 WHD in the heterodimer complex (a similar, albeit more subtle, extended protection can be detected for the G120D/L121D mutant). Interestingly, close examination of the footprint reveals that Orc1-1 G120L/L121D also produces patterns observed for its paralog, Orc1-3, including the formation of a hypersensitive site at position 27 of the bottom strand (40% intensity increase) and an additional hypersensitivity immediately 3′ to the sequences normally contacted by the Orc1-3 AAA+ site (position 41 of the top strand, 70% intensity increase). Thus, with the exception of more strongly protecting the mORB locus of oriC2, the Orc1-1 G120L/L121D mutant protein shows a Cu-OP reactivity pattern resembling that produced by the binding of both Orc1-1 and Orc1-3 together.
For Orc1-3, the effects of select mutations are more nuanced and varied. For instance, the G126L/I127D and G126D/I127D ISM mutations extend their protection to the neighboring mORB site (on average, a near 40% intensity decrease for positions 15-20 in the top strand). In this sense, these substitutions recapitulate to an extent the behavior exhibited by Orc1-1 ISM mutants. However, these mutations also diminish protection of the local AAA+ site and the distant Orc1-3 WHD binding site, while concomitantly reducing modification of the global DNA bend point (position 27 of the bottom strand shows only a 10% intensity increase for G126L/I127D and <5% for G126D/I127D versus 40% for wild-type Orc1-3). By contrast, one wing mutant of Orc1-3, R355A, shows a bottom strand footprint similar to that of the Orc1-3 ISM mutants, but an upper-strand protection pattern more closely related to that of wild-type Orc1-3. The R331A mutant behaves fairly similarly to WT Orc1-3, protecting both AAA+ and WHD sites, but shows a slightly lessened modification of the global bend site than the WT protein (30% intensity increase versus 40% for WT), whereas the N352A wing mutation shows decreased WHD site protection, but maintains strong AAA+ site protection (20% intensity decrease versus 30% for Orc1-3 wild-type). Mutation of N352, whose base-specific interaction occurs most closely to the global bend point, similarly abolishes Cu-OP modification of the site and induces protection of this area.
How are these deformations affected in the context of both initiators? The structure of the ternary Orc1-1/Orc1-3/DNA complex suggests that the AAA+ domain of WT Orc1-1 engages in minor groove contacts due to steric occlusion of the major groove by Orc1-3 (17). Cu-OP footprints measured in the presence of both WT initiators reflect this arrangement, which decreases DNA modification/distortion overall, and increases protection compared to either initiator alone (Figure 5). The only region that maintains a relatively substantial distortion is the global DNA bend site, which is situated where the binding sites for the two initiators overlap; however, even this region’s structure is less deformed in the presence of Orc1-1 and Orc1-3 (a 20% intensity increase over background), as compared to binding of either Orc1 protein alone (150% intensity increase for Orc1-1 and 40% for Orc1-3). The binding of both paralogs together also diminishes the effects of specific mutations in either initiator alone (Figure 5), suggesting that the wild-type binding partner may compensate for defects in the mutant initiator to help maintain a relatively constant ‘co-footprint’.
Though complex, some important general trends in the recognition of replication origin sites by Orc1 proteins can be garnered from this analysis. First, ISM residues that do not engage in base-specific contacts can nonetheless affect origin deformation as much as, or even more than, WHD residues that do directly interact with certain bases. These findings, together with our specificity studies, support the hypothesis that archaeal initiators distinguish replication origins through a combination of DNA shape recognition and direct sequence read-out. Evidence of shape discrimination is particularly apparent in the case of Orc1-1, which despite having a lower specificity of binding as compared to Orc1-3, has a more distinctive footprinting profile than Orc1-3. Although seemingly contradictory, this behavior merely reflects the complementary information derived from the fluorescence anisotropy and footprinting measurements; namely, whereas fluorescence anisotropy provides an absolute measure of affinity, CuOP footprinting supplies information regarding the physical impact of the interaction arising from protein-induced DNA deformations. Thus, these observations are in agreement with the proposal that Orc1-1, which makes one lone base-specific contact, relies heavily on DNA shape and deformability to drive specific origin recognition.
Second, we note that distortion effects often propagate beyond the local initiator-origin contact targeted by a particular mutation. For example, Orc1-1 WHD mutants abrogate Cu-OP modification of the Orc-1-1 AAA+ site. By contrast, ISM mutants often show an unexpectedly broad DNA protection pattern that extends to neighboring Orc1 binding sites, possibly indicating the promiscuous association of a second Orc1 protomer. Such an interaction would be consistent with our fluorescence anisotropy studies, which show that ISM substitutions can diminish DNA binding specificity (Figure 3C). Along these lines, a recent DNaseI footprinting effort performed with Pyrococcus furiosus Cdc6/Orc1 has revealed that ATP binding, which is known to promote the co-association of bacterial AAA+ initiators (20), similarly alters ISM/DNA contacts (26).
In sum, our mutational analysis of WHD and AAA+ residues located in the initiator/DNA interface reveals that disrupting individual domain/origin interactions affects binding affinity and DNA deformation by the protein as whole. Altering the grip of one domain also can influence the DNA-binding behavior of the other domain, as well as change the overall specificity of an initiator for origin sequences. One interesting example of this phenomenon is the Orc1-3 G126D/I127D mutant, the only initiator variant to show increased specificity for origin DNA in our studies (Figure 3C). In this case, disrupting the Orc1-3 ISM interaction with DNA alters the position of a severe origin distortion normally centered within the Orc1-3 WHD binding site (position 27, bottom stand). While this change does not substantially compromise binding affinity to the C3 site oligo, G126D/I127D binding to a random DNA sequence is severely weakened. Thus, removal of the ISM interaction appears to shift the balance between recognizing sequence context and direct sequence read-out towards the latter, allowing binding to be dictated by the three direct side chain-base contacts found in the WHD-origin interface to increase specificity of the mutant for origin DNA overall.
A continuum of origin recognition and activation strategies between cellular organisms
As expected from their sequence homology (∼35% identity) and the perfect conservation of key amino acids in their respective WHD and ISM elements, the two Sulfolobous Orc1 paralogs used in this study show some general trends in their recognition of DNA. Nonetheless, these same residues also play some unique and distinct roles in controlling how the two Orc1 proteins associate with a replication origin (Figure 5). We find that even a single amino acid substitution can alter the specificity and deformation properties of either initiator without necessarily compromising DNA binding in general. This property likely endows Orc1 proteins with an innately high level of plasticity with respect to origin recognition. This behavior also may factor into the ability of archaea to utilize more than one functional origin, or to accommodate different degrees of conservation between ORB binding sites within and between origins (indeed, many archaea utilize only subset of the ORB consensus—the ‘mORB’—or fail to have discernable ORBs at all) (10,22,27). Plasticity likewise may play a useful role in the diversification of origins; for example, allowing a mutated initiator to still bind DNA, but with an altered specificity that would help define a new origin site.
Notably, many of the functional elements responsible for DNA recognition are conserved between archaeal Orc1 initiators and their eukaryotic counterparts. For example, many eukaryotic initiator subunits possess an ISM insertion within their resident AAA+ domains (3,28). As with archaea, a conserved glycine is frequently found in the ISM of eukaryotic Orc1, occupying a region that corresponds to a tight turn within the element (17). Orc1 and Cdc6, as well as Orc4 and Orc5, similarly have been predicted to possess a WHD C-terminal to their AAA+ domains (6,8). Like most archaeal Orc1 proteins (6,29), the wing of these eukaryotic WHDs tends to contain one or more basic amino acids, typically arginine. At present, it remains to be determined whether these residues in eukaryotic initiators play analogous roles to those seen in archaea. Nonetheless, these patterns have led to the idea that certain factors involved in eukaryotic initiation, in particular Orc1 and Cdc6, may contact DNA in a manner analogous to that seen for archaeal Orc1 proteins (4,6,17,18,30). For its part, the ISM of bacterial DnaA has been implicated in binding ssDNA, as well as in fostering inter-protomer contacts that stabilize its ATP-dependent assembly (20,31).
Interestingly, an inspection of nucleotide-sequence determinants and structure-perturbation studies suggests that the relationship between archaeal/eukaryotic initiators and their respective origins may be even deeper. In particular, while analyzing archaeal origins, we noticed that several of the signature elements of ORBs—the sites recognized by Orc1-1 paralogs (10)—also are present in certain Saccharomyces cerevisiae origins (Figure 6A), such as the B1 box of ARS1 (32). Moreover, this region of the ARS1 origin shows a general increase in Cu-OP sensitivity, with some of the more significant perturbations of DNA structure exhibited by archaeal Orc1 initiators appearing to overlap with particular ORC/Cdc6-induced deformations seen for some yeast origin DNAs (10,30,32). These observations not only suggest that there is a common mode of engaging origin DNA for archaeal and eukaryotic Orc1 proteins, but also imply that several other paralogous proteins (e.g. Orc4, Orc5 and Cdc6) may bind DNA in an analogous manner in the context of an initiator supercomplex.
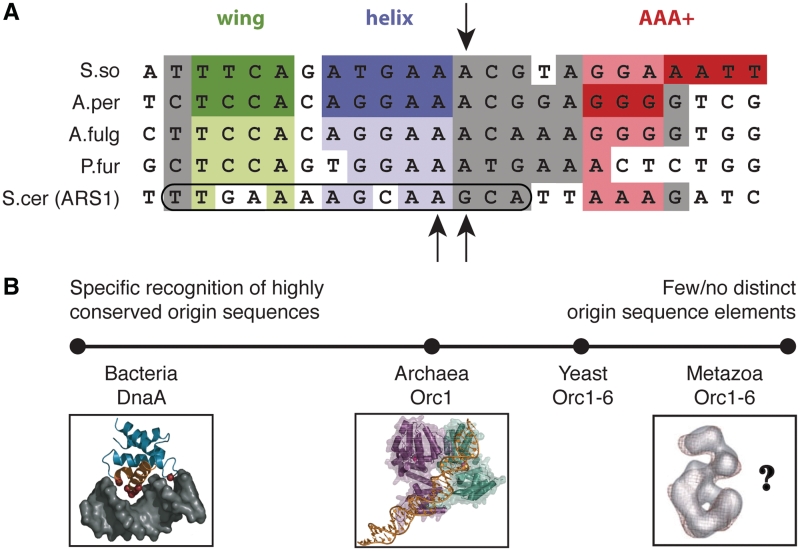
Figure 6.
Comparison of archaeal initiator and origin properties with other domains of life. (A) Alignment of archaeal and yeast origin sequences. The S. solfataricus miniORB site is shown aligned with ORB sequences from A. pernix, Archaeoglobus fulgidus, P. furiosus and to the ORC binding site of S. cerevisiae ARS1 (encircled area demarcates the B1 element). Shaded areas within the alignment denote positions of sequence homology (purine or pyrimidine conserved), with positions contacted by Orc1 wing, HTH and AAA+ elements within origin-Orc1 co-crystal structures colored dark green, dark purple and dark red, respectively (17,18). Homologous sequences are highlighted in lighter shades of these colors. The S. solfataricus sequence ‘GGA’ is colored pink to reflect the apparent site of Orc1-1 AAA+ binding in the absence of Orc1-3 (17). The global DNA bend point induced by S. solfataricus Orc1-1 is highlighted (black arrow, top), as are two particularly strong, ORC-induced DnaseI hypersensitive sites in the yeast ARS1 sequence (black arrows, bottom, 37,38). (B) A continuum of origin binding mechanisms. Archaea and yeast appear to lie in between the highly specific recognition of conserved sequence repeats by the bacterial DnaA initiator at one extreme and the absence of distinct origin sequences for ORC binding in metazoans at the other.
This congruence is also worth noting in light of the origin-recognition strategies used throughout the different cellular domains of life. Bacteria generally rely on a relatively ‘hard-wired’ approach that uses a set of common consensus DNA sequences (e.g. DnaA boxes and I-sites) for binding their initiator, DnaA (33,34). By contrast, metazoan ORC can initiate replication on nearly any piece of DNA (11,35), and appears to rely more on local DNA structure and/or cues from proteins bound at neighboring sites (14,36). Thus, there exists a continuum of discrimination tactics that have been evolved by nature around a common AAA+-ATPase core (Figure 6B). Microbial Orc proteins, which include yeast and archaeal Orc1, appear to occupy a middle ground on this scale, associating with origins that frequently contain conserved sequence hallmarks, but at the same time using sequence context and the influence of sequence on DNA shape or deformation to guide recognition (10,15,37–39). Future studies will help establish the extent to which similar factors are weighed and exploited by eukaryotic initiation systems as well.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Molecular Biology Organization Long-Term Post-Doctoral Fellowship (ALTF 869 2008 to A.C.); Edward Penley Abraham Trust; Wellcome Trust (Programme Grant 086045/Z/08/Z to S.D.B.); National Institutes of Health (GM07174 to J.M.B.). Funding for open access charge: Edward Penley Abraham Trust; Wellcome Trust (Programme Grant 086045/Z/08/Z to S.D.B.); National Institutes of Health (GM07174 to J.M.B.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the members of the Berger and Bell laboratories for helpful discussion. E.C.D. acknowledges prior support from the Miller Institute for Basic Research in Science at University of California, Berkeley. A.C. is an EMBO Fellow.
REFERENCES
- 1.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 2.Duderstadt KE, Berger JM. AAA+ ATPases in the initiation of DNA replication. Crit. Rev. Biochem. Mol. Biol. 2008;43:163–187. doi: 10.1080/10409230802058296. [DOI] [PubMed] [Google Scholar]
- 3.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Wigley DB. ORC proteins: marking the start. Curr. Opin. Struct. Biol. 2009;19:72–78. doi: 10.1016/j.sbi.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 5.Robinson NP, Bell SD. Origins of DNA replication in the three domains of life. Febs J. 2005;272:3757–3766. doi: 10.1111/j.1742-4658.2005.04768.x. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Smith CL, DeRyckere D, DeAngelis K, Martin GS, Berger JM. Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control. Mol. Cell. 2000;6:637–648. doi: 10.1016/s1097-2765(00)00062-9. [DOI] [PubMed] [Google Scholar]
- 7.Erzberger JP, Pirruccello MM, Berger JM. The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 2002;21:4763–4773. doi: 10.1093/emboj/cdf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarey MG, Erzberger JP, Grob P, Leschziner AE, Berger JM, Nogales E, Botchan M. Nucleotide-dependent conformational changes in the DnaA-like core of the origin recognition complex. Nat. Struct. Mol. Biol. 2006;13:684–690. doi: 10.1038/nsmb1121. [DOI] [PubMed] [Google Scholar]
- 9.Messer W. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 2002;26:355–374. doi: 10.1111/j.1574-6976.2002.tb00620.x. [DOI] [PubMed] [Google Scholar]
- 10.Robinson NP, Dionne I, Lundgren M, Marsh VL, Bernander R, Bell SD. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell. 2004;116:25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- 11.Harland RM, Laskey RA. Regulated replication of DNA microinjected into eggs of Xenopus laevis. Cell. 1980;21:761–771. doi: 10.1016/0092-8674(80)90439-0. [DOI] [PubMed] [Google Scholar]
- 12.Remus D, Beall EL, Botchan MR. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 2004;23:897–907. doi: 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 14.Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM. Conserved nucleosome positioning defines replication origins. Genes Dev. 2010;24:748–753. doi: 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller P, Park S, Shor E, Huebert DJ, Warren CL, Ansari AZ, Weinreich M, Eaton ML, MacAlpine DM, Fox CA. The conserved bromo-adjacent homology domain of yeast Orc1 functions in the selection of DNA replication origins within chromatin. Genes Dev. 24:1418–1433. doi: 10.1101/gad.1906410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujikawa N, Kurumizaka H, Nureki O, Terada T, Shirouzu M, Katayama T, Yokoyama S. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 2003;31:2077–2086. doi: 10.1093/nar/gkg309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dueber EL, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317:1210–1213. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- 18.Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural Basis of DNA Replication Origin Recognition by an ORC Protein. Science. 2007;317:1213–1216. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- 19.King JY, Ferrara R, Tabibiazar R, Spin JM, Chen MM, Kuchinsky A, Vailaya A, Kincaid R, Tsalenko A, Deng DX, et al. Pathway analysis of coronary atherosclerosis. Physiol. Genomics. 2005;23:103–118. doi: 10.1152/physiolgenomics.00101.2005. [DOI] [PubMed] [Google Scholar]
- 20.Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- 21.Grainge I, Scaife S, Wigley DB. Biochemical analysis of components of the pre-replication complex of Archaeoglobus fulgidus. Nucleic Acids Res. 2003;31:4888–4898. doi: 10.1093/nar/gkg662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capaldi SA, Berger JM. Biochemical characterization of Cdc6/Orc1 binding to the replication origin of the euryarchaeon Methanothermobacter thermoautotrophicus. Nucleic Acids Res. 2004;32:4821–4832. doi: 10.1093/nar/gkh819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majernik AI, Chong JP. A conserved mechanism for replication origin recognition and binding in archaea. Biochem. J. 2008;409:511–518. doi: 10.1042/BJ20070213. [DOI] [PubMed] [Google Scholar]
- 24.Schneider TD. Strong minor groove base conservation in sequence logos implies DNA distortion or base flipping during replication and transcription initiation. Nucleic Acids Res. 2001;29:4881–4891. doi: 10.1093/nar/29.23.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tullius TD. Physical studies of protein-DNA complexes by footprinting. Annu. Rev. Biophys. Biophys. Chem. 1989;18:213–237. doi: 10.1146/annurev.bb.18.060189.001241. [DOI] [PubMed] [Google Scholar]
- 26.Akita M, Adachi A, Takemura K, Yamagami T, Matsunaga F, Ishino Y. Cdc6/Orc1 from Pyrococcus furiosus may act as the origin recognition protein and Mcm helicase recruiter. Genes Cells. 15:537–552. doi: 10.1111/j.1365-2443.2010.01402.x. [DOI] [PubMed] [Google Scholar]
- 27.Robinson NP, Bell SD. Extrachromosomal element capture and the evolution of multiple replication origins in archaeal chromosomes. Proc. Natl Acad. Sci. USA. 2007;104:5806–5811. doi: 10.1073/pnas.0700206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cunningham EL, Berger JM. Unraveling the early steps of prokaryotic replication. Curr. Opin. Struct. Biol. 2005;15:68–76. doi: 10.1016/j.sbi.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Singleton MR, Morales R, Grainge I, Cook N, Isupov MN, Wigley DB. Conformational changes induced by nucleotide binding in Cdc6/ORC from Aeropyrum pernix. J. Mol. Biol. 2004;343:547–557. doi: 10.1016/j.jmb.2004.08.044. [DOI] [PubMed] [Google Scholar]
- 30.Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat. Struct. Mol. Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozaki S, Kawakami H, Nakamura K, Fujikawa N, Kagawa W, Park SY, Yokoyama S, Kurumizaka H, Katayama T. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 2008;283:8351–8362. doi: 10.1074/jbc.M708684200. [DOI] [PubMed] [Google Scholar]
- 32.Lee DG, Bell SP. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol. Cell. Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Speck C, Weigel C, Messer W. ATP- and ADP-dnaA protein, a molecular switch in gene regulation. EMBO J. 1999;18:6169–6176. doi: 10.1093/emboj/18.21.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGarry KC, Ryan VT, Grimwade JE, Leonard AC. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl Acad. Sci. USA. 2004;101:2811–2816. doi: 10.1073/pnas.0400340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DePamphilis ML. Eukaryotic DNA replication origins: reconciling disparate data. Cell. 2003;114:274–275. doi: 10.1016/s0092-8674(03)00604-4. [DOI] [PubMed] [Google Scholar]
- 36.Lipford JR, Bell SP. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell. 2001;7:21–30. doi: 10.1016/s1097-2765(01)00151-4. [DOI] [PubMed] [Google Scholar]
- 37.Diffley JF, Cocker JH. Protein-DNA interactions at a yeast replication origin. Nature. 1992;357:169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- 38.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 39.Grainge I, Gaudier M, Schuwirth BS, Westcott SL, Sandall J, Atanassova N, Wigley DB. Biochemical Analysis of a DNA Replication Origin in the Archaeon Aeropyrum pernix. J. Mol. Biol. 2006;363:355–369. doi: 10.1016/j.jmb.2006.07.076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.