Abstract
The nonsense-mediated mRNA decay (NMD) pathway is a highly conserved surveillance mechanism that is present in all eukaryotes. It prevents the synthesis of truncated proteins by selectively degrading mRNAs harbouring premature termination codons (PTCs). The core NMD effectors were originally identified in genetic screens in Saccharomyces cerevisae and in the nematode Caenorhabditis elegans, and subsequently by homology searches in other metazoans. A genome-wide RNAi screen in C. elegans resulted in the identification of two novel NMD genes that are essential for proper embryonic development. Their human orthologues, DHX34 and NAG/NBAS, are required for NMD in human cells. Here, we find that the zebrafish genome encodes orthologues of DHX34 and NAG/NBAS. We show that the morpholino-induced depletion of zebrafish Dhx34 and Nbas proteins results in severe developmental defects and reduced embryonic viability. We also found that Dhx34 and Nbas are required for degradation of PTC-containing mRNAs in zebrafish embryos. The phenotypes observed in both Dhx34 and Nbas morphants are similar to defects in Upf1, Smg-5- or Smg-6- depleted embryos, suggesting that these factors affect the same pathway and confirming that zebrafish embryogenesis requires an active NMD pathway.
INTRODUCTION
The nonsense-mediated decay (NMD) pathway plays a central role in the control of gene expression. It is an elaborate surveillance mechanism that triggers the degradation of mRNAs containing premature termination codons (PTCs). By doing so, it prevents the accumulation of truncated proteins that may interfere with cellular function [reviewed by refs. (1,2)]. The importance of NMD is highlighted by the fact that it not only regulates transcripts harboring nonsense mutations but also regulates an important fraction of the transcriptome. Gene expression profiles in Saccharomyces cerevisae, Drosophila melanogaster and human cells-lacking individual NMD factors indicated that up to 10% of cellular transcripts are regulated by NMD (3–6). Approximately, a third of all inherited genetic disorders are caused by frameshift or nonsense mutations giving rise to PTCs. Thus, NMD may act to modulate the phenotypic outcome of this type of diseases with NMD factors being potential targets for therapeutic intervention (7,8).
NMD effectors were originally identified in genetic screens conducted in both S. cerevisae and Caenorhabditis elegans. Seven genes with a central role in NMD were identified in nematodes. In addition to their NMD phenotype, mutations of these genes cause abnormal morphogenesis of the male bursa and the hermaphrodite vulva; for this reason they were named smg-1-7 (for suppressor with morphological effect on genitalia) (9–11). In S. cerevisae, three NMD genes, termed UPF1-3 (for up-frameshift) are orthologues of the C. elegans smg-2, smg-3 and smg-4 (12,13). Subsequently, orthologues for the smg genes have been identified in several species [reviewed by ref. (14)]. Only recently, four additional NMD factors have been identified in C. elegans and in human cells: NAG/NBAS, DHX34, SMG-8 and SMG-9 (15). NAG/NBAS (for neuroblastoma amplified gene/neuroblastoma amplified sequence) and DHX34 are the human homologues of C. elegans smgl-1 and smgl-2, two essential NMD genes that were identified in a genome-wide RNAi screen. Both the nematode and human genes have been shown to function in the NMD pathway (16).
A central component of the NMD pathway is the protein UPF1/SMG-2, an ATP-dependent RNA helicase that undergoes cycles of phosphorylation/dephosphorylation that are essential for NMD. Phosphorylation of UPF1/SMG-2 is carried out by SMG-1, a member of the phosphatidylinositol kinase superfamily of protein kinases (17,18). Both SMG-8 and SMG-9 were identified as subunits of the SMG-1 kinase complex and shown to be required for NMD in both humans and nematodes (19). All other known SMG genes affect the state of SMG-2 phosphorylation, with SMG-1, SMG-3 and SMG-4 being required for SMG-2 phosphorylation. By contrast, SMG-5, SMG-6 and SMG-7 have a role in SMG-2 dephosphorylation in a process that also involves the catalytic and structural subunits of protein phosphatase 2A (PP2A) (20,21). Recently, it was found that SMG-6 initiates endonucleolytic cleavage in the vicinity of the PTC in both D. melanogaster and in humans. These studies highlighted that endonucleolytic cleavage is a conserved feature in metazoan NMD (22,23). A link between the NMD machinery and the general decay complex may be provided by the human proline-rich nuclear receptor coregulatory protein 2 (PNRC2), which was shown to interact with both hyperphosphorylated UPF1- and the decapping activator, DCP1a (24). Furthermore, SMG-7 links mRNA surveillance and mRNA decay by forming a complex with SMG-5 and UPF1 and targeting bound mRNAs for mRNA decay (25). A surveillance complex, termed SURF, comprising SMG-1, UPF1 and the eukaryotic release factors eRF1and eRF3 is formed on mRNAs during recognition of a PTC. This step is followed by the interaction of the SURF complex with UPF2, UPF3 and components of the EJC that are bound to a downstream exon–exon boundary. This results in the formation of a decay-inducing complex (DECID) that triggers UPF1 phosphorylation and the dissociation of eRF1,3 (26) [reviewed by ref. (27)]. Interestingly, UPF1 can interact with the exon junction complex (EJC) either through UPF2 or UPF3b, indicating the existence of alternative branches of the NMD pathway (28,29).
A critical aspect of the NMD pathway is how to distinguish a premature from a normal termination codon. Although for the most part NMD effectors are conserved throughout evolution, different strategies have been found for PTC definition. In mammalian cells, pre-mRNA splicing and NMD are intimately linked, and the position of a termination codon relative to the last intron will determine whether it is interpreted as a PTC (30) [reviewed by ref. (31)]. This positional information is determined by the EJC, a multiprotein complex that is deposited on every exon junction following pre-mRNA splicing, marks the position of exon–exon boundaries and recruits NMD factors (32,33) [for a review see ref. (34)]. However, an intron-independent NMD mechanism that measures the distance between the termination codon and the poly(A)-binding protein PABPC1 acts to determine whether the stop codon will be recognized as a PTC in human cells (35–37). The requirement of exon boundaries to define a PTC has also been found in zebrafish and in plants (38–40) [for a review see ref. (41)]. In contrast, in invertebrates definition of a PTC occurs independently of exon boundaries as shown in yeast, in C. elegans and in Drosophila (16,42,43).
In this study, we focus on the functional characterization of two novel NMD factors in the zebrafish, Danio rerio. We found that the zebrafish genome encodes orthologues of NAG/NBAS/smgl-1 and DHX34/smgl-2. These factors were identified in a genome-wide RNAi screen in C. elegans and shown to function in the NMD pathway in both nematodes and human cells. Here, we show that both genes are required for zebrafish development and function in the NMD pathway in Danio rerio. The phenotypes observed in both nbas and dhx34 morphants are similar to those described when depleting NMD core factors, such as Upf1, Smg-5- or Smg-6 (38). This strongly suggests that these genes affect the same pathway and confirm that zebrafish embryogenesis requires an active NMD pathway.
MATERIALS AND METHODS
Animal husbandry
Zebrafish embryos and adults were raised and maintained at 28.5°C on a 12 h light/dark cycle. Embryos were collected by pair matings of a wild-type stain (AB-TPL) and the Slc24a5−/− (Golden) line and were staged as described previously (44).
Microinjection of embryos
Injections were performed on wild-type and slc24a5−/− zebrafish embryos using a nitrogen-powered Picospritzer III microinjector (Intracel) conjugated to a Nikon SMZ 1000 stereomicroscope. One- to four-cell stage embryos were injected with 4 nl of 250 µM translation block (TB) morpholino oligonucleotides (MOs) against upf1 (38), dhx34 and nbas, splice-site MOs against dhx34 and nbas, as well as their respective 5 bp-mismatch control MOs (Gene-Tools LLC). MO sequences used are provided on Supplementary Table S1. For the mRNA rescue experiments, 250 µM of TB and splice-site Dhx34 and Nbas MOs were co-injected with 35 pg of capped DHX34 and NAG/NBAS human mRNA, respectively, in a total volume of 4 nl. Capped human DHX34 and NAG/NBAS mRNAs were synthesized using the mMessage mMachine T7 kit (Ambion) according to the manufacturer's instructions.
Wholemount in situ hybridization of morphants
Zebrafish embryos [24 hours post-fertilization (hpf)] were dechorionated manually and fixed overnight in 4% parafolmaldehyde/PBS at 4°C. The embryos were serially dehydrated and stored overnight in methanol at −20°C. Antisense and sense dhx34 and nbas mRNA probes were synthesized and digoxigenin-labelled in vitro (Roche) and whole-mount in situ hybridization was carried out as previously described (45). An anti-digoxigenin antibody was incubated in a 1:5000 dilution overnight and the samples were stained in BM Purple alkaline phosphatase (Roche) for 2 h. The reaction was stopped in 20 mM EDTA/PBS. Processed embryos were mounted in 100% glycerol and were imaged using a Nikon SMZ1500 stereomicroscope in conjunction with a Q-imaging camera.
RNA isolation and RT- PCR
Control and MO-injected embryos (24 hpf) were manually dechorionated and directly homogenized in 500 µl TRIZOL (Invitrogen) using a 25G needle. RNA was isolated following manufacturer's instructions. Real-time RT–PCR was performed using target-specific primers and SuperScriptIII Platinum SYBR Green One-Step qRT-PCR Kit (Invitrogen). Real-time RT–PCT was run on Bio-Rad CFX96 cycler. Total RNA (300 ng) was used in 20 µl reaction, and each sample was run in triplicates. Data analysis were performed using the Bio-Rad CFX manager software. For the analysis of dhx34 and nag splicing defects induced by the splice-site targeting MOs, RNA was amplified using target-specific primers and SuperScriptIII One-Step RT-PCR kit (Invitrogen). PCR products were resolved on 1.5% agarose gel. Oligonucleotide sequences are provided on Supplementary Table S2.
RESULTS
Dhx34 and Nbas, which are highly conserved between zebrafish and humans, are expressed throughout zebrafish development
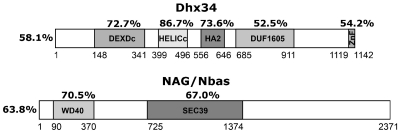
We have previously shown that DHX34 and NAG/NBAS are highly conserved between C. elegans and humans and function as NMD factors in both organisms (16). Here, we identified the zebrafish homologues of these NMD factors and found that the zebrafish proteins harbour the same functional domains of their human counterparts and share a high degree of conservation (Figure 1). The human DHX34 protein is a member of the ATP-dependent RNA helicase family and displays four protein domains typically associated with ATP-dependent RNA helicases, which are present and conserved in zebrafish (Figure 1) [for reviews see refs. (46,47)]. Human NAG/NBAS was originally identified as a gene amplified in human neuroblastomas (48,49). It contains WD40 repeats (β-propeller domain; PFAM domain PF00400) in the N-terminal part of the protein, and a SEC39 domain (PFAM domain PF08314) found in proteins involved in the secretory pathway. Both of these domains are also conserved in the zebrafish protein (Figure 1).
Figure 1.
Dhx34 and Nbas are conserved between zebrafish and human. Schematic representation of the zebrafish Dhx34 and Nbas proteins. The shaded boxes represent the regions of sequence conservation within proteins; the level of conservation is indicated. Functional domains associated with ATP-dependent RNA helicases in Dhx34 (DEXDc, HELICc, PFAM:HA2 and PFAM:DUF 1605) are indicated. Predicted functional domains of the Nbas protein are indicated.
To address the role of dhx34 and nbas in vertebrate development, we first examined their RNA expression patterns by wholemount RNA in situ hybridization. We found dhx34 and nbas to be expressed during early development (1000-cell stage) prior to gastrulation, as well as following gastrulation cell movements at 10 hpf when the embryo form starts to become apparent (Figure 2). Expression of dhx34 appeared throughout the embryo at these stages, while we observed a slight gradient of nbas expression towards the posterior end of the embryo at 10 hpf. No expression of dhx34 or nbas was detected in the yolk of the zebrafish, nor with the sense mRNA control, indicating the specificity of RNA expression in the embryo. Thus, like other NMD effectors in zebrafish, dhx34 and nbas are expressed ubiquitously during early development (38).
Figure 2.
Dhx34 and Nbas are ubiquitously expressed during zebrafish embryogenesis. Wholemount in situ hybridization against dhx34 and nbas using antisense riboprobes revealed ubiquitous expression of dhx34 and nbas transcripts (seen as purple staining) during early zebrafish development. No significant staining was detected using the control sense probes.
Dhx34 and Nbas are essential for zebrafish development
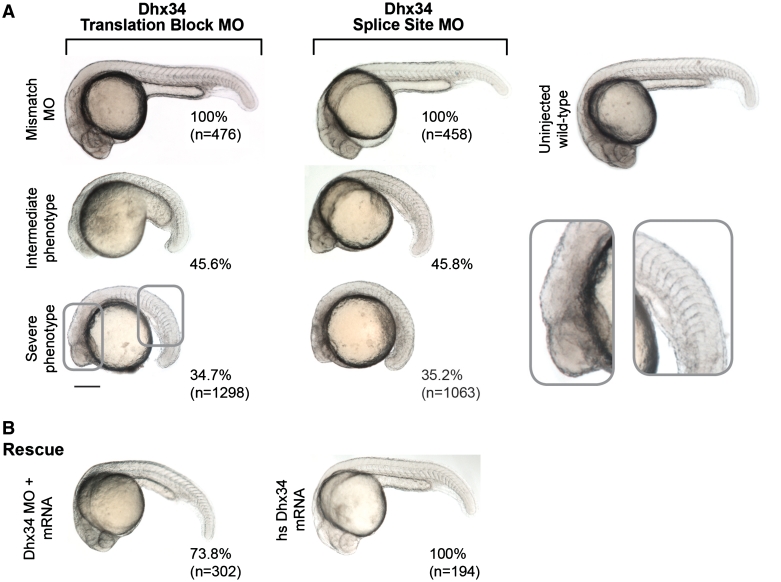
In the developing zebrafish, the core NMD factors (Upf1, Upf2, Smg5, Smg6 and Smg7) have an essential role during zebrafish embryogenesis (38). However, the role of the recently identified NMD factors Dhx34 and Nbas in vertebrate development is unknown. To address this question, we injected zebrafish embryos with TB or splice-site MOs to prevent the production of functional Dhx34 or Nbas during zebrafish development. First, we recapitulated previous experiments and showed that TB MO that prevent the production of functional Upf1 severely affected zebrafish development. As reported previously, morphants with intermediate and severe phenotypes were observed (Supplementary Figure S1) (38). As a control, we used MOs containing the same sequence as the Upf1, Dhx34 or Nbas MOs, but including a 5 bp mismatch as indicated in Supplementary Table S1. We tested the efficiency of the splice-site MOs by PCR of the injected zebrafish embryos, and confirmed their efficiency in blocking pre-mRNA splicing of the targeted gene (data not shown). We found that depletion of Dhx34 resulted in embryonic defects similar to those observed upon Upf1 depletion (Figure 3 and Supplementary Figure S1) demonstrating that this gene is essential for development. The Dhx34 morphants could be grouped into two main classes, those with an intermediate phenotype or with a severe phenotype (Figure 3A). Those with an intermediate phenotype were morphologically developmentally delayed, with aberrant formation of head and tail; however, these animals developed eye placodes and somites (TB MO n = 592/1298; splice-site block MO n = 487/1063). In contrast, animals in the severe phenotype category were severely developmentally arrested, displaying yolk extension defects, abnormal somite formation and areas of opaque tissue in the eye and brain, which is indicative of necrotic tissue (TB MO n = 451/1298; splice-site block MO n = 374/1063) (Figure 3A).
Figure 3.
Dhx34 is required for proper embryonic development in zebrafish. (A) Translation block and splice site MOs against zebrafish Dhx34 cause a distinctive range of developmental phenotypes at 24hpf characterized by shortening of the anteroposterior axis, somite malformation and significant reduction in head size. Morphants injected with the mismatch control MOs are indistinguishable from uninjected wild-type embryos. (B) Co-injection of Dhx34 translation block MO and human full-length DHX34 mRNA rescued the morphant phenotype in 73.8% of the injected embryos at 24 hpf. Expression of the mRNA alone has no overt effect on development. The numbers beside each panel indicate the percentage of embryos with the observed phenotype. Numbers in brackets correspond to the total of analysed embryos.
Next, we used MOs designed to target Nbas to determine whether this gene is required for vertebrate development. Nbas control MOs had no effect on embryonic development (5 bp mismatch TB MO n = 512/512; 5 bp mismatch splice-site MO n = 506/506). In contrast, both Nbas TB-MO and Nbas splice-site MO caused strong developmental phenotypes that were grouped into two categories (Figure 4A). The intermediate phenotype showed areas of necrosis in the brain and neural structures, and like the Dhx34 morphants, showed abnormal somites assembled like a ‘stack of pennies’ (TB MO n = 399/808; splice-site MO n = 402/866). Nbas morphants with a severe phenotype revealed embryos with few recognizable structures that had completed gastrulation stages and formed a small body on top of the yolk sac, with necrotic tissues in the anterior and posterior regions (TB MO n = 278/808; splice-site MO n = 315/866). A summary of morphant phenotypes and viability for both DhX34 and Nbas is indicated on Table 1.
Figure 4.
Nbas is essential for zebrafish embryonic development. (A) Injections with translation block and splice site MOs against zebrafish nbas reproduced the developmental phenotype observed in Upf1 and Dhx34 morphants. (B) Co-injection of Nbas translation block MO and human full length NAG/Nbas mRNA rescued the morphant phenotype in the majority of the embryos at 24 hpf. Expression of the mRNA alone had no effect on embryonic development. The numbers beside each panel represent the percentage of embryos showing each phenotype and in brackets is the total number of the analysed embryos.
Table 1.
Summary of morphant phenotypes and viability
| Severe | Intermediate (%) | Normal (%) | Morphant survival (5 dpf) (%) | Control survival (5 dpf) (%) | |
|---|---|---|---|---|---|
| Percentage of injected embryos (n) | |||||
| Dhx34 TB MO | 34.7 (1298) | 45.60 | 19.60 | 69.20 | 99.2 (476) |
| Dhx34 SS MO | 35.6 (1063) | 45.80 | 19.00 | 72.50 | 99.3 (458) |
| Dhx34 mRNA | – | – | 100 (194) | – | 92.80 |
| Dhx34 TB MO + mRNA | – | 26.10 | 73.8 (302) | 81.10 | – |
| Nbas TB MO | 34.4 (808) | 49.40 | 16.20 | 67.30 | 99.4 (512) |
| Nbas SS MO | 36.4 (866) | 46.40 | 17.20 | 73.30 | 99.0 (506) |
| Nbas mRNA | – | – | 100 (96) | – | 97.90 |
| Nbas TB MO + mRNA | – | 26.20 | 73.8 (103) | 98.70 | – |
| Upf1 TB MO | 39.2 (495) | 46.10 | 14.70 | – | – |
| Uninjected control (%) | – | – | – | – | 100.00 |
Values in parenthesis represent n.
Thus, our data demonstrated essential functions for Dhx34 and Nbas during zebrafish embryonic development. The similar embryonic phenotypes associated with both the TB and splice-site MOs for both genes strongly suggest that the phenotypes can be directly attributed to loss of gene function, rather than non-specific effects of the MOs. Notably, even though the embryonic phenotypes do not point towards specific developmental targets for Dhx34 and Nbas, the morphants share overall similarity with the Upf1, Upf2, Smg5 and Smg6 morphants, which is most apparent in the somite structures and brain necrosis (Supplementary Figure S1). This is in contrast to the somewhat more specific developmental phenotypes observed for the Smg7 and Upf3a/b morphants (38). We suggest that the pleiotropic morphant phenotypes indicate that Nbas and Dhx34 are involved in the control of multiple biological targets, and this is most likely associated with their common role with upf1, upf2, smg5 and smg6 in the NMD pathway.
Human DHX34 and NAG/NBAS compensate for loss of Dhx34 and Nbas in zebrafish development
Human DHX34 and NAG/NBAS share 58.1 and 63.8% identity to the zebrafish Dhx34 and Nbas proteins (Figure 1). We hypothesized that the function of the human and zebrafish genes are similar. To address the functional conservation between the two species, we investigated whether expression of human mRNA encoding either DHX34 or NAG/NBAS was able to rescue the developmental defects induced by MOs. As an important control, we observed no developmental abnormalities in embryos that expressed human mRNA for DHX34 (n = 194) or NAG/NBAS (n = 96) (Figures 3B and 4B, respectively). Next, we expressed human DHX34 or NAG/NBAS in zebrafish Dhx34 or Nbas morphant animals, respectively. Sufficient differences in the translation start site ensure that the Dhx34 or Nbas MOs would not be active against the human DHX34 or NAG/NBAS mRNA species. We found that human DHX34 was able to completely rescue the developmental malformations found in Dhx34 morphants (n = 223/302) (Figure 3B). A small proportion of injected embryos showed intermediate rescue with minor developmental defects (n = 79/302). Likewise, human NAG/NBAS was also able to partially (n = 27/103) and completely rescue (n = 76/103) the Nbas morphant phenotypes (Figure 4B). These experiments demonstrate that zebrafish dhx34 and nbas are the functional homologues of human DHX34 and NAG/NBAS, respectively. The ability of human DHX34 and NAG/NBAS mRNAs to rescue the severe embryonic phenotypes of the morphants further indicates that the phenotypes described are specific to the loss of Dhx34 or Nbas, rather than non-specific effects of the MO.
Dhx34 and Nbas are required for NMD in developing zebrafish
The zebrafish goldenb1 mutant carries a nonsense mutation in the fifth exon of the slc24a5 mRNA and as a result the expression of slc24a5 transcript is undetectable in the melanocyte precursor cells due to NMD-mediated degradation. Consequently, goldenb1 mutant fish are defective in melanocyte production and exhibit a hypopigmentation phenotype (50). It was previously shown that depletion of Upf1 results in upregulation of the slc24a5 nonsense transcript in goldenb1 mutant embryos (38). To conclusively demonstrate whether Dhx34 and Nbas are required for NMD in zebrafish, we tested whether MO-mediated depletion of zebrafish Dhx34 and Nbas resulted in an increase in the level of slc24a5 endogenous transcript in the golden mutant strain. The expression of slc24a5 mRNA was analysed by real-time RT–PCR and normalized to the level of the melanin biosynthesis marker dct mRNA which is also expressed in melanocyte precursor cells. First, we found that the level of slc24a5 was not affected by Dhx34 or Nbas knockdown in a wild-type strain (Figure 5A). In the goldenb1 mutant strain, we confirmed that Upf1 knockdown resulted in an ∼5-fold up-regulation of slc24a5 mRNA, whereas Upf1 mismatch control had no significant effect. Importantly, MO-induced depletion of Dhx34 and Nbas also restored the expression of the mutant transcript in a manner similar to the one observed following Upf1 depletion (Figure 5B). Injections of the corresponding mismatch control MOs did not change the level of the aberrant transcript. These results indicate that zebrafish Dhx34 and Nbas are required for the degradation of nonsense transcripts.
Figure 5.
Both Dhx34 and Nbas are required for the in vivo degradation of the nonsense slc25a5 transcript in zebrafish. Expression of slc25a5 mRNA was measured in wild-type (A) and goldenb1 mutant (B) uninjected embryos, or embryos injected with indicated MOs, by real-time RT–PCR. The level of slc25a5 transcript was normalized to dct mRNA. The normalized values were then divided by values for uninjected samples (no MO) and expressed as relative mRNA levels.
In summary, we have shown that zebrafish Dhx34 and Nbas are required for embryonic development in this vertebrate. Three lines of evidence strongly suggest a role for these factors in the NMD pathway. First, their depletion leads to phenotypes very similar to those observed with core NMD factors. Second, the ability of human NAG/NBAS or DHX34 mRNAs to rescue the phenotypes observed in zebrafish embryos depleted of Dhx34 or Nbas highlights their functional conservation. Finally, we have shown that both proteins are required for the degradation of a PTC-containing mRNA in zebrafish embryos. Altogether, this suggests that Dhx34 and Nbas are novel NMD factors in zebrafish and confirms the requirement of an active NMD pathway for proper development.
DISCUSSION
NMD is a highly conserved mechanism that is present in all eukaryotes. Whereas the central NMD effectors are conserved across species, the mechanism by which a PTC is recognized presents important differences throughout evolution. The main divergence is whether NMD is coupled or not to pre-mRNA splicing. In humans, zebrafish and other vertebrates, as well as in plants, NMD is tightly coupled to pre-mRNA splicing via proteins of the EJC [reviewed by ref. (8)]. By contrast, in Drosophila and S. cerevisae, NMD has been shown to operate independently of splicing, with the use of different mechanism/s to define a PTC. Interestingly, a ‘mixed system’ seems to be operating in S. pombe, since splicing enhances NMD but in an EJC-independent manner (51). It may well be that splicing and deposition of the EJC may act as ‘enhancers’ of NMD in organisms where pre-mRNA splicing is prominent. It is also of importance that NMD regulates an important fraction of the transcriptome, affecting not only mRNAs that harbour a PTC, but also naturally occurring transcripts [for a review see ref. (41)].
Here, we have identified the zebrafish homologues of the NMD factors NAG/NBAS/SMGL-1 and DHX34/SMGL-2. These NMD factors were initially identified in a reporter-based genome-wide RNAi screen in the nematode C. elegans and shown to act as NMD factors both in nematodes and also in human cells (16). Interestingly, these two genes define a novel class of nematode NMD factors that are essential for embryonic development. Mutations in the core smg-1-7 genes do not result in lethality, strongly suggesting that NMD is not an essential process in nematodes, and pointing to additional functions of SMGL-1, 2 in this organism. By contrast, targeted disruption of Rent1/UPF1 results in embryonic lethality in the mouse (52), whereas morpholino-mediated depletion of core SMG proteins in zebrafish results in severe developmental defects ((38) and this study). This could reflect the fact that NMD is not essential in C. elegans but is required for viability in vertebrates or it could point to additional cellular processes in which NMD factors are involved. For instance, SMG-2/UPF1 has been shown to be also required for genome stability in human cells (53). Furthermore, a role for SMG-2/UPF1 in alternative mRNA degradation pathways has been described. For example, in SMD (Staufen-mediated decay), UPF1 is recruited to 3′-UTRs of mRNAs bound by the RNA binding protein STAUFEN 1 and elicits mRNA degradation (54). It also functions in regulated degradation of histone mRNAs, where is recruited by the stem–loop binding protein (55).
The phenotypes observed following morpholino-mediated depletion of Nbas and Dhx34 resemble those observed for core NMD factors in zebrafish (38). This strongly suggests that NMD is indeed an essential process in this organism. Importantly, we could demonstrate a role for these two factors in NMD in vivo. Of course, we cannot rule out additional cellular functions for Nbas and Dhx34 in Danio rerio. Future studies using expression profiles in human, zebrafish and C. elegans depleted of individual NMD factors will result in the identification of RNA targets and pathways of NMD regulation throughout evolution. This could also lead to the identification of additional cellular functions, other than NMD, for Dhx34 and Nbas.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Medical Research Council (to E.E.P. and J.F.C.); Eurasnet (LSHG- CT-2005-518238) (to J.F.C.); Wellcome Trust (to E.E.P.); European Union FP7 programme ZF-CANCER (to E.E.P); Association for International Cancer Research (to E.E.P.); Medical Research Scotland (to E.E.P.). Funding for open access charge: MRC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Karthika Paranthaman for excellent zebrafish husbandry, and Nele Hug (MRC HGU) and Ian Jackson (MRC HGU) for critical reading of the manuscript.
REFERENCES
- 1.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 2.Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lelivelt MJ, Culbertson MR. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol. Cell. Biol. 1999;19:6710–6719. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 6.Wittmann J, Hol EM, Jack HM. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell. Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuzmiak HA, Maquat LE. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol. Med. 2006;12:306–316. doi: 10.1016/j.molmed.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE. NMD: RNA biology meets human genetic medicine. Biochem. J. 2010;430:365–377. doi: 10.1042/BJ20100699. [DOI] [PubMed] [Google Scholar]
- 9.Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 11.Cali BM, Kuchma SL, Latham J, Anderson P. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics. 1999;151:605–616. doi: 10.1093/genetics/151.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 13.Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muhlemann O, Eberle AB, Stalder L, Zamudio OR. Recognition and elimination of nonsense mRNA. Biochim. Biophys. Acta. 2008;1779:538–549. doi: 10.1016/j.bbagrm.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 15.Nicholson P, Yepiskoposyan H, Metze S, Zamudio OR, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol. Life Sci. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longman D, Plasterk RH, Johnstone IL, Caceres JF. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev. 2007;21:1075–1085. doi: 10.1101/gad.417707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimson A, O'Connor S, Newman CL, Anderson P. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA Decay in Caenorhabditis elegans. Mol. Cell. Biol. 2004;24:7483–7490. doi: 10.1128/MCB.24.17.7483-7490.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23:1091–1105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Page MF, Carr B, Anders KR, Grimson A, Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders KR, Grimson A, Anderson P. SMG-5, required for C.elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J. 2003;22:641–650. doi: 10.1093/emboj/cdg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14:2609–2617. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 24.Cho H, Kim KM, Kim YK. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol. Cell. 2009;33:75–86. doi: 10.1016/j.molcel.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 25.Unterholzner L, Izaurralde E. SMG7 Acts as a Molecular Link between mRNA Surveillance and mRNA Decay. Mol. Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 26.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Behm-Ansmant I, Izaurralde E. Quality control of gene expression: a stepwise assembly pathway for the surveillance complex that triggers nonsense-mediated mRNA decay. Genes Dev. 2006;20:391–398. doi: 10.1101/gad.1407606. [DOI] [PubMed] [Google Scholar]
- 28.Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA, Wilkinson MF. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–1830. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 31.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 32.Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lykke-Andersen J, Shu MD, Steitz JA. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- 34.Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 35.Buhler M, Steiner S, Mohn F, Paillusson A, Muhlemann O. EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3′ UTR length. Nat. Struct. Mol. Biol. 2006;13:462–464. doi: 10.1038/nsmb1081. [DOI] [PubMed] [Google Scholar]
- 36.Eberle AB, Stalder L, Mathys H, Orozco RZ, Muhlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6:e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wittkopp N, Huntzinger E, Weiler C, Sauliere J, Schmidt S, Sonawane M, Izaurralde E. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol. Cell. Biol. 2009;29:3517–3528. doi: 10.1128/MCB.00177-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hori K, Watanabe Y. Context analysis of termination codons in mRNA that are recognized by plant NMD. Plant Cell Physiol. 2007;48:1072–1078. doi: 10.1093/pcp/pcm075. [DOI] [PubMed] [Google Scholar]
- 40.Kertesz S, Kerenyi Z, Merai Z, Bartos I, Palfy T, Barta E, Silhavy D. Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 2006;34:6147–6157. doi: 10.1093/nar/gkl737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rehwinkel J, Raes J, Izaurralde E. Nonsense-mediated mRNA decay: target genes and functional diversification of effectors. Trends Biochem. Sci. 2006;31:639–646. doi: 10.1016/j.tibs.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 42.Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 2007;26:1591–1601. doi: 10.1038/sj.emboj.7601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 45.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 46.Linder P. Dead-box proteins: a family affair–active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wimmer K, Zhu XX, Lamb BJ, Kuick R, Ambros PF, Kovar H, Thoraval D, Motyka S, Alberts JR, Hanash SM. Co-amplification of a novel gene, NAG, with the N-myc gene in neuroblastoma. Oncogene. 1999;18:233–238. doi: 10.1038/sj.onc.1202287. [DOI] [PubMed] [Google Scholar]
- 49.Scott DK, Board JR, Lu X, Pearson AD, Kenyon RM, Lunec J. The neuroblastoma amplified gene, NAG: genomic structure and characterisation of the 7.3 kb transcript predominantly expressed in neuroblastoma. Gene. 2003;307:1–11. doi: 10.1016/s0378-1119(03)00459-1. [DOI] [PubMed] [Google Scholar]
- 50.Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, Jurynec MJ, Mao X, Humphreville VR, Humbert JE, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 51.Wen J, Brogna S. Splicing-dependent NMD does not require the EJC in Schizosaccharomyces pombe. EMBO J. 2010;29:1537–1551. doi: 10.1038/emboj.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 53.Azzalin CM, Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr. Biol. 2006;16:433–439. doi: 10.1016/j.cub.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 54.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3'UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 55.Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.