Abstract
Accumulating evidences show that small non-protein coding RNAs (ncRNAs) play important roles in development, stress response and other cellular processes. The silkworm is an important model for studies on insect genetics and control of lepidopterous pests. Here, we have performed the first systematic identification and analysis of intermediate size ncRNAs (50–500 nt) in the silkworm. We identified 189 novel ncRNAs, including 141 snoRNAs, six snRNAs, three tRNAs, one SRP and 38 unclassified ncRNAs. Forty ncRNAs showed significantly altered expression during silkworm development or across specific stage transitions. Genomic comparisons revealed that 123 of these ncRNAs are potentially silkworm-specific. Analysis of the genomic organization of the ncRNA loci showed that 32.62% of the novel snoRNA loci are intergenic, and that all the intronic snoRNAs follow the pattern of one-snoRNA-per-intron. Target site analysis predicted a total of 95 2′-O-methylation and pseudouridylation modification sites of rRNAs, snRNAs and tRNAs. Together, these findings provide new clues for future functional study of ncRNA during insect development and evolution.
INTRODUCTION
A large number of genomic transcripts in both prokaryotes and eukaryotes are non-protein coding RNAs (ncRNAs). Small ncRNAs, such as miRNAs, siRNAs and piRNAs have been widely studied, and accumulating evidences show that they play important and diverse roles in development and various cellular processes. Moreover, snRNAs and snoRNAs have been showed closely related to development and human disease (1–4). With the completion of an increasing number of genome sequencing projects, numerous ncRNAs other than miRNAs, siRNAs and piRNAs, have been computationally predicted or experimentally identified, and their functions are gradually being revealed (5,6). In Drosophila, the two mlRNAs roX1 and roX2 are essential for dosage compensation (7,8). Transcription is also regulated by ncRNAs. For example, in human cells the small nuclear RNA 7SK influences the activity of RNA polymerase II (Pol II) by inhibiting the CDK9/cyclin T1 kinase (9). Espinoza et al. (10) found that the small ncRNA B2 can inhibit RNA Pol II transcription in mouse cells; and the steroid RNA receptor activator (SRA) acts to confer functional specificity upon multiprotein complexes recruited by a liganded receptor during transcriptional activation (11). Small nucleolar RNAs are involved in RNA editing or pre-mRNA splicing, like MBII-52 which specifically decreases the efficiency of ADAR2-mediated RNA editing (12). HBII-52, the human ortholog of MBII-52, has also been suggested to perform an important role in the Prader–Willi syndrome through regulation of alternative pre-mRNA splicing (13). The Drosophila Pgc RNA is specifically expressed in primordial germ cells, and required for maintenance of germ cell fate (14); whereas the bereft RNA is only expressed in the peripheral nervous system, and bereft mutant flies exhibit aberrant development of the eye and defects in other external sensory organs (15). More recently, Dinger et al. (16) have presented evidence that the expression of a large number of long ncRNAs (lncRNAs) in mice correlates well with pluripotency or specific differentiation events, probably through engagement of the epigenetic machinery. In plants, two ncRNAs have been identified as regulators of respectively leaf morphology and root growth under salt stress (17).
The silkworm Bombyx mori is the model insect for the order Lepidoptera, and an organism of considerable economic importance. The B. mori genome has a size of ∼432 Mb, and was completely sequenced in 2004 (18). Although ncRNAs have been studied extensively in D. melanogaster (19,20), less work has been carried out in B. mori. Recently, 420 novel miRNA candidates were identified in silkworm, 248 of which have either egg- or pupa-specific expression (21,22). Given the large number of ncRNAs existing in Drosophila, we assumed that there might be many more ncRNAs also in B. mori.
We constructed a B. mori cDNA library of small RNAs with lengths of 50–500 nt, and identified 189 novel, small non-messenger RNA candidates. Most of these ncRNAs were not conserved in other organisms. Microarray and northern blot analyses showed that 40 ncRNAs showed significantly altered expression during silkworm development or across specific stage transitions. Our results provide the first genome-wide survey of genomic organization, biogenesis and developmental expression of this particular complement of the non-coding transcriptome in B. mori.
MATERIALS AND METHODS
Construction of cDNA library of 50–500 nt ncRNAs in B. mori
Total RNA was extracted from four developmental stages (egg, larva, pupa and adult) of an inbred domesticated variety of B. mori, Dazao P50, by the TRIzol (Invitrogen) method. The small RNA fraction (50–500 nt) was isolated with a QIAGEN tip (QIAGEN). mRNAs and rRNAs (probes in Supplementary Table S2) were removed with the Poly(A) Purist MAG (Ambion) and MICROB Express kits (Ambion). RNAs were dephosphorylated with calf intestine alkaline phosphatase (Fermentas) and then ligated to the 3-adaptor (3AD) oligonucleotide by T4 RNA ligase (Fermentas). The ligation product was split into two aliquots: one was treated with polynucleotide kinase (PNK; Fermentas) to phosphorylate uncapped RNA, and the other was treated with tobacco acid pyrophosphatase (TAP; Fermentas) to remove 5′-end methyl-guanosine caps from capped RNA. The two differently treated aliquots were ligated to the 5′-adaptor (5AD) oligonucleotide. Small molecules and excessive adaptors were removed with the RNeasy Minelute cleanup kit (QIAGEN) and the ligation products were reversely transcribed with Thermoscript™ Reverse Transcriptase (Invitrogen) at 50°C, using oligo 3RT as the RT primer. The cDNA was PCR-amplified and digested with SphI and SacI (NEB) and cloned in pGEM-4Z (Promega), as preivousely described (23).
Microarray analysis
Total RNA was extracted from silkworm egg, larva, pupa and adult samples using the TRIzol reagent (Invitrogen). Total RNA (25 µg) from each developmental stage were reverse transcribed to cDNA and labeled with Cy3 or Cy5 at the same time. Labeled cDNA were hybridized to dual channel microarrays. SSC buffer and oligonucleotides with no homology (<9 bp identity) to the B. mori genome were used as negative hybridization controls, and three silkworm protein coding genes (actinA3, rpl3 and ph56) were used as positive controls. Oligonucleotide probes of different ncRNAs (Supplementary Table S3) were printed in triplicate on the slides by a PE SpotArray72, then UV crosslinked at 65 mJ, and hybridized to the labeled cDNAs. We applied the LOWESS normalization method for dual-channel microarray analysis.
To investigate the functional roles of ncRNAs through developmental transitions, total RNA was isolated from four developmental stages (egg, the first and fifth instar larva, and the pupa) of silkworm by the TRIzol reagent (Invitrogen). Total RNA (25 µg) from egg and the first instar larva were reverse transcribed to cDNA and labeled with Cy3 or Cy5, respectively, before being hybridized to dual channel microarrays. Subsequently, 25 µg of total RNA from the fifth instar larva and pupa were treated similarly. The hybridization procedure was same to the above. The data used in hierarchical clustering was the ratios of the egg to the first instar larva and of pupa to the fifth instar larva.
Northern blot
RNA probes were synthesized and labeled by in vitro transcription of plasmid with SP6 RNA polymerase (Fermentas) and Dig-11-UTP (Roche). Total RNA extracted from four different developmental stages and five instars and eight time points over pupation were analyzed. Northern blot was performed with standard protocols. Blots were hybridized in Dig Easy Hyb buffer (Roche) at 68°C overnight, and then treated with Blocking and Washing buffer (Roche) and detected by CDP-star (Roche). Chemiluminescence signals were recorded with Kodak films, and pictures were obtained with Epson scan at 600 bits resolution ratio.
5′- and 3′-RACE
RACE was performed by PCR amplification of the RT products (RT methods same to the above for small RNA library construction), with one primer specific to the ncRNA sequence and the other primer specific to the 5′-adaptor (5CD) and complementary to 3′-adaptor (3RT) for 5′- and 3′-RACE, respectively. All primer sequences are given in Supplementary Table S4.
Semi-quantitative RT–PCR analysis
Total RNA was extracted from four developmental stages, namely egg, larva (mixture of the first to the fifth instar larva), pupa and adult, of silkworm B. mori. Equal amount of total RNA were used to synthesize the first strand of cDNA with the M-MLV reverse transcriptase (Promega). PCR was performed at an annealing temperature of 55°C (see Supplementary Table S5 for primer sequences). actinA3 was used as an internal control.
Gel mobility shift assay
DNA templates of Bm-15 and BGIBMGA011962-TA carrying a T7 promoter sequence were generated by RT–PCR (primers were listed in Supplementary Table S7). The transcript of BGIBMGA011962-TA has a length of 257 nt containing 12 nt matching the antisense elements of Bm-15. In vitro synthesis of both RNAs was carried out with T7 RNA polymerase (Fermentas). One picomole RNA of Bm-15 was dephosphorylated with 10 units of FastAP™ Thermosensitive Alkaline Phosphatase (Fermentas) in a 20 μl reaction at 37°C for 1 h. The dephosphorylated RNA was subsequently 5′-end-labeled with 1 unit of T4 Polynucleotide Kinase (Fermentas) and 1 mCi of γ-32P-ATP. Unincorporated nucleotides were removed by using Microspin G-50 Colums (GE Healthcare).
Binding assays were performed in 1× TMN buffer (20 mM Tris–acetate at pH 7.6, 100 mM sodium acetate, 5 mM magnesium acetate) as follows: 5′-end-labeled RNA of Bm-15 (0.1 pmol) and 1 μg of carrier yeast tRNA (Ambion) were incubated with increasing concentrations of unlabeled RNA of BGIBMGA011962-TA in 10 μl at 37°C for 30 min (experiment in Figure 1). The concentrations of the unlabeled BGIBMGA011962-TA RNA were 0, 3, 15, 30, 90 μM. The 5′-end-labeled antisense RNA of Bm-15 (0.05 pmol) was used as negative control to bind with an RNA of BGIBMGA011962-TA in the concentration of 30 μM. The binding reactions were run on native 5% polyacrylamide gels in 0.5× TBE buffer at 200 V in a cold room for 1 h. Gel was dried and analyzed using a PhosphorImager.
Figure 1.
Gel mobility shift assay for Bm-15 and BGIBMGA011962-TA RNA. Labeled Bm-15 RNA [marked with Bm-15 (+)*], was incubated for 30 min at 37°C with increasing concentrations of unlabeled BGIBMGA011962-TA RNA. The concentrations of the unlabeled BGIBMGA011962-TA RNA were 0, 3, 15, 30, 90 μM. Labeled antisense strand RNA of Bm-15 [Bm-15(−)*] was used as a negative control to incubate with unlabeled BGIBMGA011962-TA RNA in the concentration of 30 μM.
Bio-informational analysis
RNA secondary structures were predicted with mfold (version 3.2) (24). Internal or upstream and downstream motifs of ncRNA were detected with MEME version 3.5.7 (25). We used snoScan and snoGPS softwares to predict the potential targets of snoRNAs, prediction criteria of C/D box snoRNA set as follows: (i) the lowest score of the complementary region was 13.0 bits; (ii) besides the first mismatch next to box D or D′, the maximum mismatch was 1 nt; (iii) the maximum GU pairs were 3 bp; and (iv) the minimum complementary length was 10 nt (26). The promoters of the ncRNA genes were predicted by the web server of Neural network promoter prediction software (http://www.fruitfly.org/seq_tools/promoter.html). The sequences of U3 snoRNAs were aligned with clustal W2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html).
RESULTS
Construction of the silkworm ncRNA library
To ensure the ncRNA library we obtained was as complete as possible, total RNA was extracted from four developmental stages (egg, larva, pupa and adult). Altogether, 3027 clones were sequenced, of which novel ncRNA candidates constituted 55.1% of the sequenced clones, known ncRNAs made up 4.6%, and the rest were rRNAs, tRNAs, mtRNAs (mitochondrial tRNAs and 12S rRNA), SRP RNAs, mRNA degradation products and plant RNA contaminations (Figure 2A). The high percentage of novel transcripts in our library demonstrates that the construction strategy was efficient.
Figure 2.
Clonal and functional distributions. (A) Distribution of the sequenced clones on different RNA species and categories. (B) Functional distribution of all novel ncRNAs.
We identified 189 unique novel ncRNA candidates among the sequenced clones (Supplementary Table S1). The sequences were classified by alignment with known ncRNAs. The length of the sequence reads agreed well with the size estimates obtained from the northern blots (Figures 3 and 4 and data not shown). The sequences of 25 ncRNAs determined by 5′- and 3′-RACE were different from the sequence reads by one or two terminal nucleotides. Of the 189 novel ncRNAs, 141 (74.6%) were characterized as snoRNAs, six as snRNAs, three as novel tRNAs and one as a novel SRP RNA, while 20.1% novel transcripts could not be assigned to any known group owing to lack of sequence motifs or secondary-structure hallmarks, and will henceforth be referred to as ‘unclassified’ ncRNAs. Among the snoRNAs, 82 had characteristic sequence elements and secondary-structures resembling the H/ACA box snoRNA class, 57 could be classified as C/D box snoRNAs; whereas two showed both H/ACA and C/D box features and therefore could be regarded as C/D-H/ACA box ‘hybrid’ snoRNAs (27), and secondary structures of the two C/D-H/ACA box hybrid snoRNAs are shown in Supplementary Figure S2. Among the C/D box snoRNAs, five of which had conserved boxes and secondary structures resembling U3 snoRNAs in human and yeast, and thus are regarded as U3 snoRNAs. Alignment of these novel U3 snoRNAs to the human and Drosophila U3 snoRNAs are shown in Supplementary Figure S1. The functional distribution of the novel ncRNAs is shown in Figure 2B.
Figure 3.
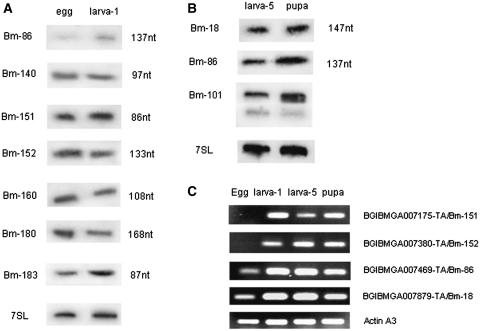
Northern blot analysis of differentially expressed ncRNAs and semi-quantitative RT–PCR results of the host gene or sense–antisense gene pairs of ncRNAs during the four developmental stages. The clone designation is indicated on the left of each panel, and the estimated size of the ncRNA on the right. Expression of 7SL RNA/SRP RNA is included as an internal control at the bottom of each panel. (A) Differential expression of nine ncRNAs in the four developmental stages: egg (E), larva (L), pupa (P) and adult (A). (B) Expression of two transposon-overlapping ncRNAs. (C) Expression of ncRNAs in different larva instars. (D) Expression of ncRNAs at different time point during pupation. (E) Semi-quantitative RT–PCR results of coding gene partner in sense–antisense gene pairs or ncRNA-host gene pairs. Sense–antisense gene pairs: BGIBMGA007175-TA and Bm-151; BGIBMGA007380-TA and Bm-152. BGIBMGA000829-TA is the host gene of Bm-51. BGIBMGA007469-TA is the host gene of Bm-86. The actinA3 gene was used as an internal control.
Figure 4.
Northern blot results of differentially expressed ncRNAs and semi-quantitative RT–PCR results of the host gene or sense–antisense gene pairs of ncRNAs during the two major developmental transitions. (A) Differential expression of ncRNAs from egg to the first instar larva (larva-1). (B) Differential expression of ncRNAs during the transition from fifth instar larva (larva-5) to the pupa. 7SL was used as an internal control. (C) Semi-quantitative RT–PCR results of coding gene partner in sense–antisense gene pairs or ncRNA-host gene pairs. Sense–antisense gene pairs: BGIBMGA007175-TA and Bm-151; BGIBMGA007380-TA and Bm-152. BGIBMGA007469-TA is the host gene of Bm-86; BGIBMGA007879-TA is the host gene of Bm-18. The actinA3 gene was used as an internal control.
Silkworm-specific ncRNAs
To analyze the evolutionary conservation of novel ncRNAs identified in this study, we searched for homologs of the 189 ncRNAs against known ncRNAs in other organisms. Except for the highly conserved (sequence identity >77%) snRNAs and SRP, the majority of the novel ncRNAs were not conserved or exhibited only very low nucleotide similarities to ncRNAs from other organisms. Analyzing for snoRNA antisense element and secondary-structure characteristics, we could identify 19 potential C/D box snoRNAs (including five U3 snoRNAs) and 37 potential H/ACA box snoRNAs with conserved homologues in other species.
Surprisingly, Blast searches against many other genomic sequences (including six insect species) could not identify any homologs or orthologs for 38 C/D box snoRNAs, 45 H/ACA box snoRNAs, two C/D-H/ACA box ‘hybrid’ snoRNAs and 38 unclassified ncRNAs, indicating that a large fraction of the novel ncRNA loci are potentially silkworm-specific.
Moreover, six of novel ncRNAs were found to perfectly match the small RNAs cloned by Kawaoka et al. (28), and the number increased to nine when one to three mismatches were allowed. Close inspection of the silkworm genome showed that these matched small RNAs either uniquely located within ncRNA genes identified in this study, or matched repetitive sequences (Supplementary Table S6 and Figure S8).
Genomic organization of the novel ncRNAs
Mapping of the 189 novel ncRNAs to the genome showed no particular chromosomal bias. The ncRNA loci are distributed on all but four (chrs 12, 17, 18 and 25) of the 28 B. mori chromosomes. The majority of the loci was either intergenic (74 loci) or located in the sense orientation in introns of coding genes (105 loci). A small number of loci overlapped either exons of coding genes (four loci) or were located on the complementary (antisense) strand of introns of coding loci (four loci). The two remaining loci overlapped with predicted transposon regions.
In consistent with a previous report in Drosophila (19), analysis of functionally annotated coding genes hosting snoRNAs showed that 16 of 40 intronic C/D box snoRNA loci are nested in genes encoding proteins associated with ribosome processing and assembly. The remaining 24 intronic snoRNA loci were hosted by genes related to RNA binding, damaged DNA binding, zinc ion binding or unknown functions. The snoRNA loci Bm-1, Bm-2 and Bm-3 are distributed on individual introns of BGIBMGA000074-TA, an RNA binding protein expressed in the embryo 100 h after fertilization, ovary and the prothoracic gland and silk gland of fifth-instar (Day 3) larva. In contrast to D. melanogaster, where almost all snoRNA loci are intronic (19,20), a substantial fraction (32.62%) of the B. mori snoRNA loci are intergenic. Moreover, in Drosophila only C/D box snoRNA loci follow a one-snoRNA-per-intron pattern (H/ACA box snoRNA loci generally being clustered) (19), whereas in the silkworm genome both C/D and H/ACA box snoRNAs have adopted the one-snoRNA-per-intron pattern.
The intergenic C/D box snoRNA loci Bm-42, Bm-43, Bm-44 and Bm-45 are clustered in a region downstream to the BGIBMGA014094-TA, which encodes the ribosomal protein L14. Bm-44 and Bm-45 are spaced by 251 bp and exhibit high nucleotide identity (71.90%), indicating tandem duplication during evolution. Although the 100 bp upstream regions of the transcription start sites (TSSs) of both Bm-44 and Bm-45 consist of the two conserved motifs TTGGTG and CTTC, their developmental expression patterns were distinct according to the microarray results (Figure 5A), indicating that they have evolved differential regulatory mechanisms since their divergence.
Figure 5.
Hierarchical clustering of ncRNA expression profiling at different developmental stages and during two major developmental transitions. (A) ncRNA expression profiling at different developmental stages, red and green indicates high and low expression, respectively. (B) ncRNA expression profiling during two major developmental transitions. In each panel, the left column indicates the relative difference in expression between the egg and the first instar larva; and the right column represents the relative difference in expression between the fifth instar larva and the pupa. Green indicates relatively low expression in the egg and the pupa; red indicates high expression.
The U3 snoRNA, Bm-101, is peculiar in that it is transcribed from a locus overlapping the first exon of BGIBMGA009516-TA by 64 bp. Analysis of the upstream sequence of BGIBMGA009516-TA identified a possible promoter located 634 bp upstream to the TSS of BGIBMGA009516-TA (Supplementary Figure S3). Closer scrutiny of the sequence found a putative H/ACA box snoRNA locus of 136 bp located between the promoter and Bm-101. Polycistronic snoRNAs are found in some organisms; however, the overlap of Bm-101 with a putative coding gene renders this a specific case. EST database search showed that BGIBMGA009516-TA is only expressed in the posterior silkgland of fourth molt, at a time when Bm-101 is also expressed as revealed by the northern blot result (Figure 4B). It is possible that the primary transcript has two processing routes, either cleaved into mature snoRNAs, or spliced into a protein-coding mRNA in silkgland of fourth molt. However, a Blast search found no homologous protein corresponding to the 85 amino acid long predicted protein sequence encoded by BGIBMGA009516-TA, suggesting that this locus may not represent an active gene.
Of the 38 unclassified ncRNAs, more than 68% (26 loci) are located in intergenic regions, seven are transcribed from intronic loci, four from the complementary region of introns, and one overlapped the predicted transposon region in the silkworm genome. Five of the intronic loci are found in protein-coding genes associated with ribosome biogenesis, nucleic acid binding or ion binding, while the rest located in introns of genes encoding proteins with various functions.
Northern blot were used to validate the facticity of 38 unclassified ncRNAs. Results showed that 12 of them were highly expressed in silkworm, while the other 26 had undetectable hybridization signals, probably due to their low expression level. To exclude the possibility of these low abundance ncRNAs originating from artificial genomic DNA, we examined their expression by RT–PCR, and found that they are true novel ncRNAs (Supplementary Figure S4 and data not shown).
The genomic organization of the novel loci hints at the mode of ncRNA biogenesis. The 74 intergenic loci are most likely independently transcribed under their own promoters, as observed in other species (20,29). The same probably also applies to the four antisense ‘intronic’ loci (Bm-146, Bm-150, Bm-151, Bm-152), which may form ‘sense–antisense’ gene pairs with the protein-coding genes with which they overlap. The sense-oriented intronic loci may either be independently transcribed from yet unidentified promoter sequences located within the intron or in the upstream exon, or be co-transcribed with the host gene and released by either splicing or pre-mRNA processing (30,31). The loci overlapping exons or transposons may share transcriptional features with their overlapping genomic element, or be independently transcribed.
Predicted targets of the novel snoRNAs
Since the B. mori 28S rRNA sequence is not complete, we used the 28S rRNA sequence from Drosophila and human (32) together with the B. mori sequences of 18S rRNA, 5.8S rRNA, snRNAs and tRNAs to predict potential targets of the novel snoRNAs. Twenty-six C/D box snoRNAs were predicted to guide 2′-O-methylation at 31 sites, of which 13 sites were conserved between silkworm, Drosophila and human, while the remaining sites were all specific to B. mori. The 45 H/ACA box snoRNAs were likewise predicted to guide pseudouridylation at 64 sites, of which 48 sites were common to silkworm, Drosophila and human. Targeted sites of 18S rRNA in B. mori were shown in Supplementary Figures S5 and S6. Detailed comparison showed that the 18S and 5.8S rRNA sequences are 64.3 and 62.9% identical, respectively, between B. mori and D. melanogaster. Seven out of 14 methylation sites and four out of five pseudouridylation sites in the 18S rRNA were specific to B. mori (Supplementary Figure S7A), whereas the silkworm and Drosophila shared no predicted target sites in the 5.8S rRNA (Supplementary Figure S7B). Furthermore, the majority of the silkworm-specific modification sites were found to be dispersed over the phylogenetically conserved regions of rRNA sequences (Supplementary Figure S7A and data not shown), with only one methylation and one pseudouridylation sites falling outside these regions.
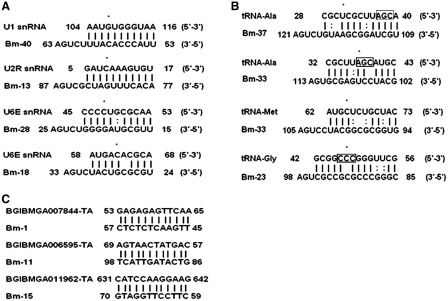
Among the C/D box snoRNAs not predicted to guide 2′-O-methylation of rRNAs, four were predicted to guide methylation of snRNAs (Figure 6A), similar to observations in Drosophila and plants (19,29). Another three novel C/D box snoRNAs were predicted to direct methylation of four sites in tRNAs, a phenomenon thus far only experimentally demonstrated in Archaea. The predicted interactions between the snoRNAs and their tRNA targets are shown in Figure 6B, from which it can be seen that Bm-23 and Bm-33 have the potential to guide the methylation of the anti-codons of Gly-tRNA and Ala-tRNA, respectively, whereas the other potential target sites are located in the stems adjacent to the anti-codon regions.
Figure 6.
Predicted interactions between novel C/D box snoRNAs and their potential targets. Interactions between C/D box snoRNAs and their targets in snRNAs (A), tRNAs (B) and mRNAs (C). Anti-codon is indicated by square.
Moreover, three C/D box snoRNAs were predicted to target three different mRNAs through 12 nucleotide long and perfectly matching antisense elements (Figure 6C). Of these three snoRNAs, Bm-1 and Bm-11 targeted genes encoding hypothetical proteins, while Bm-15 might target a member of the notch family, which is annotated with transmembrane receptor activity, transcription activator activity, and involvement in organ morphogenesis and developmental processes. Bm-15 had higher expression in the egg and adult stage (Figure 5A), while its predicted target gene BGIBMGA011962-TA is undetectable in egg. To probe the interaction between Bm-15 and BGIBMGA011962-TA, we performed EMSA as previously described (33,34). Gel mobility shift assay revealed that Bm-15 specifically bound to BGIBMGA011962-TA in vitro, whereas the antisense strand of Bm-15 exhibited no binding activity (Figure 1).
Expression profiling of the novel ncRNAs
ncRNAs are frequently expressed in specific tissues or developmental stages in eukaryotes, and are involved in replication, translation initiation and metabolic processes in prokaryotes (13,35–38), indicating their diverse roles in various cellular processes. In order to analyze the expression patterns of the novel ncRNAs in four developmental stages (egg, larva, pupa and adult), we prepared a microarray of 132 oligo probes spotted on a glass slide, and hybridized this with total RNA extracts from the four developmental stages. Good hybridization signals were successfully obtained for 117 of the 132 ncRNAs. The microarray analysis did not identify any transcript that exhibited strict stage-specific expression in any of the four developmental stages, however, 36 ncRNAs exhibited significantly altered expression between certain stages (Figure 5A, P ≤ 0.05), including 13 unclassified ncRNAs, 10 C/D and 13 H/ACA box snoRNAs. Most of the dynamically expressed ncRNAs accumulated more in the egg and adult stages. This suggests that changes in expression levels of the ncRNAs (when observed) occurred mainly during the transitions from egg to larva, and from pupa to adult, while remaining relatively stable through the larva to pupa stages.
To validate the microarray results, 15 of the 36 ncRNAs with significant differences in expression were selected for verification by northern blotting. Although the difference in expression for three of the ncRNAs appeared less pronounced than in the microarray data, the northern blot results for all 15 were consistent with microarray data (Figure 3A and data not shown). Among the ncRNAs validated by northern blot, Bm-100, Bm-162 and Bm-183 showed highest accumulation in egg. Bm-158 showed markedly reduced expression in the larva, while Bm-51 showed low expression levels in the pupa and adult stages (Figure 3A).
Among the ncRNAs with elevated expression in the larva, six ncRNAs were selected for a more detailed analysis of expression patterns within the larval stages. The expression of the selected ncRNAs was monitored by northern blots from the first to the fifth instar larval stages, but only minor differences in expression was observed from first to fifth instar larva (Figure 3C). In addition, six differentially expressed ncRNAs were analyzed during pupation. The results showed that the expression of Bm-51, Bm-86, Bm-152 and Bm-160 gradually decreased over this developmental stage (Figure 3D).
Developmental transitions are conspicuous in the silkworm life cycle. Therefore we further examined whether specific ncRNAs are involved in the transition between major developmental stages of the insect. To this end, we compared samples of the 132 ncRNAs extracted from egg and the first instar larva stage, and from the fifth instar larva and the pupa stage. Hybridization signals were obtained for 123 of the 132 ncRNAs. There was no distinct overall tendency towards increased expression of ncRNAs in any of the tested stages (i.e. approximately equally many ncRNAs showed increased expression in either direction across a developmental transition; Figure 5B). However, compared to other classes of ncRNAs, a larger number of H/ACA box snoRNAs accumulated during the transition from egg to the first instar larva, followed by a decrease in concentration from the fifth instar larva to pupa (Figure 5B). Moreover, six of the 30 unclassified ncRNAs showed higher expression in the first instar larva, while the rest showed higher expression in egg.
Twelve different ncRNAs showed significant (P ≤ 0.05) changes in expression, at least during one transition, among which eight were common to the 36 ncRNAs exhibiting significant differential expression at four developmental stages. Nine transcripts decreased significantly from the egg to the first instar larva stage, of which one was H/ACA box snoRNA and the other eight were unclassified ncRNAs. One C/D box snoRNA Bm-18 and an U3 snoRNA Bm-101 showed a significant increase in expression during transition from the fifth instar larva to the pupal stage (Figures 4A, B and 5B). The remaining transcript (H/ACA box snoRNA Bm-86) showed significant increases in expression across both transitions (Figures 4A, B and 5B). Ten of the above 12 ncRNAs were chosen for further validation by northern blot. The expression pattern observed for these ncRNAs were consistent with our microarray results (Figures 4A, B and 5B).
The H/ACA box snoRNA Bm-86 was predicted to direct pseudoridylation of the 18S and 28S rRNAs, and is transcribed from the second intron of BGIBMGA007469-TA. The host gene encodes translation initiation factor 5A, and shows elevated expression in the silkgland, as revealed by EST database analysis. Semi-quantitative RT–PCR showed that the expression of BGIBMGA007469-TA increased during the transition from egg to the first instar larva, but decreased from the fifth instar larva to pupa (Figure 4C). The uncorrelated expression profiles of the intronic snoRNA and host transcript during the transition from the fifth instar larva to the pupa suggest that Bm-86 may be transcribed independently of its host gene under specific developmental context.
A few of the ncRNAs showing significant decrease in expression from egg to the first instar larva also appear of particular interest. The Bm-151 locus is transcribed from the antisense strand of the fourth intron of BGIBMGA007175-TA, which encodes a protein involved in cystoblast division, spermatogonial cell division, spermatogenesis and oogenesis. During the transition from the egg to the first instar larva, BGIBMGA007175-TA showed an opposite expression pattern relative to Bm-151 (Figure 4C), suggesting a negative regulatory relationship between host and ncRNA gene, possibly through a natural antisense RNA mechanism (39,40). However, during the developmental transition from the fifth instar larva to pupa, Bm-151 and BGIBMGA007175-TA showed similar expression patterns, suggesting that regulation of these two genes may be more complex. Bm-152 is located on the antisense strand of the fourth intron of BGIBMGA007380-TA, encoding a gartenzwerg protein implicated in ER to Golgi vesicle-mediated transport, phagocytosis, engulfment and regulation of ADP ribosylation factors (ARF) protein signal transduction. At both developmental transitions, Bm-152 and BGIBMGA007380-TA exhibited opposite expression patterns (Figure 4A and C) indicating a negative regulatory relationship between the two genes.
Two snoRNAs showed significant increase in expression during the transition from the fifth instar larva to the pupa. Bm-18 is a C/D box snoRNA (with a predicted target site in U6 snRNA) located in sense orientation relative to the first intron of BGIBMGA007879-TA, the silkworm homolog of the ribosomal protein L5. Similar to Bm-86, Bm-18 and its host gene showed differential expression profiles when both transition are considered (Figure 4B and, C), indicating independent transcription of the intronic Bm-18. The Bm-101 U3 snoRNA overlaps the first exon of BGIBMGA009516-TA by 64 bp. Analysis of the upstream sequence of BGIBMGA009516-TA identified a putative promoter sequence of 50 bp located 634 bp upstream to the TSS of BGIBMGA009516-TA (Supplementary Figure S3). However, EST database analysis showed that the expression profiles of these two genes were quite different. Bm-101 showed its highest expression level in the pupa, while BGIBMGA009516-TA is uniquely expressed in the posterior silkgland of the fourth molt.
DISCUSSION
Increasing number of ncRNAs has been identified from various organisms by experimental RNomics combined with bioinformatics analysis (41,42). In this study, nearly 200 silkworm ncRNAs have been cloned, of which many appear to be specific to this organism, and 95 potential modification sites in various RNAs guided by novel snoRNAs were predicted. Furthermore, analysis of the expression profiles of the novel ncRNAs showed that 40 ncRNAs changed significantly during silkworm development or across specific stage transitions, suggesting roles for these ncRNAs in silkworm development.
Silkworm specific ncRNAs
Most of the novel ncRNAs identified in this study were not conserved in other organisms, indicating that there are a large number of silkworm-specific ncRNA loci in the genome, which is consistent with previous results in other organisms. Expression profiles, structural analysis and target prediction showed that these were bona fide ncRNAs, rather than transcriptional noise. Although many ncRNAs are conserved in different species (43), there also exists a large number of species-specific ncRNAs (20,23,44).
Based on secondary structures and functional antisense elements, the novel snoRNAs were predicted to direct modification of a total of 95 methylation and pseudouridylation sites in rRNAs, snRNAs and tRNAs, providing the first list of potential RNA modification sites guided by snoRNAs in B. mori. Consistent with previous results (45,46), the majority of the silkworm-specific snoRNAs were predicted to direct modifications within the phylogenetically most conserved regions of the rRNA sequences. Although rRNA modifications may be vital to rRNA function (47), single site modification generally only has small beneficial effect on the folding of the rRNA (37,48), and apparent phenotypes generally appear only after modifications of three or more sites are blocked (49). Thus, most modification sites might serve synergistically to maintain accurate and efficient ribosomal function, rather than each exerting appreciable effects alone.
The reason for the high number of apparently species-specific snoRNAs in the silkworm is not immediately clear. An intriguing possibility is that the large amount of proteins required for the formation of cocoon lead to the evolution of efficient ribosomes with high protein output through novel modification sites within the rRNA sequences. The microarray results showed that 7 of 15 silkworm-specific snoRNAs with predicted targets showed higher expression during pupation than in the larva and adult stages, possibly suggesting their involvement in optimizing the rRNAs for cocoon production. Alternatively, the explanation may be found in the habitat environment of the Bombyx ancestral species. It has been shown that plants and trypanosomes that are exposed to large temperature changes, harbor more species-specific snoRNAs and correspondingly more novel modifications sites (43,50). B. mori was domesticated ∼5000 years ago from the Chinese wild-silkworm, B. mandarian, and both B. mori and its ancestor B. mandarian were subject to large temperature changes during their evolution, which may have been the driving force of evolution of a larger number of silkworm-specific snoRNAs.
Consistent with findings in Drosophila, Arabidopsis and mouse (20,29,51), there were 70 orphan snoRNAs (31 C/D box and 37 H/ACA box snoRNAs and two C/D-H/ACA hybrid snoRNAs) for which no target could be predicted in B. mori. A number of these showed differential expression across various stages, indicating roles in the developmental or regulatory program of the silkworm other than RNA modification.
Interestingly, among the novel ncRNAs, we found that nine ncRNAs matched small RNAs either perfectly or by one to three mismatches when aligned to a recently reported small RNA library (Supplementary Table S6 and Figure S8). These matched small RNAs are either uniquely located within ncRNA gene identified in this study, or matched to repetitive sequences according to the newly released silkworm genome database, which indicated that several ncRNAs may turn out to be precursors of small RNAs and stably exist in cells. With respect to the classification by Kawaoka et al. (28), these small RNAs may be piRNAs specifically expressed in ovary, and it is reasonable that some stable precursor to piRNAs could present in our library due to the usage of mixed tissues. These ncRNAs, like Bm-3 and Bm-6, Bm-31, Bm-33, Bm-34, Bm-49, Bm-113 and Bm-137, are predicted snoRNAs, reminiscent of previously reported miRNAs processed through snoRNAs (52–54). Among these ncRNAs, we confirmed that Bm-31and Bm-34 are bona fide ncRNAs by northern blot. ncRNAs have been predicted or experimentally proved to be precursors to small RNAs, therefore, adopting ncRNA as precursor seems a general mechanism for different small RNAs, although most of the mechanism remains unclear (52–59). These matched small RNAs (probably piRNAs) are more likely to arise from ncRNAs and function to silence the selfish genetic elements in silkworm, and if so, this may contribute to the diverse biogenesis pathway of piRNAs in animal.
Novel snoRNAs potentially guides tRNA and mRNA modification
Of particular interest were a few snoRNAs with a potential to target either tRNAs or mRNAs. Two canonical C/D box snoRNAs and one U3 snoRNA were predicted to direct 2′-O-methylation of four tRNAs. Methylation of tRNAs is generally catalyzed by methyltransferases, however, recent evidence suggests that some Archaean tRNA methylation events are mediated both by methyltransferases and by snoRNA guided action (60–62). Also in C. elegans, five tRNAs were predicted to be modified by snoRNAs (63). Furthermore, in human and yeast, tRNAs were found to pass transiently through the nucleolus during its maturation, further indicating involvement of snoRNPs in the modification or other processing events of tRNAs in eukaryotes (64,65). Among the three novel snoRNAs carrying 12 nt long-sequence elements that are perfectly complementary to three different mRNAs (Figure 6C), Bm-15 potentially targets a notch family member (BGIBMGA011962-TA), which could participate in organ morphogenesis and developmental processes. Our results showed that Bm-15 interacted with BGIBMGA011962-TA in vitro, which suggested the regulatory role of Bm-15. Small non-coding RNAs have been shown to regulate gene expression by means of mediating mRNA decay, repressing mRNA translation, involving in pre-mRNA splicing or alternative splicing (13,33,34,66–69). Although we could not detect the expression of BGIBMGA011962-TA in egg, larva and pupa stage, whereas it was expressed in adult stage during which Bm-15 exhibited high expression level (data not shown). Moreover, experiments with RT–PCR showed that BGIBMGA011962-TA had no splicing variants in various developmental stages analyzed (data not shown). Taken together, our results suggested that Bm-15 might be involved in regulating the expression of BGIBMGA011962-TA in silkworm development; however, the mechanism through which Bm-15 may regulate the expression of BGIBMGA011962-TA remains to be determined.
Two major genomic distribution patterns
The genomic organization of snoRNA genes displays great diversity among different eukaryotes (19,70–73). In yeast, the majority of the snoRNAs originates from single genes with independent promoters (74,75), while almost all Drosophila and vertebrates snoRNAs are transcribed from nested loci within introns of protein coding genes (19,20,51). The B. mori snoRNAs exhibit a more complex genomic organization, in that 32.62% of B. mori snoRNA loci are located in intergenic regions. The intronic snoRNAs in B. mori follow the pattern of one-snoRNA-per-intron, which is prevalent in vertebrates, and distinct from the intronic clusters of H/ACA box snoRNAs seen in Drosophila. Many intronic snoRNA genes of vertebrates and Drosophila are nested within genes encoding proteins involved in ribosome biogenesis. These intronic snoRNAs are coordinately expressed with their host genes, and probably implicated in the same cellular processes (43). The B. mori intronic snoRNAs also tend to be located in genes associated with the translational machinery. Compared to the numerous intronic clusters in Drosophila, only two intergenic snoRNA clusters were found, suggesting that the genomic organization of snoRNA loci in B. mori may not include many clusters.
It was found that 22–26% of human (76,77), 14.9–29% of mouse (39,78,79), 15.4–16.8% of Drosophila (80), 8.9% of Arabidopsis (81) and 7% of rice genes (82) are transcribed as sense–antisense gene pairs, and that the ‘antisense’ transcripts can regulate the expression of the ‘sense’ genes through genomic imprinting, RNA interference and translational regulation (83–86). Moreover, transcription of RNAs located antisense to introns of coding genes have been shown to interfere with pre-mRNA splicing (87,88), and thereby to regulate the expression of coding genes. We found that four unclassified ncRNAs are located antisense to introns of protein-coding genes. For instance, Bm-152 is located antisense to the fourth intron of BGIBMGA007380-TA, a coding gene involved in ER to Golgi vesicle-mediated transport and ARFs protein signal transduction. Bm-152 showed higher expression in egg (in the microarray data), whereas BGIBMGA007380-TA was hardly detected at this stage (Figure 3E), whereas in the subsequent three stages (larva through adult), these two genes showed similar expression patterns. Another intronic antisense ncRNA, Bm-151, and its sense coding gene BGIBMGA007175-TA, also exhibited a converse expression pattern in the egg stage, but showed similar expression patterns after hatching.
Developmentally regulated expression of novel ncRNAs in B. mori
It is well accepted that ncRNAs play regulatory roles in development and stress response (5,6,89–91). Zhang et al. (22) found that the expression of many silkworm miRNAs were either egg- or pupa-specific, suggesting their participation in embryogenesis or metamorphosis. Other ncRNAs are also related to development, stress response or disease. For example, snoRNAs usually exhibit stable levels of expression during growth, yet in human and mouse, some H/ACA box and C/D box snoRNAs, like HBI-36 and HBII-52 are brain-specific and related to the Prader-Willi syndrome (4). Also in our study, of the 36 ncRNAs that exhibited dynamic expressions through silkworm development, there were 10 C/D box and 13 H/ACA box snoRNAs that showed markedly altered expression at specific stages, suggesting their involvement in silkworm development. Similarly, when the transitions between specific stages were examined, several of the snoRNAs showed significant changes in expression. Moreover, many of the unclassified ncRNAs were also dynamically expressed, and showed significant changes in expression levels during phase transitions, indicating that they may play roles during development. In Drosophila, the unclassified ncRNAs Dm-308 and Dm-65 are processed through alternative splicing, are differentially expressed, and probably play diverse roles in fly development (20). In Arabidopsis, 15 of 36 unclassified ncRNAs showed tissue-specific expression, and may participate in the regulation of plant growth (29). Our results further supported the existence of differentially expressed unclassified ncRNAs that may play important roles in silkworm development.
Of the 23 snoRNAs with markedly altered expressions across silkworm development, 14 are ‘orphan’ snoRNAs with no predicted targets. In mammals, several orphan snoRNAs are specifically expressed in brain, and mutants or knockouts were lethal (92,93). In this study, the intergenic orphan H/ACA box snoRNA Bm-102 showed significantly reduced expression in larva relative to other stages, suggesting a role for Bm-102 during embryogenesis and in the transition from pupa to adult.
Absence of known sequence motifs or structural features in a transcript does not necessarily imply absence of function. The hsrω gene in D. melanogaster, produces several non-coding transcripts in a developmentally regulated manner in nearly all cell types, and the expression of hsrω increases dramatically by a variety of cellular stresses (94,95). In Arabidopsis, over-expression of the two ncRNAs, npc48 and npc536, affected differentiation and growth in response to abiotic stresses (17); and in maize, knockdown of the mRNA-like ncRNA zm401 was found to significantly affect the expression of genes required for pollen development, leading to aberrant development of the microspore and tapetum, and finally to male-sterility (96). Thirteen unclassified ncRNAs showed significant variation in expression during silkworm development, among which, Bm-162 and Bm-183 were highly expressed in egg, and showed reduced expression through larva to adult, while Bm-158 significantly accumulated in egg, pupa and adult relative to larva. Their developmentally dynamic expression patterns suggest regulation and possibly regulatory activity.
Zhang et al. (44) found that ncRNAs accumulated differentially in chicken tissues during a specific developmental stage, and might thus be involved in the regulation of divergent muscle growth. None of the analyzed B. mori ncRNAs showed much variation in expression during the course of larva development, however, the expression of several transcripts (Bm-51, Bm-86, Bm-152 and Bm-160) gradually decreased during pupa stage (Figure 3D), and might be involved in the transition from pupa to adult.
Collectively, our results represent the first genome-wide survey of small non-coding RNAs in the size range of 50–500 nt in B. mori. Our data suggest that there are a number of species-specific ncRNA loci in the silkworm genome, many of which show differential expression and may be involved in the silkworm developmental regulation. The data enabled the prediction of 95 modification sites in rRNAs, snRNAs and tRNAs. Analysis of the genomic organization suggested that nearly 33% of the identified novel snoRNAs were independently transcribed from intergenic regions, and the remaining being intronic and following a pattern of one-snoRNA-per-intron. This study thus provides a basis for future investigations of the functions of ncRNAs in insects.
ACCESSION NUMBERS
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Basic Research Program of China (2009CB825400); National Science Foundation of China (30700070); National High-Tech Research and Development Program of China (2006AA10A119). Funding for open access charge: National Science Foundation of China (30700070).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Jian-kang Zhu for his critical reading of the article, Prof. Lianghu Qu for his kind discussion about the classification of snoRNAs; Dr Yijun Qi for his kind help with EMSA; Dr Muwang Li for providing the B. mori Dazao P50 strain; and Yinghao Cao, Beibei Chen and Yalong Xu for assistance with the bioinformatics analysis.
REFERENCES
- 1.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Godfrey AC, Kupsco JM, Burch BD, Zimmerman RM, Dominski Z, Marzluff WF, Duronio RJ. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis. RNA. 2006;12:396–409. doi: 10.1261/rna.2270406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Runte M, Varon R, Horn D, Horsthemke B, Buiting K. Exclusion of the C/D box snoRNA gene cluster HBII-52 from a major role in Prader-Willi syndrome. Hum. Genet. 2005;116:228–230. doi: 10.1007/s00439-004-1219-2. [DOI] [PubMed] [Google Scholar]
- 5.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 6.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Meller VH, Rattner BP. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 2002;21:1084–1091. doi: 10.1093/emboj/21.5.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelley RL. Path to equality strewn with roX. Dev. Biol. 2004;269:18–25. doi: 10.1016/j.ydbio.2004.01.039. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 10.Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat. Struct. Mol. Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 11.Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O'Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 12.Vitali P, Basyuk E, Le Meur E, Bertrand E, Muscatelli F, Cavaille J, Huttenhofer A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J. Cell Biol. 2005;169:745–753. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura A, Amikura R, Mukai M, Kobayashi S, Lasko PF. Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science. 1996;274:2075–2079. doi: 10.1126/science.274.5295.2075. [DOI] [PubMed] [Google Scholar]
- 15.Hardiman KE, Brewster R, Khan SM, Deo M, Bodmer R. The bereft gene, a potential target of the neural selector gene cut, contributes to bristle morphogenesis. Genetics. 2002;161:231–247. doi: 10.1093/genetics/161.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ben Amor B, Wirth S, Merchan F, Laporte P, d'Aubenton-Carafa Y, Hirsch J, Maizel A, Mallory A, Lucas A, Deragon JM, et al. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 2009;19:57–69. doi: 10.1101/gr.080275.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, Zhao P, Zha X, Cheng T, Chai C, et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- 19.Huang ZP, Zhou H, He HL, Chen CL, Liang D, Qu LH. Genome-wide analyses of two families of snoRNA genes from Drosophila melanogaster, demonstrating the extensive utilization of introns for coding of snoRNAs. RNA. 2005;11:1303–1316. doi: 10.1261/rna.2380905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan G, Klambt C, Bachellerie JP, Brosius J, Huttenhofer A. RNomics in Drosophila melanogaster: identification of 66 candidates for novel non-messenger RNAs. Nucleic Acids Res. 2003;31:2495–2507. doi: 10.1093/nar/gkg361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu X, Zhou Q, Li SC, Luo Q, Cai Y, Lin WC, Chen H, Yang Y, Hu S, Yu J. The silkworm (Bombyx mori) microRNAs and their expressions in multiple developmental stages. PLoS ONE. 2008;3:e2997. doi: 10.1371/journal.pone.0002997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Zhou X, Ge X, Jiang J, Li M, Jia S, Yang X, Kan Y, Miao X, Zhao G, et al. Insect-Specific microRNA Involved in the Development of the Silkworm Bombyx mori. PLoS One. 2009;4:e4677. doi: 10.1371/journal.pone.0004677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng W, Zhu X, Skogerbo G, Zhao Y, Fu Z, Wang Y, He H, Cai L, Sun H, Liu C, et al. Organization of the Caenorhabditis elegans small non-coding transcriptome: genomic features, biogenesis, and expression. Genome Res. 2006;16:20–29. doi: 10.1101/gr.4139206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 26.Yang JH, Zhang XC, Huang ZP, Zhou H, Huang MB, Zhang S, Chen YQ, Qu LH. snoSeeker: an advanced computational package for screening of guide and orphan snoRNA genes in the human genome. Nucleic Acids Res. 2006;34:5112–5123. doi: 10.1093/nar/gkl672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jady BE, Kiss T. A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 2001;20:541–551. doi: 10.1093/emboj/20.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawaoka S, Hayashi N, Katsuma S, Kishino H, Kohara Y, Mita K, Shimada T. Bombyx small RNAs: genomic defense system against transposons in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2008;38:1058–1065. doi: 10.1016/j.ibmb.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 29.Marker C, Zemann A, Terhorst T, Kiefmann M, Kastenmayer JP, Green P, Bachellerie JP, Brosius J, Huttenhofer A. Experimental RNomics: identification of 140 candidates for small non-messenger RNAs in the plant Arabidopsis thaliana. Curr. Biol. 2002;12:2002–2013. doi: 10.1016/s0960-9822(02)01304-0. [DOI] [PubMed] [Google Scholar]
- 30.Hirose T, Shu MD, Steitz JA. Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol. Cell. 2003;12:113–123. doi: 10.1016/s1097-2765(03)00267-3. [DOI] [PubMed] [Google Scholar]
- 31.Ooi SL, Samarsky DA, Fournier MJ, Boeke JD. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. RNA. 1998;4:1096–1110. doi: 10.1017/s1355838298980785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hertel J, Hofacker IL, Stadler PF. SnoReport: computational identification of snoRNAs with unknown targets. Bioinformatics. 2008;24:158–164. doi: 10.1093/bioinformatics/btm464. [DOI] [PubMed] [Google Scholar]
- 33.Udekwu KI, Darfeuille F, Vogel J, Reimegard J, Holmqvist E, Wagner EG. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, Vogel J. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szell M, Bata-Csorgo Z, Kemeny L. The enigmatic world of mRNA-like ncRNAs: their role in human evolution and in human diseases. Semin. Cancer Biol. 2008;18:141–148. doi: 10.1016/j.semcancer.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 36.Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu. Rev. Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- 37.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- 39.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 40.Okada Y, Tashiro C, Numata K, Watanabe K, Nakaoka H, Yamamoto N, Okubo K, Ikeda R, Saito R, Kanai A, et al. Comparative expression analysis uncovers novel features of endogenous antisense transcription. Hum. Mol. Genet. 2008;17:1631–1640. doi: 10.1093/hmg/ddn051. [DOI] [PubMed] [Google Scholar]
- 41.Pang KC, Stephen S, Dinger ME, Engstrom PG, Lenhard B, Mattick JS. RNAdb 2.0–an expanded database of mammalian non-coding RNAs. Nucleic Acids Res. 2007;35:D178–D182. doi: 10.1093/nar/gkl926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown JW, Echeverria M, Qu LH. Plant snoRNAs: functional evolution and new modes of gene expression. Trends Plant Sci. 2003;8:42–49. doi: 10.1016/s1360-1385(02)00007-9. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Wang J, Huang S, Zhu X, Liu J, Yang N, Song D, Wu R, Deng W, Skogerbo G, et al. Systematic identification and characterization of chicken (Gallus gallus) ncRNAs. Nucleic Acids Res. 2009;37:6562–6574. doi: 10.1093/nar/gkp704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reddy R, Busch H. Small nuclear RNAs and RNA processing. Prog. Nucleic Acid Res. Mol. Biol. 1983;30:127–162. doi: 10.1016/s0079-6603(08)60685-6. [DOI] [PubMed] [Google Scholar]
- 46.Maden BE. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 47.Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 48.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 49.Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 50.Liang XH, Uliel S, Hury A, Barth S, Doniger T, Unger R, Michaeli S. A genome-wide analysis of C/D and H/ACA-like small nucleolar RNAs in Trypanosoma brucei reveals a trypanosome-specific pattern of rRNA modification. RNA. 2005;11:619–645. doi: 10.1261/rna.7174805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huttenhofer A, Kiefmann M, Meier-Ewert S, O'Brien J, Lehrach H, Bachellerie JP, Brosius J. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 2001;20:2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput. Biol. 2009;5:e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 55.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Luo J, Zhou H, Liao JY, Ma LM, Chen YQ, Qu LH. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008;36:6048–6055. doi: 10.1093/nar/gkn596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Renalier MH, Joseph N, Gaspin C, Thebault P, Mougin A. The Cm56 tRNA modification in archaea is catalyzed either by a specific 2′-O-methylase, or a C/D sRNP. RNA. 2005;11:1051–1063. doi: 10.1261/rna.2110805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh SK, Gurha P, Tran EJ, Maxwell ES, Gupta R. Sequential 2′-O-methylation of archaeal pre-tRNATrp nucleotides is guided by the intron-encoded but trans-acting box C/D ribonucleoprotein of pre-tRNA. J. Biol. Chem. 2004;279:47661–47671. doi: 10.1074/jbc.M408868200. [DOI] [PubMed] [Google Scholar]
- 62.Singh SK, Gurha P, Gupta R. Dynamic guide-target interactions contribute to sequential 2′-O-methylation by a unique archaeal dual guide box C/D sRNP. RNA. 2008;14:1411–1423. doi: 10.1261/rna.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zemann A, op de Bekke A, Kiefmann M, Brosius J, Schmitz J. Evolution of small nucleolar RNAs in nematodes. Nucleic Acids Res. 2006;34:2676–2685. doi: 10.1093/nar/gkl359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bertrand E, Houser-Scott F, Kendall A, Singer RH, Engelke DR. Nucleolar localization of early tRNA processing. Genes Dev. 1998;12:2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ko YG, Kang YS, Kim EK, Park SG, Kim S. Nucleolar localization of human methionyl-tRNA synthetase and its role in ribosomal RNA synthesis. J. Cell Biol. 2000;149:567–574. doi: 10.1083/jcb.149.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Semenov DV, Vratskih OV, Kuligina EV, Richter VA. Splicing by exon exclusion impaired by artificial box C/D RNA targeted to branch-point adenosine. Ann. NY Acad. Sci. 2008;1137:119–124. doi: 10.1196/annals.1448.037. [DOI] [PubMed] [Google Scholar]
- 67.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol. Cell. 2008;32:827–837. doi: 10.1016/j.molcel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 69.Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 2009;16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 70.Qu LH, Meng Q, Zhou H, Chen YQ. Identification of 10 novel snoRNA gene clusters from Arabidopsis thaliana. Nucleic Acids Res. 2001;29:1623–1630. doi: 10.1093/nar/29.7.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu Y, Zhou H, Zhou W, Zhu Y, Qu L. A novel snoRNA gene cluster in yeast is transcribed as polycistronic pre-snoRNAs. Sci. China C Life Sci. 1999;42:529–537. doi: 10.1007/BF02881777. [DOI] [PubMed] [Google Scholar]
- 72.Liang D, Zhou H, Zhang P, Chen YQ, Chen X, Chen CL, Qu LH. A novel gene organization: intronic snoRNA gene clusters from Oryza sativa. Nucleic Acids Res. 2002;30:3262–3272. doi: 10.1093/nar/gkf426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shao P, Yang JH, Zhou H, Guan DG, Qu LH. Genome-wide analysis of chicken snoRNAs provides unique implications for the evolution of vertebrate snoRNAs. BMC Genomics. 2009;10:86. doi: 10.1186/1471-2164-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li SG, Zhou H, Luo YP, Zhang P, Qu LH. Identification and functional analysis of 20 Box H/ACA small nucleolar RNAs (snoRNAs) from Schizosaccharomyces pombe. J. Biol. Chem. 2005;280:16446–16455. doi: 10.1074/jbc.M500326200. [DOI] [PubMed] [Google Scholar]
- 75.Schattner P, Decatur WA, Davis CA, Ares M, Jr, Fournier MJ, Lowe TM. Genome-wide searching for pseudouridylation guide snoRNAs: analysis of the Saccharomyces cerevisiae genome. Nucleic Acids Res. 2004;32:4281–4296. doi: 10.1093/nar/gkh768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, et al. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 77.Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y, Liu XS, Liu QR, Wei L. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res. 2006;34:3465–3475. doi: 10.1093/nar/gkl473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kiyosawa H, Yamanaka I, Osato N, Kondo S, Hayashizaki Y. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 2003;13:1324–1334. doi: 10.1101/gr.982903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS, Hradecky P, Huang Y, Kaminker JS, Millburn GH, Prochnik SE, et al. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 2002;3:RESEARCH0083. doi: 10.1186/gb-2002-3-12-research0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang XJ, Gaasterland T, Chua NH. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 2005;6:R30. doi: 10.1186/gb-2005-6-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Osato N, Yamada H, Satoh K, Ooka H, Yamamoto M, Suzuki K, Kawai J, Carninci P, Ohtomo Y, Murakami K, et al. Antisense transcripts with rice full-length cDNAs. Genome Biol. 2003;5:R5. doi: 10.1186/gb-2003-5-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Knee R, Murphy PR. Regulation of gene expression by natural antisense RNA transcripts. Neurochem. Int. 1997;31:379–392. doi: 10.1016/s0197-0186(96)00108-8. [DOI] [PubMed] [Google Scholar]
- 84.Kumar M, Carmichael GG. Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev. 1998;62:1415–1434. doi: 10.1128/mmbr.62.4.1415-1434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rougeulle C, Heard E. Antisense RNA in imprinting: spreading silence through Air. Trends Genet. 2002;18:434–437. doi: 10.1016/s0168-9525(02)02749-x. [DOI] [PubMed] [Google Scholar]
- 86.Brantl S. Antisense-RNA regulation and RNA interference. Biochim. Biophys. Acta. 2002;1575:15–25. doi: 10.1016/s0167-4781(02)00280-4. [DOI] [PubMed] [Google Scholar]
- 87.Volloch V, Schweitzer B, Rits S. Inhibition of pre-mRNA splicing by antisense RNA in vitro: effect of RNA containing sequences complementary to introns. Biochem. Biophys. Res. Commun. 1991;179:1600–1605. doi: 10.1016/0006-291x(91)91757-4. [DOI] [PubMed] [Google Scholar]
- 88.Munroe SH. Antisense RNA inhibits splicing of pre-mRNA in vitro. EMBO J. 1988;7:2523–2532. doi: 10.1002/j.1460-2075.1988.tb03100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.St Laurent G, 3rd, Faghihi MA, Wahlestedt C. Non-coding RNA transcripts: sensors of neuronal stress, modulators of synaptic plasticity, and agents of change in the onset of Alzheimer's disease. Neurosci. Lett. 2009;466:81–88. doi: 10.1016/j.neulet.2009.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brookheart RT, Michel CI, Listenberger LL, Ory DS, Schaffer JE. The non-coding RNA gadd7 is a regulator of lipid-induced oxidative and endoplasmic reticulum stress. J. Biol. Chem. 2009;284:7446–7454. doi: 10.1074/jbc.M806209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nakamura T, Naito K, Yokota N, Sugita C, Sugita M. A cyanobacterial non-coding RNA, Yfr1, is required for growth under multiple stress conditions. Plant Cell Physiol. 2007;48:1309–1318. doi: 10.1093/pcp/pcm098. [DOI] [PubMed] [Google Scholar]
- 92.Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Huttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cavaille J, Vitali P, Basyuk E, Huttenhofer A, Bachellerie JP. A novel brain-specific box C/D small nucleolar RNA processed from tandemly repeated introns of a noncoding RNA gene in rats. J. Biol. Chem. 2001;276:26374–26383. doi: 10.1074/jbc.M103544200. [DOI] [PubMed] [Google Scholar]
- 94.Mallik M, Lakhotia SC. The developmentally active and stress-inducible non-coding hsr{omega} gene is a novel regulator of apoptosis in Drosophila. Genetics. 2009;183:831–852. doi: 10.1534/genetics.109.108571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Savvateeva-Popova E, Medvedeva A, Popov A, Evgen'ev M. Role of non-coding RNAs in neurodegeneration and stress response in Drosophila. Biotechnol. J. 2008;3:1010–1021. doi: 10.1002/biot.200800120. [DOI] [PubMed] [Google Scholar]
- 96.Ma J, Yan B, Qu Y, Qin F, Yang Y, Hao X, Yu J, Zhao Q, Zhu D, Ao G. Zm401, a short-open reading-frame mRNA or noncoding RNA, is essential for tapetum and microspore development and can regulate the floret formation in maize. J. Cell Biochem. 2008;105:136–146. doi: 10.1002/jcb.21807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.