Abstract
In Drosophila, siRNAs are classified as endo- or exo-siRNAs based on their origin. Both are processed from double-stranded RNA precursors by Dcr-2 and then loaded into the Argonaute protein Ago2. While exo-siRNAs serve to defend the cell against viruses, endo-siRNAs restrict the spread of selfish DNA in somatic cells, analogous to piRNAs in the germ line. Endo- and exo-siRNAs display a differential requirement for double-stranded RNA binding domain proteins (dsRBPs): R2D2 is needed to load exo-siRNAs into Ago2 while the PD isoform of Loquacious (Loqs-PD) stimulates Dcr-2 during the nucleolytic processing of hairpin-derived endo-siRNAs. In cell culture assays, R2D2 antagonizes Loqs-PD in endo-siRNA silencing and Loqs-PD is an inhibitor of RNA interference. Loqs-PD can interact via the C-terminus unique to this isoform with the DExH/D-helicase domain of Drosophila Dcr-2, where binding of R2D2 has also been localized. Separation of the two pathways is not complete; rather, the dicing and Ago2-loading steps appear uncoupled, analogous to the corresponding steps in miRNA biogenesis. Analysis of deep sequencing data further demonstrates that in r2d2 mutant flies, siRNAs can be loaded into Ago2 but not all siRNA classes are equally proficient for this. Thus, the canonical Ago2-RISC loading complex can be bypassed under certain circumstances.
INTRODUCTION
Small non-coding RNAs play a pivotal role in several aspects of gene regulation and genome defense: Piwi-interacting RNAs (piRNAs) counteract the mobilization of transposable elements in the germ line (1,2), microRNAs (miRNAs) regulate gene expression and exogenous small interfering RNAs (exo-siRNAs) mediate anti-viral defense (3,4). Endogenous siRNAs (endo-siRNAs) also suppress selfish DNA such as transposable elements in both soma and germ line. In addition, certain hairpin-derived endo-siRNAs can regulate specific mRNAs in Drosophila (5–8). Installing a mechanism to repress selfish DNA requires a reliable means to identify these sequences within the genome, essentially a self versus non-self recognition problem on the level of genomic DNA. The piRNA system relies heavily on inheritance of a pool of maternally transmitted piRNAs (9), together with specific loci that carry inactive remnants of transposon sequences and that give rise to corresponding antisense transcripts. These then engage in an amplification loop, leading to rapid repression of transposons (1,10–12). The piRNA system is very efficient and poised to react against the previously encountered transposons due to the maternally provided pool of piRNAs, but it is slow to adapt towards a new transposon threat. If a naïve female fly is crossed with a male fly carrying a new transposon the offspring is sterile, while a cross in the opposite orientation has no detrimental effect (9,13). The endo-siRNA response, on the other hand, can initiate a de novo response to foreign DNA even upon transient transfection (7,14). While the exact mechanisms that generate the double-stranded RNA precursor for these siRNAs are still unknown, copy-number dependent silencing that depends on antisense transcripts was demonstrated for a cell culture model system (14). An alternative model proposed the involvement of an atypical putative RNA-dependent RNA polymerase (15), but there are a number of experimental observations which currently appear inconsistent with this model [discussed in ref. (16)].
Three of the small RNA classes, miRNAs, exo-siRNAs and endo-siRNA, depend on a mechanistically similar nucleolytic processing step in the cytoplasm carried out by a complex of Dicer and a double-stranded RNA binding domain protein (dsRBP) [reviewed in ref. (3,4)]. In Drosophila Dicer-1 (Dcr-1) together with the PB isoform of the dsRBP Loquacious (Loqs-PB) processes pre-miRNAs, which are then loaded into the effector protein Ago1. In contrast, exo-siRNA precursors are processed by Dicer-2 (Dcr-2), then loaded by a complex of Dcr-2 and R2D2 into Ago2 (17–19). Endo-siRNA biogenesis depends on Dcr-2 paired with a different isoform of Loqs, Loqs-PD (20–22). Although these complexes can be used to define the different small RNA classes by their biogenesis pathways, the biochemical basis for this specificity remains unclear. In addition to their RNA-binding activity, double-stranded RNA binding domains (dsRBDs) can mediate protein–protein interactions (23). R2D2 contains two dsRBDs and a C-terminal part, the latter mediating association with Dcr-2 (17,24). The complex of Dcr-2 and R2D2 does not have enhanced dsRNA processing activity; instead, it serves as the RISC loading complex (RLC) that loads exo-siRNAs into Ago2 (18,19,25). There are four known splice variants of Loquacious (20,26). Loqs-PA and Loqs-PB both have three dsRBDs (L1L2L3) (26,27); while the role of Loqs-PA is still largely uncharacterized, Loqs-PB increases the efficiency of Dcr-1 processing (26,28–30). Loqs-PC and Loqs-PD both lack the third dsRBD and instead carry short peptide sequences at their C-termini (20,26). A recent study by Carthew and colleagues proposed a model of sequential action where Loqs-PD is involved in the dicing step of endo- as well as exo-siRNAs and both types of siRNAs are then loaded into Ago2 with the help of R2D2/Dcr-2 (31). This model contrasts the situation in cultured cells. Here, R2D2 is required for loading of certain miRNAs into Ago2, but no contribution of R2D2 to the endo-siRNA pathway could be demonstrated yet (6–8,20,32).
In this study we characterize the interaction of Loqs-PD with Dcr-2 and the complexes required for endo-siRNA mediated silencing in S2 cells. We show that the PD-specific amino acids can mediate an interaction with the N-terminal helicase domain of Dcr-2. Loqs-PD and R2D2 appear as functional antagonists during both endo- and exo-siRNA mediated silencing in S2-cells, arguing that they compete for a common factor. Both Loqs-PD and R2D2 contribute to silencing by inverted-repeat transgenes, but a quantitative analysis suggests that neither protein is absolutely required; instead, their effects appear to be additive suggesting parallel pathways rather than sequential action. This model is further supported by deep sequencing experiments showing that endo-siRNAs are still detectable and retain their thermodynamic asymmetry in the absence of R2D2.
MATERIALS AND METHODS
Cell culture, RNAi and FACS analysis
Drosophila Schneider 2 (S2) cells were cultured in Schneider’s medium (Bio & Sell, Nürnberg, Germany) supplemented with 10% fetal calf serum (Thermo). The miR-277 perfect match (33) and endo-siRNA (20) reporter cell lines were described before. Transfection and RNAi treatment were conducted essentially as explained in (34). The dsRNA triggers were previously described (20,26). Cells were seeded at 0.5 × 106 cells/ml and 20 μg/ml dsRNA were added to the medium. On Day 2, the cells were split 1:5 into a fresh culture dish and the dsRNA treatment was repeated. On Day 5 or 6, GFP fluorescence was quantified in a Becton Dickinson FACSCalibur flow cytometer. For transfections cells were seeded at 0.5 × 106 cells/ml and split on Day 3 after transfection.
Molecular cloning
The myc-tagged constructs for Loqs-PB/-PA/-PD, R2D2 and GFP were described previously (20,26). The myc-Loqs-PC construct (pEH38) was made analogously with 5′-CAA AGC GGC CGC CTA CTG CGG GGC TGT AAA TAA G-3′ as the antisense primer. The myc-tagged C-terminus of Loqs-PB was PCR-amplified with primers 5′-AGC GGA TCC ATG GAA CAA AAA CTT ATT TCT GAA GAA GAC TTG GAA CAA AAA CTT ATT TCT AAA GAA GAC TTG CCC CGC AGT AGT GAA AAT TAT TAT G-3′ and 5′-TTA AGC GGC CGC CTA CTT CTT GGT CAT GAT CTT CAA GTA CTC-3′, then cloned BamHI-NotI into the pKF63 backbone, yielding plasmid pKF112.
Nomenclature of dsRBP constructs is as follows: ‘L’ and ‘R’ stand for Loqs and R2D2, respectively, numbers mark dsRBDs. The Loqs-PC-specific and Loqs-PD-specific sequences are abbreviated ‘L3PC’ and ‘L3PD’, respectively; the part C-terminal to the second dsRBP of R2D2 was dubbed ‘R3′. Ligation dependent addition of two codons (glycine and serine) is signified by a ‘:’.
To create fragments of Loqs and R2D2 the following primer pairs were used:
5′-CAA GGA TCC AAG AAC ACC ATG GAC CAG GAG-3′ and 5′-CAA AGA TCT TTC GCC CTC CAA CTC GCC GCA G-3′ for Loqs L1L2 as well as 5′-CAA GGA TCC CTT GAA CTC ATG GAT AAC AAG-3′ and 5′-CAA AGA TCT TTT CAG GGT AGG ATA GAA GTT CTT GAA-3′ for R2D2 R1R2. 5′-CAA GGA TCC AAG GAG GCC ATT GAG GCC ATC-3′ and 5′-CAA AGC GGC CGC TTA TAC GCA TTA AAT CAA-3′ for R3 of R2D2, 5′-CAA GGA TCC AAC GAA TCT GTA AAG CAC CT-3′ and 5′-CAA AGC GGC CGC CTA CTG CGG GGC TGT AAA TAA G-3′ for the L3PC sequence as well as 5′-CAA GGA TCC GTG AGT ATC ATT CAA GAC ATC-3′ and 5′-CAA AGC GGC CGC TTA GAT CTT GAT GAA CTC-3′ for the L3PD sequence. Fragments were amplified by PCR from Drosophila cDNA, subsequently BglII/BamHI digested and ligated. The fused constructs were BamHI/NotI digested and ligated into the pKF63 expression plasmid as described before (26). To create the truncated Loqs (L1L2), the following primer pair was used: 5′-CAA GGA TCC AAG AAC ACC ATG GAC CAG GAG-3′ and 5′-CAA AGC GGC CGC CTA TTC GCC CTC CAA CTC GCC GCA G-3′. PCR amplification products were BamHI/NotI cloned into the pKF63 expression vector.
The GFP scaffold for fusion proteins was constructed analogously with the following primer pairs: 5′-CAA GGA TCC ATG GTG AGC AAG GGC GAG GAG CTG-3′ (or for the myc-tagged form: 5′-AGC GGA TCC ATG GAA CAA AAA CTT ATT TCT GAA GAA GAC TTG GAA CAA AAA CTT ATT TCT GAA GAA GAC TTG GCC GTG AGC AAG GGC GAG GAG CTG-3′) and 5′-CAA AGA TCT CTT GTA CAG CTC GTC CAT G-3′.
The Flag-Dcr-2 constructs were produced as follows: A 2 kb promoter fragment was excised from JB17 vector (35) with EcoRI/KpnI and ligated into pCasper5 (36) cloning vector. A tubulin 3′-UTR was subsequently amplified, using 5′-CAA GCT AGC ATT CGA ATC GGA AAT CAA TCG AAT TC-3′ and 5′-CAA CCT AGG AGA CTT GTG AAC AAA ATT GGA TCC G-3′, and NheI/AvrII ligated into the pCasper5 construct. The full-length Flag-Dcr-2 was PCR amplified using 5′-CAA GCG GCC GCA TGG ATT ATA AAG ATG ATG ATG ATA AAG AAG ATG TGG AAA TCA AGC CTC GC-3′ and 5′-CAA GCT AGC TTA GGC GTC GCA TTT GCT TAG CTG CTG-3′, the Δhel-Flag-Dcr-2 by 5′-CAA GCG GCC GCA TGG ATT ATA AAG ATG ATG ATG ATA AAC AGG ACG ATA TTG ACC CTT TTA CCA-3′ and 5′-CAA GCT AGC TTA GGC GTC GCA TTT GCT TAG CTG CTG-3′. Both Dcr-2 amplification products were NotI/NheI cloned into the pCasper5 expression system. All constructs were sequenced to ensure correctness.
Protein extraction, co-immunoprecipitation and western blotting
Cells were harvested and washed twice in PBS. The pellet was resuspended in lysis buffer [30 mM HEPES, 100 mM KOAc, 2 mM MgCl2, 1 mM fresh DTT, 1% (v/v) Triton X-100 (Sigma), 2× protease inhibitor cocktail (Complete® without EDTA, Roche Molecular Biochemicals; Basel, Switzerland)] and frozen in liquid nitrogen. Samples were thawed on ice and cell debris was pelleted in a refrigerated microcentrifuge at 15 000g (Eppendorf). Protein concentrations were determined by Bradford Assay (BioRad, Hercules, CA, USA).
Immunoprecipitation was performed essentially as described (20). Protein G Plus/Protein A Agarose beads (IP05, Calbiochem) were washed three times in 1 ml lysis buffer and agitated for 30 min at 4°C with the respective antibody. Beads were then washed four times and incubated with 1–5 mg of total protein in lysis buffer at 4°C for 1 h on an overhead rotator. For α-Flag immunoprecipitation, α-Flag affinity agarose (A2220, Sigma), washed three times in 1 ml of lysis buffer, was used. For IP experiments in Figures 2A, B and 4C a Triton X-100 concentration of 0.4% (v/v) was used. GFP-fusion constructs were precipitated using GFP-Trap®_A beads (Chromotek, Planegg-Martinsried, Germany).
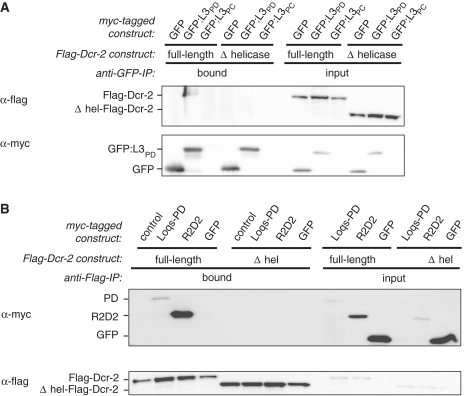
Figure 2.
Loqs-PD interacts with the N-terminal helicase domain of Dcr-2 for endo-siRNA silencing. (A) Co-immunoprecipitation from Drosophila S2 cell extract co-expressing GFP-fusion proteins together with either Flag-Dcr-2 or Δhel-Flag-Dcr2; the GFP proteins in this experiment also contained an N-terminal myc-tag that was used for detection, but GFP-trap beads were used for immunoprecipitation. (B) Co-immunoprecipitation from Drosophila S2 cell extract co-expressing myc-tagged Loqs-PD, R2D2 or a pUC18 control together with either Flag-Dcr-2 or Δhel-Flag-Dcr-2; α-Flag agarose was used for IP; myc-GFP served as a control.
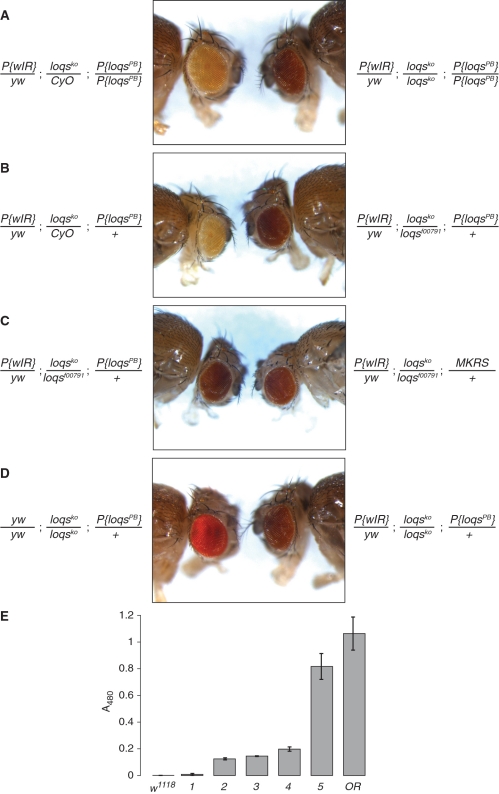
Figure 4.
- P{wIR}/yw; loqsko/CyO; P{loqs-PB}/+
- P{wIR}/yw; loqsko/loqsf00791; +/+
- P{wIR}/yw; loqsko/loqsf00791; P{loqs-PB}/+
- P{wIR}/yw; loqsko/loqsko; P{loqs-PB}/+
- yw/yw; loqsko/loqsko; P{loqs-PB}/+
(note that the P{wIR} transgene is carried on an X-chromosome with a wild-type w gene; sample 5 is therefore a slightly underestimated reference point).
Flow-through and beads were separated by spin columns (MoBiTec, Göttingen, Germany) and washed four times with 500 µl lysis buffer. Bound proteins were eluted by applying 15 μl 1× SDS sample buffer and heating to 95°C for 5 min. Western blotting was performed as previously described (33); α-Loqs antibody (27) was diluted 1:1000, α-Dcr-2 antibody (Abcam, ab4732-100) was diluted 1:1000, α-myc antibody (Sigma, M4439) was diluted 1:1000, α-Flag (Sigma, F1804) was diluted 1:2000, α-Dcr-1 (33) was diluted 1:1000, α-GFP (Santa Cruz Biotechnology, sc-9996) was diluted 1:4000, α-β-tubulin (DSHB, E7). Rabbit IgG was purchased from Sigma (I5006).
α-Loqs-PD antibody production: Loqs-PD specific peptide was synthesized, coupled to KLH and used to immunize rabbits (Davids Biotechnologie, Regensburg, Germany). Affinity purification was performed as described (37) using Loqs-PD peptide coupled to NHS-Sepharose (Sigma, H8280).
RNA extraction and northern blotting
RNA was extracted with Trizol (Invitrogen) and probes for detection of bantam miRNA, miR-277 miRNA and 2S rRNA were as described (26). We used a DNA antisense probe for the detection of CG4068B endo-siRNA (5).
Test of exo-siRNA mediated RNA interference
S2 cells were treated twice (days 1 and 3) with dsRNA directed against small RNA silencing factors or luciferase as a control. Ago2-RISC was then primed with dsRNA directed against GFP or again luciferase as control (day 6). On day 8, all cells were transfected with a GFP-expression plasmid (pKF63) and 3 days later GFP fluorescence was measured by flow cytometry.
Fly husbandry, preparation and analysis of deep sequencing libraries
Flies were reared on standard cornmeal/yeast/molasses medium. Quantification of eye pigments was performed as previously described (26). For deep sequencing, RNA was extracted from the head + thorax portion of 5–10 female flies using Trizol (Invitrogen, Carlsbad, CA, USA). Subsequent steps were performed as described (14) with the exception that the sequence barcodes required for multiplexing were introduced only during the final PCR step. Deep sequencing was performed by Fasteris S.A. (Geneva/CH) on an Illumina GAIIx instrument. Data processing was performed with Perl scripts (available upon request) and alignment of the reads to the indicated references was performed using BOWTIE (38). The accession numbers for the published deep sequencing libraries were: GSM278708 (wt untr.), GSM578573 (wt ox.), GSM278707 (white-IR untr.), GSM278575 (white-IR ox.), GSM278705 (dcr2/CyO untr.), GSM278703 (dcr2/dcr2 untr.), GSM278692 (dcr2/dcr2 ox.), GSM278704 (ago2/ago2 untr.), GSM278693 (ago2/ago2 ox.), GSM466494 (r2d2/CyO untr.), GSM466495 (r2d2/CyO ox.), GSM466496 (r2d2/r2d2 untr.) and GSM466497 (r2d2/r2d2 ox.). The data generated in this publication can be accessed under GSE26230.
RESULTS
The PD-specific C-terminus is essential for endo-siRNA function
Since Loqs-PB binding to Dcr-1 is mediated via the C-terminus containing the third dsRBD (24,26), we wondered whether the Loqs-PD specific C-terminus, placed at an equivalent position, is functionally important and interacts with Dcr-2. We fused the unique sequences of Loqs-PD (L3PD) as well as Loqs-PC (L3PC) to GFP and performed anti-GFP immunoprecipitations (Figure 1A). Addition of the Loqs-PC specific amino acids resulted in much lower expression levels (see also Figure 2A). Given that endogenous Loqs-PC is not detectable by western blotting (14), this may indicate active degradation of the fusion protein by the cell. A small amount of Dcr-2 co-precipitated reproducibly in case of the GFP-L3PD fusion (Figure 1A), demonstrating that the PD-specific part can be sufficient for interaction with Dcr-2. This interaction appears much weaker than what can be observed using full-length Loqs-PD [Supplementary Figure S1 and (14)], indicating that other parts of Loqs contribute further to Dcr-2 binding or to the stability of the complex. On the other hand, co-overexpression of Dcr-2 resulted in a robust interaction with GFP-L3PD (Figure 2A). We observed no interaction between Dcr-2 and the GFP-L3PC fusion, but due to the low expression levels this should be interpreted with caution.
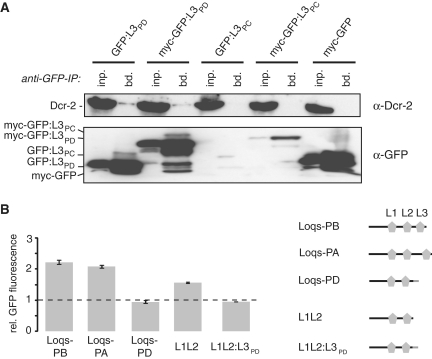
Figure 1.
Dcr-2 interacts with the C-terminal 22 amino acids specific to Loqs-PD. (A) Top panel: Co-immunoprecipitation of endogenous Dcr-2 with GFP-fusion constructs in S2-cells; bottom panel: α-GFP western blot to determine expression levels and successful immunoprecipitation. Fusion of the Loqs-PC specific amino acids to GFP reduce the expression levels; this appears to be generally the case with GFP-PC constructs (see also Figure 2A). (B) Effect of Loqs isoform overexpression on GFP expression levels of the endo-siRNA cell culture reporter; protein isoforms are depicted on the right (dsRBDs are represented by symbols; Loqs-PD-specific sequence is colored in grey); measurement values represent the mean ± SD (n = 3) and were normalized to a pUC18 transfected control (dashed line).
To test whether the Loqs-PD specific amino acids are required for endo-siRNA dependent silencing, we employed an S2 cell line where a stably integrated GFP-expression construct has triggered the production of corresponding endo-siRNAs (20). In these cells, the efficiency of endo-siRNA silencing can be measured via GFP-fluorescence. We transfected this cell line, henceforth called ‘endo-siRNA cell culture reporter’, with expression vectors coding for individual Loqs isoforms. Overexpression of the protein variants not involved in endo-siRNA silencing (Loqs-PA and Loqs-PB) caused an increase in GFP expression, while Loqs-PD had no effect. Since overexpressed Loqs-PA and Loqs-PB interact with Dcr-2 (14), their dominant-negative effect may be due to the formation of artificial complexes that are not functional (see also Figures 3 and 5). Transfection of a version of Loqs truncated at the start of the PD-specific amino acids resulted in impaired endo-siRNA silencing to an extent comparable with Loqs-PA or Loqs-PB, suggesting that the truncated protein is non-functional in the endo-siRNA pathway. Re-addition of the Loqs-PD specific sequence to the truncated Loqs variant to reconstitute Loqs-PD (L1L2:L3PD, containing two ligation-dependent extra amino acids) completely reverted the dominant-negative effect. This is consistent with data from Miyoshi and colleagues (21) who also described that truncation of Loqs-PD weakens the interaction with Dcr-2. Taken together, our results indicate that the C-terminus of Loqs-PD is required for endo-siRNA silencing, either by mediating the interaction with Dcr-2 or by modulating the affinity or activity (i.e. KM or kcat) of Dcr-2 to process dsRNA.
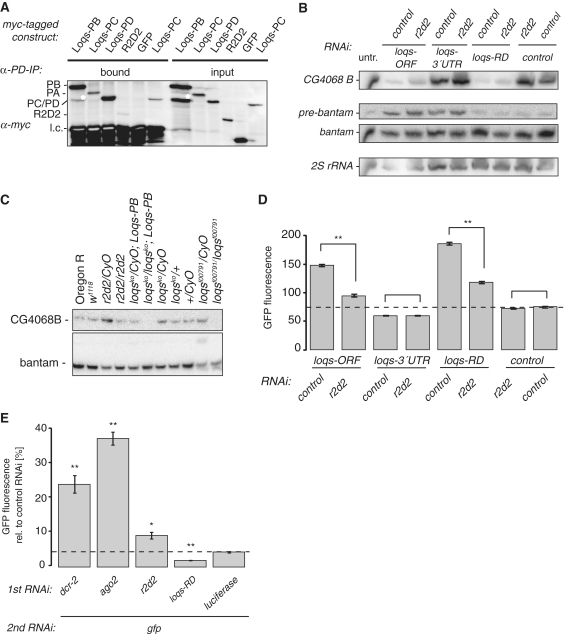
Figure 3.
R2D2 acts as an inhibitor of Loqs-PD in endo-siRNA silencing. (A) Co-immunoprecipitation with Drosophila S2 cell extract prepared after transfection of myc-tagged Loqs isoforms or myc-tagged R2D2; α-Loqs-PD-specific antibody was used to immunoprecipitate endogenous Loqs-PD. The white asterisk marks a faster migrating, myc-tag containing band in the lanes from myc-Loqs-PB transfected cells. This band is found consistently but to a varying extent in transfections with this construct (14); it likely represents a degradation product of the overexpressed Loqs-PB. (B) Northern blot using RNA extracted from Drosophila S2 cells after RNAi treatment with combinations of Loqs isoforms and R2D2; DsRed dsRNA served as a control for RNAi; a DNA probe against the long hairpin-derived endo-siRNA CG4068B (66) and 2′-OMe oligonucleotide probes against bantam miRNA were used; 2S rRNA served as a control for loading. (C) Northern blot using RNA isolated from flies carrying mutant alleles of loqs and r2d2. Note that the particularly strong expression of CG4068B endo-siRNA in r2d2/CyO flies is consistent with deep sequencing data obtained from an independent RNA preparation from the same fly strain (Table 1). (D) Effect of RNAi treatment with combinations of Loqs isoforms and R2D2 on GFP expression of the endo-siRNA cell culture reporter; double-stranded RNA directed against DsRed served as a control, RNAi triggers are indicated below the bars; measurement values represent the mean ± SD (n = 3) and were normalized to a DsRed/DsRed control (dashed line); asterisk: P < 0.005 (two-tailed t-test, unequal variance). (E) Effect of R2D2 and Loqs-PD on exo-siRNA mediated silencing; RNAi triggers are indicated below the bars. GFP fluorescence was calculated relative to a control treatment with dsRNA directed against luciferase during the second RNAi step. The dashed line marks GFP fluorescence levels under control conditions (dsRNA against luciferase, followed by dsRNA against GFP); measurement values represent the mean ± SD (n = 3); *P < 0.02, **P < 0.005 (two-tailed t-test, unequal variance).
Figure 5.
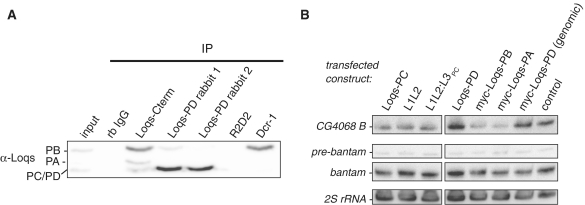
Dominant-negative effects on endo-siRNA biogenesis by overexpression of Loqs isoforms. (A) Detection of endogenous Loqs protein from S2-cell extract after immunoprecipitation with α-Dcr-1, α-Loqs-PB C-terminus and α-Loqs-PD; rb IgG and α-R2D2 served as controls; α-Loqs monoclonal antibody was used for detection of endogenous Loqs isoforms. (B) Northern blot from Drosophila S2 cell extract overexpressing Loqs isoforms; transfection with pUC18 served as a control; L1L2 = Loqs truncation lacking the PD-specific amino acid sequence, L1L2:L3PC = reconstituted Loqs-PC; myc-loqs-PD (genomic) = expression construct derived from genomic DNA; DNA probes against the long hairpin-derived endo-siRNA CG4068B (66) and against bantam miRNA were used; 2S rRNA served as a control for loading; the two parts were extracted from a single blot using the same exposure and image processing settings.
Loqs-PD interacts with the N-terminal helicase domain of Dcr-2 during endo-siRNA biogenesis
Our results suggest that the 22 amino acids unique to Loqs-PD influence association with Dcr-2 and are an essential part of its functionality during endo-siRNA silencing. We therefore proceeded to characterize the corresponding interaction domain in Dcr-2. Recently, an EM-model proposed an L-shaped arrangement for human Dicer with the catalytic domain residing in the long branch (39,40). The N-terminal DExH/D helicase domain of human Dicer, situated in the short branch of the structure, mediates binding to the mammalian Loqs homologue TRBP (39–41). We therefore tested if Drosophila Dcr-2 also interacts through its N-terminal helicase domain with Loqs-PD by co-expressing Flag-tagged full-length or truncated Dcr-2 together with myc-GFP-L3PD/-L3PC fusions in S2 cells. Full-length overexpressed Flag-Dcr-2 efficiently co-precipitated with the myc-GFP-L3PD fusion proteins, corroborating the observations for endogenous Dcr-2 in Figure 1A. When we substituted the full-length Flag-Dcr-2 with a construct lacking the N-terminal helicase region (Δ1-551 = Δhel), myc-GFP-L3PD failed to enrich Δhel-Flag-Dcr-2 in comparison to the input and showed only background binding similar to that observed for myc-GFP alone (Figure 2A and data not shown). Again, although no interaction between myc-GFP-L3PC and Flag-Dcr-2 was observed, the low expression levels of the myc-GFP-L3PC fusion protein do not allow firm conclusions. We also probed the interaction in the opposite direction by immunoprecipitating Flag-Dcr-2 and detecting associated Loqs-variants by western Blot. Both myc-Loqs-PD and myc-R2D2 were almost completely lost when Δhel-Flag-Dcr-2 was immunoprecipitated (Figure 2B), confirming that both interact with the helicase-domain of Dcr-2.
R2D2 acts as an antagonist of Loqs-PD in endo-siRNA silencing
A possible consequence of the association of both Loqs-PD and R2D2 with Dcr-2 is competition and commitment of the enzyme to either the endo-siRNA or the exo-siRNA pathway depending on the association partner. Binding of Loqs-PD and R2D2 could be mutually exclusive if the same binding site is used; alternatively, Loqs-PD and R2D2 could simultaneously bind Dcr-2 but induce different conformations. Reduced co-precipitation of both Loqs-PD and R2D2 with the Δhel-Flag-Dcr-2 construct (Figure 2B) substantiates the competition hypothesis as both proteins seem to interact with the helicase region of Dcr-2 (24,42). On the other hand, a recent finding by the Siomi lab (21) reported association of R2D2 with the Loqs-PD/Dcr-2 complex upon overexpression of Loqs-PD. To repeat this experiment at the endogenous Loqs-PD protein level, we immunized rabbits with the 22 amino acid peptide that is unique to the Loqs-PD isoform (20) and affinity purified the resulting antiserum. The specificity of the affinity-purified antibodies was confirmed in immunoprecipitations and western blotting (Supplementary Figure S2). While other Loqs isoforms—when overexpressed—co-immunoprecipitate to some extent with endogenous Loqs-PD, association of overexpressed R2D2 was marginal (Figure 3A). Based on this result, we favor the hypothesis that Loqs-PD and R2D2 interact with the same site on Dcr-2 and that formation of a trimeric complex is inefficient at endogenous levels.
We previously demonstrated that depletion of R2D2 results in an increased repression of our GFP-based endo-siRNA reporter (14). Here, we further tested whether silencing of the reporter depends on the relative levels of Loqs-PD and R2D2. To this end we employed a double knock down strategy. As described, depletion of all Loqs isoforms or only Loqs-PD leads to de-repression of the reporter, while depletion of Loqs-PA, -PB and -PC via the common 3′-UTR but not Loqs-PD, which has a different 3′-UTR, leads to hyper repression (14). Figure 3D shows that an additional depletion of R2D2 partially restored silencing of the reporter only when the Loqs-PD levels had been concomitantly lowered. There is a slight trend for increased silencing upon knock-down of R2D2 in the control case, consistent with our previous report. The effect is presumably weaker in the double knock down experiment because two independent targets are silenced concomitantly. We conclude that the ratio of Loqs-PD to other dsRBD proteins is critical for optimal endo-siRNA biogenesis.
Loqs-PD and R2D2 function independently during RNAi
Since R2D2 antagonizes Loqs-PD in endo-siRNA silencing, we tested whether the inverse is true during exo-siRNA mediated silencing. In a two-step RNAi assay [analogous to (15)] we tested the efficiency of GFP knock-down after previous depletion of RNAi factors. We saw a clear impairment of RNAi following depletion of Dcr-2, Ago2 and R2D2, whereas depletion of Loqs-PD increased RNAi efficiency (Figure 3E). Thus, R2D2 and Loqs-PD are functional antagonists during both endo- and exo-siRNA mediated silencing in cultured cells. RNA interference can also be assayed in vivo using inverted-repeat transgenes, which produce dsRNA upon transcription. A convenient assay using the white gene as readout has been established by the Carthew lab (43). Silencing activity of the white-IR transgene depends to a large extent on r2d2, but a small contribution of loqs has been described using the hypomorphic allele loqsf00791 (26). It was recently proposed that a null mutant of loqs strongly impaired the activity of the white-IR transgene (31), but the eye pigment level was not quantified.
We therefore repeated the assay using the same knock-out allele of loqs (44) in combination with a white-IR transgene located on the X-chromosome. Since miRNAs play an important role in eye biogenesis (45), we included a transgene that restores expression of Loqs-PB (44) to avoid potential indirect effects. Consistent with published results, we saw that silencing by the white-IR transgene is perturbed in loqsko homozygous mutant flies when compared to heterozygous controls (Figure 4A). However, both qualitative comparison as well as photometric quantification of the produced eye pigments revealed that this impairment represents only a fraction of the eye color produced when no silencing trigger is present (Figure 4D and E, 25% of controls). Furthermore, trans-heterozygous mutant animals carrying one knock-out allele and one hypomorphic allele of loqs showed only a marginal change in silencing efficiency when compared with homozygous loqsko flies, indicating that the hypomorphic allele loqsf00791 has essentially the same phenotype in this assay. Using this hetero-allelic combination we compared flies with and without the loqs-PB transgene (Figure 4C and E). Again, no major difference was detected, consistent with the notion that the Loqs-PB does not participate in siRNA-mediated silencing.
Multimerization and competition of Loqs isoforms for Dcr-2 binding
As shown in Figure 1B, overexpression of Loqs-PA and Loqs-PB led to impaired endo-siRNA silencing. This effect was dose dependent (Supplementary Figure S2B). In addition to the known interaction of Loqs-PA and Loqs-PB with Dcr-2 upon overexpression (14), the dominant-negative effect could further be explained by multimerization of overexpressed Loqs isoforms together with endogenous Loqs-PD, resulting in the formation of dsRBP complexes unsuitable for silencing. Figure 3A indicates that overexpressed Loqs isoforms can be co-immunoprecipitated with endogenous Loqs-PD. However, this interaction is much weaker or absent when we probed for co-immunoprecipitation of endogenous Loqs-PB and loqs-PA (Figure 5A). If the Loqs multimers induced by overexpression had a dominant-negative effect, this should be visible at the level of endo-siRNA biogenesis. Loqs-PD overexpression from both cDNA- and genomic DNA-derived expression constructs did not change the levels of the hairpin-derived endo-siRNA CG4068B. In contrast, overexpression of full-length myc-Loqs-PB/-PA or Loqs-PC (either full-length or reconstituted L1L2:L3PC) led to a reduction of mature CG4068B (Figure 5B). Consistent with our cell culture reporter experiments (Figure 1B and C) truncated Loqs, lacking only the PD-specific amino acid sequence (L1L2), caused an impairment of long hairpin endo-siRNA biogenesis. In summary, overexpression of individual Loqs isoforms perturbs the function of the small RNA silencing system, possibly via the formation of non-physiological multimers.
Processing and loading of small RNAs into Ago2 can occur independently of R2D2
To test whether a functional interaction between Loqs-PD and R2D2—as previously proposed (31)—can be observed at the level of endo-siRNA biogenesis, we treated S2 cells with RNAi against different Loqs isoforms together with RNAi against R2D2. Subsequently, we isolated RNA and probed Northern blots for a long hairpin-derived endo-siRNA [CG4068B; (5)] as well as the miRNA bantam. As published (20), we could observe impaired endo-siRNA biogenesis of CG4068B (also referred to as esi2.1) after treatment with the dsORF construct against all Loqs isoforms and—even more pronounced—with dsRNA against Loqs-PD specifically (Figure 3B). Simultaneous knock down of R2D2 did not reduce the levels of mature endo-siRNA further in all combinations; if anything, endo-siRNA biogenesis improved upon knock down of R2D2. We confirmed these conclusions with RNA extracted from flies carrying mutant alleles of loqs or r2d2 (Figure 3C). As before, biogenesis of the CG4068B hairpin-derived siRNA depends on loqs [see also refs (5,7,8)] and the processing cannot be rescued by a loqs-PB transgene, confirming that Loqs-PB does not participate in hairpin-derived endo-siRNA biogenesis in vivo. This analysis can be extended to transposon-targeting endo-siRNAs by deep sequencing. To avoid contamination by piRNA degradation products [a potential problem in the libraries analyzed in (31)], we prepared libraries with RNA derived from only the head and thorax of female flies. In the case of loqsko mutants we restored miRNA biogenesis with a loqs-PB transgene. Transposon-targeting endo-siRNAs were detectable in all four libraries (Table 1), though unfortunately we obtained only a very low number of reads for homozygous loqsko flies in our multiplexing experiment, making even qualitative comparisons difficult. Nonetheless, transposon-matching endo-siRNAs were present among those reads, indicating that Loqs-PD may not be required for dicing of all endo-siRNA precursors. The size distribution of the transposon-matching reads showed a peak at 21 nt in all cases (Supplementary Figure S4). A comparison of the number of transposon-matching reads relative to the number of genome-matching reads in each library revealed roughly comparable amounts for the r2d2/Cyo, r2d2/r2d2 and loqs/Cyo; Loqs-PB genotypes (Table 1). In addition to technical variability, the differences may reflect distinct transposon contents between the balancer chromosome and the chromosomes carrying the mutant alleles. In summary, we could validate the previous finding that the nucleolytic processing of both hairpin-derived and transposon-targeting endo-siRNAs does not depend on r2d2. In addition, it appears that Loqs-PD may not be essential for the dicing of transposon-derived endo-siRNAs either.
Table 1.
Analysis of deep sequencing libraries generated in this study
| Library | Total no. of inserts 19–25 nt | Inserts matching the genome (percentage of total) | Inserts matching mature miRNAs (percentage of genome matching) | Inserts matching transposons (percentage of genome matching) | Inserts matching CG4068-hairpin (percentage of genome matching) | Inserts CG4068B Sense strand | Inserts CG4068B Star strand |
|---|---|---|---|---|---|---|---|
| r2d2/CyO | 1 771 480 | 911 748 (51.5) | 479 462 (52.6) | 23 365 (2.6) | 519 (0.06) | 207 | 12 |
| r2d2/r2d2 | 6 195 408 | 4 439 743 (71.7) | 2 729 007 (61.5) | 53 616 (1.2) | 505 (0.01) | 340 | 77 |
| loqsko/CyO; P{Loqs-PB} | 2 405 256 | 1 720 875 (71.5) | 842 232 (48.9) | 9269 (5.4) | 127 (0.007) | 91 | – |
| loqsko/ loqsko; P{Loqs-PB} | 47 532 | 35 972 (75.7) | 23 414 (65.1) | 205 (0.6) | – | – | – |
Several features can be exploited to determine whether loading has occurred: Ago2-bound siRNAs are 2′-O-methyl modified on their terminal nucleotide by the methyl transferase hen1 (46), which renders them resistant to oxidation of the vicinal diol-specific oxidation reagent sodium periodate. Since oxidized terminal nucleotides cannot be ligated to the cloning adapter, a comparison of deep sequencing libraries before and after oxidation allows assessing the extent to which small RNAs are 2′-O-methyl modified. Furthermore, upon loading of siRNAs into Ago2 the strand with lower base-pairing stability on its 5′-end is preferentially incorporated, while the passenger strand is destroyed (47). An excess free energy of hybridization at the 3′-end of siRNAs is therefore another indicator of loading, since the duplex intermediates should—on average—be thermodynamically symmetric. Finally, in the case of hairpin-derived endo-siRNAs, the cloning frequency of guide and passenger strands can be directly counted and thus an asymmetry value can be calculated.
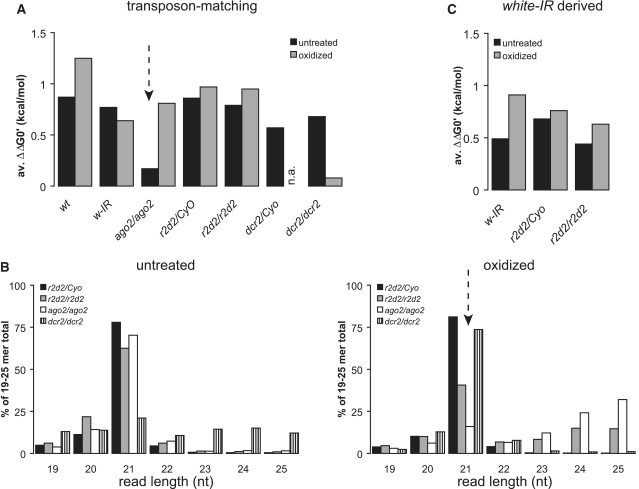
We therefore analyzed a collection of deep sequencing libraries from fly heads before and after oxidation (6,48) for the presence, modification and asymmetry of the CG4068B endo-siRNA (Supplementary Table S1). Consistent with our northern blot results and our own sequencing libraries (Table 1), the r2d2/Cyo strain appears particularly efficient in producing this endo-siRNA. In all strains proficient for processing, an excess of CG4068B-matching reads over the passenger strand could be detected. Several recent insights need to be further considered: This particular hairpin-derived endo-siRNAs is initially loaded into both, Ago1 and Ago2, but then cleared from Ago1 by tailing and degradation because a target with nearly perfect complementarity exists (49). The Ago1-loading is intensified in the absence of R2D2 (50), a situation that may be analogous to the one described for miRNAs that partition into both, Ago1 and Ago2 (33). It was also demonstrated that most of the hairpin-derived endo-siRNAs become vulnerable to sodium periodate oxidation in r2d2 mutants (31,50). Nonetheless we searched for indications that some of this endo-siRNA remains associated with Ago2 in r2d2 mutants in the deep sequencing data. Consistent with our own libraries, the abundance of CG4068B was roughly comparable between r2d2/CyO and r2r2/r2d2 flies, while a strong reduction could be seen when comparing dcr2/CyO with dcr2/dcr2 flies. An excess of guide over passenger strand was also detectable in all cases, indicating that loading into an Ago-protein had occurred. In the libraries that were prepared from r2d2/CyO heads after oxidation, the guide-strand excess was stronger than in the non-treated library, arguing that a certain amount of un-loaded duplex intermediate is normally present and/or that the Ago1-loaded portion is unwound slowly. CG4068B matching reads were present in the library from r2d2/r2d2 heads after oxidation and the guide-strand excess was only slightly lower than in the corresponding library from r2d2/Cyo heterozygotes. Furthermore, the abundance of CG4068B matching reads relative to the total number of reads in the library was >2-fold higher in r2d2 mutants than in ago2 mutants. This implies that a fraction of the CG4068B endo-siRNAs is associated with Ago2 in the soma of r2d2 mutant animals. Our analysis describes only one small RNA product of only one hairpin locus (CG4068/esi2), certainly an oversimplification of the situation. Okamura and colleagues present a thorough description about how the r2d2 mutation affects processing of the CG4068 hairpin (50) and Ameres and colleagues describe sorting of different hairpin-derived endo-siRNAs in detail (49).
We proceeded to analyze the extent to which transposon-matching endo-siRNAs are loaded into Ago2 in the absence of R2D2. It had previously been noted that in the absence of Ago2 a small amount of 2′-O-methyl modified small RNAs in the size range of piRNAs is generated (6), we therefore limited our scope to siRNAs of 21 nt length. We note that while this approach focuses on the size preferred by Ago2, it will make changes in the overall size distribution in a mutant library invisible. We refer to the studies by Okamura and colleagues as well as Ameres and colleagues for a discussion of this aspect (49,50). An analysis of the relative abundance revealed that r2d2 mutant flies do not show a gross reduction of transposon-matching endo-siRNAs relative to heterozygous animals, consistent with the results obtained from our own libraries (Supplementary Table S2, 3.63 versus 2.51%). As previously reported, their abundance also did not change in the untreated ago2 mutant library (3.59%). We calculated the free energy of base pairing across the first 5 nt of each read at either end of the presumed duplex siRNA (47,51) as a surrogate for direct counting of guide and passenger strand pairs, then determined the difference between the 5′- and the 3′-end of each presumed endo-siRNA precursor. A comparison between two different untreated wild-type libraries and the untreated ago2 mutant library revealed that in the absence of Ago2 the thermodynamic asymmetry of transposon-matching endo-siRNAs is lost (Figure 6A and Supplementary Table S2). Presumably, the duplexes are produced but then no longer loaded and accumulate as double-stranded intermediates. This does not occur in r2d2 mutants, because the thermodynamic asymmetry is maintained in this library. Transposon-matching endo-siRNAs partition at least partially into Ago1 in ovaries from r2d2 mutant animals (50), yet their relative amount can still be enriched by oxidation of the small RNAs (from 3.63 to 11.1%). Thus, they do retain the 2′-O-methyl modification, indicating that again loading of Ago2 can occur in the absence of R2D2. A comparison of the length distribution for transposon-matching reads in the libraries from oxidized RNA further demonstrates that a greater population of 2′-O-methyl modified 21 nt transposon-matching endo-siRNAs is present in r2d2 mutants compared with ago2 mutants (Figure 6B, dashed arrow). In addition, the generation of larger, piRNA-sized products is also observable in the oxidized library from r2d2 mutant animals.
Figure 6.
Analysis of strand asymmetry and read length distribution in deep sequencing data. (A) We calculated the difference in the free energy of duplex formation at either end of the presumed siRNA precursor for each sequence read using the nearest neighbor method (52), then calculated the difference (ΔΔG0′). A positive value indicates that on average the 5′-ends of the reads were less stably base paired than the opposite ends. The thermodynamic asymmetry was calculated for transposon-matching endo-siRNAs of the indicated genotypes; n.a.: not available in the series. (B) Read length distribution of transposon-matching endo-siRNAs in deep sequencing libraries from r2d2/CyO, r2d2/r2d2 and ago2414/ago2414 mutant fly heads. Left side: sequencing libraries from untreated RNAs; right side: sequencing libraries prepared from oxidized RNA to enrich for 2′-O-methyl modified RNA species. (C) Calculation of the thermodynamic asymmetry for white-IR derived siRNAs from the indicated genotypes.
The published libraries even allowed us to extend our analysis to siRNAs derived from the white-IR transgene, which we analyzed functionally in Figure 4. As previously described, the untreated r2d2 mutant library showed a more than 10-fold reduction of white-IR derived siRNAs. However, the remaining siRNAs retain their thermodynamic asymmetry, indicating that they are loaded into an Ago-protein (Figure 6C and Supplementary Table S3). Oxidation of the RNA prior to library generation strongly enriched for the white-IR derived siRNAs, in particular in the r2d2/CyO heterozygotes. However, the enrichment seen in the absence of r2d2 is comparable to the enrichment observed in a library from wild-type flies, indicating that while their abundance is clearly reduced, the remaining white-IR derived siRNAs in r2d2 mutant animals are loaded into Ago2. This is fully consistent with functional data showing that r2d2 mutant animals retain about one-fifth of the white-IR directed silencing capacity (26). In contrast to the transposon-derived endo-siRNAs, we did not observe any piRNA-sized small RNAs in the absence of R2D2 among the white-IR derived siRNAs (data not shown). In summary, our analysis demonstrates that hairpin-derived, transposon-derived and transgenic inverted-repeat derived siRNAs can be loaded—though to a reduced extent—into Ago2 in the absence of R2D2. We noted that some oxidation resistant, 21 nt long siRNAs also persist in libraries prepared from dcr2 mutant heads (Supplementary Tables S1 and S2). It is unclear at this point how these small RNAs are processed. In the case of transposon-matching endo-siRNAs, this 21 nt population has lost the thermodynamic asymmetry (Supplementary Table S2)—a speculative interpretation could be that in the absence of Dcr-2 loading into Ago2 can occur (because the siRNAs resist oxidation), but the sensing of thermodynamic asymmetry is no longer possible. However, the preference for a 5′-adenosine or uracil remains (Supplementary Figure S5).
DISCUSSION
The small RNA silencing system of Drosophila has evolved to cope with a remarkable range of small RNA abundances: Deep sequencing libraries uncovered variations in the cloning frequency of up to five orders of magnitude between abundant miRNAs and individual transposon-targeting endo-siRNAs. On top of this great dynamic range, the system keeps considerable capacities in spare since a large pool of siRNAs can be deployed rapidly upon infection with an RNA virus. Functional specialization of the effector proteins Ago1 and Ago2 for miRNAs and siRNAs, respectively, likely plays a key role for this phenomenon and the underlying biogenesis machinery that channels precursors into either pathway must be able to recognize which kind of trigger molecule it is dealing with.
In this study we describe that Loqs-PD likely interacts with Dcr-2 at an equivalent position as its homolog R2D2. Functional analysis showed that the two double-stranded RNA binding domain (dsRBD) proteins define distinct pathways, which can distinguish silencing triggers produced by high-copy sequences, such as transposons, from experimentally introduced dsRNA, a model for replicating RNA viruses. The two dsRBD proteins participate in two independent events during siRNA biogenesis: First the nucleolytic processing by assisting Dcr-2 and second the loading of Ago2. It was demonstrated that for miRNAs, these two events are uncoupled; this appears to be true for endo-siRNAs as well.
Antagonism of small RNA biogenesis pathways
All studies concerned with endo-siRNA silencing in Drosophila unanimously found a dependence on Ago2 and Dcr-2 (6–8,22,32). The surprising finding was the requirement of Loqs instead of the canonical RNAi factor R2D2. R2D2 had no influence on the biogenesis of endo-siRNAs. Later on, a specific isoform of Loqs (Loqs-PD) was identified that separates the function of Loqs during miRNA and endo-siRNA biogenesis (14,22). Miyoshi and coworkers proposed that a trimeric complex consisting of Dcr-2, Loqs and R2D2 can form (21). Such a complex would be compatible with functional specialization of the dsRBD proteins for the dicing or the Argonaute-loading reaction, and furthermore suggests that the two events may be coupled. The experimental evidence for this trimeric complex was obtained upon overexpression of tagged proteins, though, and may not reflect the physiological situation. We now present evidence that Loqs-PD and R2D2 compete for Dcr-2 binding. The interaction of Loqs-PD and Dcr-2 maps to the helicase domain of Dcr-2, the same region that was determined for R2D2 (Figure 2). While this region may be large enough to accommodate more than one protein, we demonstrate that decreasing the level of either R2D2 or Loqs-PD resulted in an increased silencing activity of the other pathway. This indicates that competition for a distinct, common factor—most likely Dcr-2—is the rate-limiting under normal circumstances. Indeed, the overexpression of Dcr-2 but not R2D2 can enhance the efficiency of transgenic RNAi (52). Furthermore, we find little evidence for an association between endogenous Loqs-PD and overexpressed R2D2, while the other Loqs isoforms do seem to associate with Loqs-PD upon overexpression (Figures 3A and 5A). A parallel phenomenon has been reported for the competition between Loqs-PA and -PB in miRNA biogenesis (28). Here, the authors proposed that the two isoforms compete for binding to Dcr-1 and induce different conformations (28). Possibly the Loqs-PB induced conformation increases Dcr-1 affinity for miRNA precursors, thus enhancing dicing efficiency (29). We speculate that Loqs-PD and R2D2 may similarly induce different states of Dcr-2 or may otherwise enable the complex to preferentially act in either endo- or exo-siRNA silencing.
Parallel pathways versus sequential action of Loqs-PD and R2D2
The hypothesis that Loqs-PD and R2D2 act sequentially (31) was proposed based on three lines of evidence: First, RNAi is impaired in both loqs and r2d2 mutants as measured by injection of dsRNA into embryos, or by silencing induced with the help of a white inverted repeat construct. Second, a reduction of siRNA density was measured by deep sequencing in loqs mutant flies and third, biochemical data probing for binding and processing activities in mutant embryo extracts suggested a function of loqs in dsRNA-triggered silencing. Generally speaking, the interpretation of these results could be hampered by the fact that a null allele of loqs was employed, resulting in loss of all protein isoforms. As a consequence, miRNA biogenesis was impaired in addition to the effects on siRNA generation and some of the observed effects may be indirect. The miRNA pathway plays an important role in eye development (43,45) and loqs mutant flies have visible morphological changes of the eye. This may have resulted in a perturbed tissue balance and partially explain a reduction of siRNA production from the white-IR transgene. Furthermore, a dcr-1 mutation also leads to partial loss of silencing by the white-IR transgene (43), and the need to form a very large complex containing both Dcr-1 and Dcr-2 was proposed to be the underlying cause (18). It may well be that the miRNA-pathway specific Loqs-PB isoform is also present in this putative complex, resulting in indirect effects on siRNA accumulation downstream of the biogenesis cascade. We did not observe a significant change in white-IR silencing activity upon re-addition of Loqs-PB (Figure 4C and E), but this was only in a trans-heterozygous configuration between the loqsko and the hypomorphic loqsf00791 allele. We are unfortunately unable to obtain homozygous mutant loqsko flies in our laboratory, despite several attempts to outcross the existing stock. The reduced efficiency of silencing by injected dsRNA in loqs mutant embryos may likewise have been affected indirectly, since Loqs-PB and miRNAs are important for germ-line stem cell renewal (44) and multiple downstream steps. Consequently, changes in tissue balance or protein composition of loqs mutant embryos may lead to indirect effects on dsRNA-triggered silencing. This view is also consistent with the reported defects of RNA interference in ago1k08121 mutant embryos (53).
Our analysis of deep sequencing libraries in conjunction with two recently published studies (49,50) further demonstrates that the different siRNA classes are not affected equally by mutation of r2d2. While the white-IR derived small RNAs are clearly reduced in abundance, the amount of hairpin-derived as well as transposon-derived endo-siRNAs is unchanged in libraries form untreated RNA. This is inconsistent with a model where dicing and Ago2-loading are coupled in an obligatory manner. Furthermore, we could demonstrate that R2D2 is not always required for Ago2 loading, indicating that the Dcr-2/R2D2 RISC loading complex (RLC) can be bypassed for some substrates. This argument relies strongly on the presence of corresponding 21-mer reads in the deep sequencing libraries from oxidized RNA. Could these simply be Ago1-loaded small RNAs that have escaped the sodium periodate treatment rather than an indication of R2D2-independent loading? We think that this is unlikely because the different siRNA classes are affected unequally by the r2d2 mutation. A comparison of the oxidized libraries from r2d2/CyO and r2d2/r2d2 animals reveals that the relative abundance of CG4068B matching, hairpin-derived endo-siRNAs decreases ∼3-fold (Supplementary Table S1) that white-IR derived siRNAs decrease about 19-fold (Supplementary Table S3), while the transposon-derived endo-siRNAs increase >2-fold (Supplementary Table S2). This is consistent with our finding that depletion of r2d2 increases the efficiency of endo-siRNA mediated silencing of a high-copy transgene in S2-cells (14) and argues that the biogenesis of exo-siRNAs and transposon-derived endo-siRNAs occurs by a distinct mechanism. Furthermore, Ago2-loaded small RNAs from the white-IR transgene were reported to have a bias for cytosine as their 5′ nt, but it was unclear whether this reflects a sequence preference of Ago2 itself or the RISC loading complex (RLC) (6). In contrast, transposon-derived, Ago2-loaded endo-siRNAs have a preference for adenosine or uracil at their 5′-end [Supplementary Figure S5A and (50)]. This bias is even more pronounced in r2d2 mutants, indicating that it may correspond to the sequence preference of a distinct RLC that does not require R2D2. Finally, Ago1 is reported to unwind duplex siRNAs only slowly (49,50). Indeed, the library generated from untreated RNA of ago2 mutant flies shows a loss of thermodynamic asymmetry which may reflect their re-direction into Ago1 (50). In contrast, the corresponding library from r2d2 mutants retained asymmetry (Figure 6), indicating that loss of r2d2 and loss of ago2 have distinct effects on the transposon-derived siRNA population.
Separation of the endo- and exo-siRNA pathway is not clear cut and intermediate states exist: We repeated the white-IR silencing assay in flies with restored miRNA function (Figure 4) and our quantitative analysis revealed that loqs-PD mutant flies de-repress the white reporter by only ∼25%. This is in agreement with a previous study where the hypomorphic allele loqsf00791 was employed. Furthermore, de-repression of the white reporter by only 70–80 % was observed in r2d2 mutants (26). Since neither null mutation leads to complete loss of silencing, and Ago2-loaded white-IR derived siRNAs remain detectable in r2d2 mutant flies, the simplest interpretation is that two parallel pathways exist. In this model, the Dcr-2/Loqs-PD complex processes and/or loads the white-IR derived, double-stranded RNA in one-fifth of the cases, while the Dcr-2/R2D2 complex takes care of the rest (Figure 7). Sequential action of Loqs-PD and R2D2 is also difficult to reconcile with the mutually antagonistic activities seen in our cell culture experiments [Figure 3E and (14)].
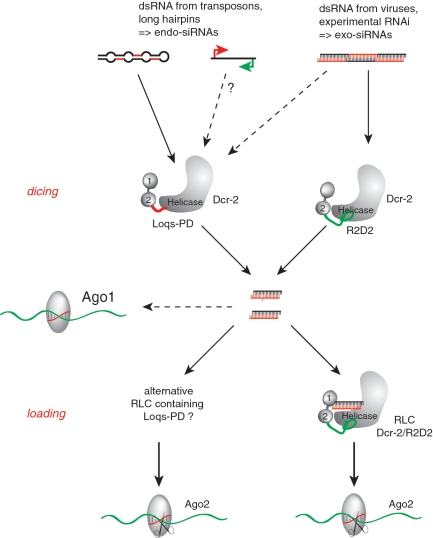
Figure 7.
Distinct biogenesis pathways for exo- and endo-siRNAs. Loqs-PD and R2D2 both contain two N-terminal double-stranded RNA binding domains (labeled 1 and 2) and associate via their C-terminal domain with the N-terminal helicase domain of Dcr-2. This is analogous to the situation of Dcr-1/Loqs-PB and of human Dicer/TRBP or PACT. Despite the similar architecture, the two Drosophila complexes are functionally distinct: Our results indicate that R2D2 and Loqs-PD compete for association with Dcr-2, and that parallel pathways for processing and loading of siRNAs exist. Analogous to the situation for miRNA biogenesis, they appear to be uncoupled after the dicing step. Thus, it is possible that a siRNA is processed by Dcr-2/Loqs-PD and loaded by Dcr-2/R2D2, but this sequence of events is not obligatory. R2D2 may not be required for the dicing reaction; since competition for Dcr-2 exists we propose that the presence of “free” Dcr-2 unlikely. Two recent publications demonstrate that endogenous siRNAs can also be loaded into Ago1 to some extent (49,50).
Assuming that two parallel biogenesis pathways exist, and that the separation is not always perfect (as judged by the white-IR assay), it becomes difficult to interpret the biochemical data provided by Carthew and colleagues in favor of sequential action versus parallel pathways. In fact, the authors acknowledge that some dsRNA processing activity remains in loqs mutant embryos or embryo extracts (39 and 14%, respectively), but also r2d2 mutant extracts showed only 53% of wild-type processing activity. Furthermore, loqs mutant extracts showed a strong reduction of RISC Loading Complex (RLC) formation, consistent with a potential role of loqs in both dicing and loading of endo-siRNAs. UV-crosslinking of siRNA to Dcr-2 was dependent on r2d2 but not loqs, but this may only reflect the situation of exo-siRNA biogenesis.
What feature of the substrate determines processing by Dcr-2/Loqs-PD or Dcr-2/R2D2?
Our model (Figure 7) is consistent with the experimental data, but raises an important question: How can the same substrate molecule—double-stranded RNA—be sorted into two distinct pathways? It seems reasonable that our current definition of endogenous versus exogenous origin is unlikely to be shared by the cellular machinery. The transgenic silencing triggers demonstrate that dsRNA of nuclear origin is—at least for the greater portion—processed by the same machinery as experimentally introduced, in vitro transcribed dsRNA. A more likely scenario is that the substrates are distinguished based on their abundance, with dsRNA derived from transposons being much rarer than the dsRNA produced during replication of an RNA virus or experimental RNAi. If one accepts the amount of siRNAs produced as a surrogate measure for the concentration of their precursors, then this hypothesis is not unreasonable (54). Given that transposons are far more abundant in the oocyte, a consequence of this model might even be that endo-siRNAs are differentially processed between germ line and soma. Also, within the somatic tissue, individual transposable elements might be targeted the Dcr-2/Loqs-PD and Dcr-2/R2D2 pathways to a different extent depending on their transcriptional activity. The endo-siRNA products of hairpin-generating loci may likewise be affected by differential promoter activities between soma and germ line or between different somatic tissues.
On the other hand, a difference in size, with e.g. endo-siRNA precursors being shorter on average than exo-siRNA precursors, could be a distinguishing feature. Preferential binding of Caenorhabditis elegans RDE-4, a homolog of Loqs and R2D2, to long dsRNA has been described (55). Alternatively, a specific factor that associates with nuclear and/or cytoplasmic dsRNA may also be responsible for the selectivity. In this case, the dsRNA derived from an inverted repeat transgene (such as white-IR) should be of intermediate size with respect to the selection cutoff or less efficient in retaining/recruiting this specific protein factor and thus enter either pathway with a certain probability. Further studies can now be designed to address these questions.
Conserved relations between Dicer and dsRBD proteins
The single Dicer enzyme found in humans can interact with TRBP and PACT, both of which are dsRBD proteins and homologs of Drosophila Loqs and R2D2 (41,56–58). This interaction could be mapped to 69 amino acids at the C-terminal end of TRBP and 165 amino acids in the N-terminal domain of Dicer (59). Thus, the positions of the interacting domains are conserved not only between Drosophila Dcr-1 and Dcr-2, but also in vertebrate Dicer enzymes. In C. elegans, RDE-4 interacts with Dcr-1 as well as Drh-1 (60). While the Dicer-interacting domain of RDE-4 has not been mapped, the C-terminal third dsRBD mediates dimerization of RDE-4 and thereby increases the affinity to long dsRNA (55). It is intriguing that TRBP and PACT have antagonistic effects on human Dicer: TRBP stimulates miRNA dicing and stabilizes Dicer, while PACT inhibits miRNA processing (61). In addition to their function in RNAi, the two proteins are involved in the antagonistic regulation of the dsRNA activated Protein Kinase (PKR). Whereas TRBP inhibits PKR function both by direct dsRBD-mediated binding of PKR and sequestering dsRNA (62,63), PACT binds to PKR through its dsRBDs 1 and 2 and activates the enzyme via the C-terminus (64,65). Thus, the modulation of enzyme activity by mutually exclusive dsRBD protein partners may be a conserved strategy even between otherwise unrelated protein complexes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
DFG-grant FO-360/2; the Munich Center for integrated Protein science (CiPSM); the DFG Sonderforschungsbereich 646; an ‘LMU excellent’ grant (to K.F.); Boehringer-Ingelheim Fonds PhD fellowship (to J.V.H.); LMU-Mentoring program (to J.V.H.); Career Development Award from the International Human Frontier Science Program Organization (to K.F.). Funding for open access charge: DFG-grant FO-360/2.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Romy Böttcher for technical assistance in the preparation of deep sequencing libraries, M. Siomi for a gift of monoclonal anti-Loqs antibody and Gunter Meister as well as Michael Sattler for critical comments on the article.
REFERENCES
- 1.Hartig JV, Tomari Y, Forstemann K. piRNAs–the ancient hunters of genome invaders. Genes Dev. 2007;21:1707–1713. doi: 10.1101/gad.1567007. [DOI] [PubMed] [Google Scholar]
- 2.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell. Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 5.Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 9.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 11.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 12.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 13.Blumenstiel JP, Hartl DL. Evidence for maternally transmitted small interfering RNA in the repression of transposition in Drosophila virilis. Proc. Natl. Acad. Sci. USA. 2005;102:15965–15970. doi: 10.1073/pnas.0508192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartig JV, Esslinger S, Bottcher R, Saito K, Forstemann K. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J. 2009;28:2932–2944. doi: 10.1038/emboj.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipardi C, Paterson BM. Identification of an RNA-dependent RNA polymerase in Drosophila involved in RNAi and transposon suppression. Proc. Natl. Acad. Sci. USA. 2009;106:15645–15650. doi: 10.1073/pnas.0904984106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Forstemann K. Transposon defense in Drosophila somatic cells: a model for distinction of self and non-self in the genome. RNA Biol. 2010;7:158–161. doi: 10.4161/rna.7.2.11059. [DOI] [PubMed] [Google Scholar]
- 17.Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 18.Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- 19.Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 20.Hartig JV, Esslinger S, Bottcher R, Saito K, Forstemann K. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J. 2009;28:2932–2944. doi: 10.1038/emboj.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyoshi K, Miyoshi T, Hartig JV, Siomi H, Siomi MC. Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila. RNA. 2010;16:506–515. doi: 10.1261/rna.1952110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou R, Czech B, Brennecke J, Sachidanandam R, Wohlschlegel JA, Perrimon N, Hannon GJ. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA. 2009;15:1886–1895. doi: 10.1261/rna.1611309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyle M, Jantsch MF. New and old roles of the double-stranded RNA-binding domain. J. Struct. Biol. 2002;140:147–153. doi: 10.1016/s1047-8477(02)00544-0. [DOI] [PubMed] [Google Scholar]
- 24.Ye X, Paroo Z, Liu Q. Functional anatomy of the Drosophila microRNA-generating enzyme. J. Biol. Chem. 2007;282:28373–28378. doi: 10.1074/jbc.M705208200. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Jiang F, Kalidas S, Smith D, Liu Q. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA. 2006;12:1514–1520. doi: 10.1261/rna.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyoshi K, Okada TN, Siomi H, Siomi MC. Characterization of the miRNA-RISC loading complex and miRNA-RISC formed in the Drosophila miRNA pathway. RNA. 2009;15:1282–1291. doi: 10.1261/rna.1541209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Park JK, Jiang F, Liu Y, McKearin D, Liu Q. Dicer-1, but not Loquacious, is critical for assembly of miRNA-induced silencing complexes. RNA. 2007;13:2324–2329. doi: 10.1261/rna.723707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marques JT, Kim K, Wu PH, Alleyne TM, Jafari N, Carthew RW. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat. Struct. Mol. Biol. 2010;17:24–30. doi: 10.1038/nsmb.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung WJ, Okamura K, Martin R, Lai EC. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr. Biol. 2008;18:795–802. doi: 10.1016/j.cub.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Förstemann KH, MichaelD, Wee LM, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by Dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah C, Forstemann K. Monitoring miRNA-mediated silencing in Drosophila melanogaster S2-cells. Biochim. Biophys. Acta. 2008;1779:766–772. doi: 10.1016/j.bbagrm.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 36.Le T, Yu M, Williams B, Goel S, Paul SM, Beitel GJ. CaSpeR5, a family of Drosophila transgenesis and shuttle vectors with improved multiple cloning sites. Biotechniques. 2007;42:164. doi: 10.2144/000112386. 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor; 1988. [Google Scholar]
- 38.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau PW, Potter CS, Carragher B, MacRae IJ. Structure of the human Dicer-TRBP complex by electron microscopy. Structure. 2009;17:1326–1332. doi: 10.1016/j.str.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang HW, Noland C, Siridechadilok B, Taylor DW, Ma E, Felderer K, Doudna JA, Nogales E. Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol. 2009;16:1148–1153. doi: 10.1038/nsmb.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim do H, Kim J, Kim S, Carthew RW, Lee YS. Functional analysis of dicer-2 missense mutations in the siRNA pathway of Drosophila. Biochem. Biophys. Res. Commun. 2008;371:525–530. doi: 10.1016/j.bbrc.2008.04.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 44.Park JK, Liu X, Strauss TJ, McKearin DM, Liu Q. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr. Biol. 2007;17:533–538. doi: 10.1016/j.cub.2007.01.060. [DOI] [PubMed] [Google Scholar]
- 45.Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123:1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 46.Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 47.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 48.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ameres SL, Hung JH, Xu J, Weng Z, Zamore PD. Target RNA-directed tailing and trimming purifies the sorting of endo-siRNAs between the two Drosophila Argonaute proteins. RNA. 2010 doi: 10.1261/rna.2498411. doi:10.1261/rna.2498411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okamura K, Robine N, Liu Y, Liu Q, Lai EC. R2D2 organizes small regulatory RNA pathways in Drosophila. Mol. Cell. Biol. 2010 doi: 10.1128/MCB.01141-10. doi:10.1128/MCB.01141-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia T, SantaLucia J, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 52.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 53.Williams RW, Rubin GM. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc. Natl. Acad. Sci. USA. 2002;99:6889–6894. doi: 10.1073/pnas.072190799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okamura K, Balla S, Martin R, Liu N, Lai EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat. Struct. Mol. Biol. 2008;15:581–590. doi: 10.1038/nsmb.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parker GS, Eckert DM, Bass BL. RDE-4 preferentially binds long dsRNA and its dimerization is necessary for cleavage of dsRNA to siRNA. RNA. 2006;12:807–818. doi: 10.1261/rna.2338706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gatignol A, Buckler-White A, Berkhout B, Jeang KT. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 59.Daniels SM, Melendez-Pena CE, Scarborough RJ, Daher A, Christensen HS, El Far M, Purcell DF, Laine S, Gatignol A. Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC Mol. Biol. 2009;10:38. doi: 10.1186/1471-2199-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 61.Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daher A, Longuet M, Dorin D, Bois F, Segeral E, Bannwarth S, Battisti PL, Purcell DF, Benarous R, Vaquero C, et al. Two dimerization domains in the trans-activation response RNA-binding protein (TRBP) individually reverse the protein kinase R inhibition of HIV-1 long terminal repeat expression. J. Biol. Chem. 2001;276:33899–33905. doi: 10.1074/jbc.M103584200. [DOI] [PubMed] [Google Scholar]
- 63.Park H, Davies MV, Langland JO, Chang HW, Nam YS, Tartaglia J, Paoletti E, Jacobs BL, Kaufman RJ, Venkatesan S. TAR RNA-binding protein is an inhibitor of the interferon-induced protein kinase PKR. Proc. Natl. Acad. Sci. USA. 1994;91:4713–4717. doi: 10.1073/pnas.91.11.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta V, Huang X, Patel RC. The carboxy-terminal, M3 motifs of PACT and TRBP have opposite effects on PKR activity. Virology. 2003;315:283–291. doi: 10.1016/s0042-6822(03)00589-0. [DOI] [PubMed] [Google Scholar]
- 65.Huang X, Hutchins B, Patel RC. The C-terminal, third conserved motif of the protein activator PACT plays an essential role in the activation of double-stranded-RNA-dependent protein kinase (PKR) Biochem. J. 2002;366:175–186. doi: 10.1042/BJ20020204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.