Abstract
Methylation of the five position of cytosine in DNA plays important roles in epigenetic regulation in diverse organisms including humans. The transfer of methyl groups from the cofactor S-adenosyl-l-methionine is carried out by methyltransferase enzymes. Using the paradigm bacterial methyltransferase M.HhaI we demonstrate, in a chemically unperturbed system, the first direct real-time analysis of the key mechanistic events—the flipping of the target cytosine base and its covalent activation; these changes were followed by monitoring the hyperchromicity in the DNA and the loss of the cytosine chromophore in the target nucleotide, respectively. Combined with studies of M.HhaI variants containing redesigned tryptophan fluorophores, we find that the target base flipping and the closure of the mobile catalytic loop occur simultaneously, and the rate of this concerted motion inversely correlates with the stability of the target base pair. Subsequently, the covalent activation of the target cytosine is closely followed by but is not coincident with the methyl group transfer from the bound cofactor. These findings provide new insights into the temporal mechanism of this physiologically important reaction and pave the way to in-depth studies of other base-flipping systems.
INTRODUCTION
The phenomenon of hyperchromicity refers to increased absorbance of nucleobases in free nucleotides or single polynucleotide strands (60–80% or 30–40%, respectively) as compared to those in DNA duplexes, which is thought to derive from the lost or weakened π–π interactions between the orderly stacked bases in the helical structures (1). A unique example of a local but dramatic distortion of the DNA helix, in which a target base is unstacked completely and the two adjacent nucleobases are unstacked on one face, is provided by enzyme-induced base flipping. A paradigm system for studies of this phenomenon is the HhaI DNA cytosine-5 methyltransferase (M.HhaI) (Figure 1A) (2), which recognizes and methylates the GCGC target sites in duplex DNA. The base flipping event in M.HhaI is accompanied by a massive movement of the catalytic loop (residues 81–100) towards the DNA which locks the flipped-out base for methylation (Figure 1A). Enzymatic catalysis involves a transient formation of a Michael adduct between the catalytic cysteine (Cys81 in M.HhaI) and C6 of the flipped out cytosine, permitting a direct SN2 transfer of the methyl group onto C5 (Figure 1B). Although many kinetic and structural aspects of this biologically relevant enzymatic reaction have been determined (3–11), the underlying kinetics of the target cytosine flipping, loop closure, covalent bond formation have not yet been established. Previous kinetic studies employing DNA substrates with fluorescent base analogs such as 2-aminopurine replacing the target cytosine or mutational variants of the protein (12–14) could not provide final answers due to substantial structural alterations to the system.
Figure 1.
Conformational transitions and covalent catalysis by M.HhaI. (A) Upon binding of DNA and cofactor, M.HhaI flips its target cytosine out of the DNA helix into the active site; the catalytic loop in the protein makes a large motion to lock the target base and the bound cofactor. M.HhaI is shown as backbone trace, the catalytic loop (residues 81–100) is red, the engineered Ile86 residue is shown as space fill, DNA and cofactor are represented as sticks models in blue and orange, respectively. (B) The mechanism of covalent target base activation and methyl group transfer by M.HhaI along with associated spectral changes of the target base. (C) General kinetic scheme of conformational transitions upon formation of an unproductive ternary complex with AdoHcy (upper) and during catalytic turnover in the presence of AdoMet (lower). C, cofactor (AdoHcy); E, enzyme; D, DNA; m, methylgroup on cofactor or DNA; F, flipped out conformation; L, locked loop conformation; dot, non-covalent association; hyphen, a covalent bond between enzyme and DNA.
Understanding the mechanism of enzymatic cytosine-5 methylation is of utmost importance as DNA methylation is a part of an intricate epigenetic regulatory network in vertebrates. In this work, we used stopped-flow kinetic analysis to follow in real-time the events of target cytosine flipping, loop closure, covalent bond formation during catalysis of the M.HhaI methyltransferase in a chemically unperturbed system. Base flipping and covalent bond formation were directly observed by monitoring UV absorbance changes that occur due to hyperchromicity and the loss of the cytosine chromophore, respectively. In parallel, conformational changes in the catalytic loop have been followed using an engineered variant of M.HhaI that contained a unique tryptophan fluorophore at the tip of the 20 residue loop. This unique set up allowed us, for the first time, to establish the temporal order and kinetic parameters of key steps in the catalytic cycle of a DNA cytosine-5 methyltransferase.
MATERIALS AND METHODS
AdoMet purification
For the stopped-flow analyses commercial AdoMet (p-toluensulphonate salt, Sigma) was purified by cation-exclusion chromatography (passage through a Dowex 1 × 8 column in 0.1 mM formic acid), concentrated on a rotary evaporator and stored in small aliquots at −20°C. The resulting AdoMet was at least 95% pure and contained no detectable AdoHcy.
Mutagenesis and protein purification
The I86W mutation in the catalytic loop was introduced as previously described (15) using the following oligonucleotide primer (the modified codon is in bold, a silent mutation is italicized): 5′-TTGTTTTCCACTCCAGGAAAACGCTTG G-3′. The double-mutant W41F/I86W was created by recombining the I86W mutation with the previously constructed single mutant W41F (11).
M.HhaI variants were expressed in Escherichia coli ER2267 (16) and purified as described previously (8). Briefly, cells were disrupted by sonication in low ionic strength buffer (10 mM Hepes pH 7.2, 5 mM EDTA, 10% glycerol, 2 mM 2-mercaptoethanol), cell debris was extracted with high ionic strength buffer (10 mM K-PO4 pH 7.4, 400 mM NaCl, 5 mM EDTA, 10% glycerol, 2 mM 2-mercaptoethanol) and extensively dialyzed to remove endogenous AdoMet. Then the solution was loaded onto a Q-Sepharose column and the flow-through containing M.HhaI was fractionated on an SP-Sepharose column with 0.1–0.5 M linear gradient of NaCl. Protein concentrations were estimated using a Coomassie G-250 assay with BSA as standard and further refined by active site titration as described previously (12). Bound endogenous AdoMet in M.HhaI preparations was under 2 mol% as determined using a DNA protection assay.
Steady-state kinetics
Reactions were carried out in Methylation buffer (50 mM Tris–HCl, pH 7.4, 50 mM NaCl, 0.5 mM EDTA, 2 mM 2-mercaptoethanol and 0.2 mg/ml of BSA) containing 2 nM MTase, 1 µM poly [dG–dC]·poly[dG–dC] (concentration of double-stranded GCGC sites) and varying [3H-methyl]-AdoMet from 0.01 to 100 µM. After incubation at 37°C for 10 min, the reactions were stopped with 0.5 N HCl and the radioactivity incorporated into DNA was measured by filter-binding assay, as described previously (12). Data were fitted into the Michaelis–Menten mechanism using Grafit 5.0.6 (Erithacus Software) or Dynafit (17) software.
Single turnover assay
Single turnover reactions were performed in Methylation buffer at 25°C in a Rapid-Quench-Flow instrument RQF-3 (KinTek). MTase preincubated with DNA was mixed rapidly with [3H-methyl]AdoMet and after a specified period (0.1–300 s) reactions were quenched in 0.7 N hydrochloric acid. The amount of radioactivity incorporated into DNA was measured by filter binding assay as described (12). For WT M.HhaI, 200 nM MTase, 100 nM GCGC/GMGC double-stranded oligonucleotide (Table 1) and 2.5 µM [3H-methyl]AdoMet (16.1 Ci/mmol) were used; for the W41F/I86W mutant, reactions contained 1 µM MTase, 0.5 µM GCGC/GMGC and 30 µM [3H-methyl]AdoMet (1.62 Ci/mmol). Data were fitted into a single-exponential function using Grafit 5.0.6 (Erithacus Software).
Table 1.
DNA duplexes used for the biochemical studies
| M.HhaI target | Target base pair | DNA sequence |
|---|---|---|
| GCGC/GMGC | C:G | 5′-TACAGTATCAGGCGCTGACCCACAA |
| TGTCATAGTCCGMGACTGGGTGTTG-5′ | ||
| GCGC/GMTC | C:T | 5′-TACAGTATCAGGCGCTGACCCACAA |
| TGTCATAGTCCTMGACTGGGTGTTG-5′ | ||
| GCGC/GMCC | C:C | 5′-TACAGTATCAGGCGCTGACCCACAA |
| TGTCATAGTCCCMGACTGGGTGTTG-5′ | ||
| GsGC/GMGC | s:G | 5′-TACAGTATCAGGsGCTGACCCACAA |
| TGTCATAGTCCGMGACTGGGTGTTG-5′ | ||
| GPGC/GMTC | P:T | 5′-TACAGTATCAGGPGCTGACCCACAA |
| TGTCATAGTCCTMGACTGGGTGTTG-5′ | ||
| GFGC/GMGC | F:G | 5′-TACAGTATCAGGFGCTGACCCACAA |
| TGTCATAGTCCGMGACTGGGTGTTG-5′ | ||
| TCGA | Non-cognate site | 5′-TACAGTATCAGTCGATGACCCACAA |
| TGTCATAGTCAGCTACTGGGTGTTG-5′ |
M, 5-methylcytosine; s, abasic site (sugar); P, 2-aminopurine; F, 5-fluorocytosine.
Stopped-flow kinetics
Measurements were performed using an Applied Photophysics SX.18MV-R spectrometer equipped with a monochromator, 150 W xenon lamp, 20 µl cell and dual detector for simultaneous acquisition of absorbance and fluorescence signals. Excitation wavelength was typically 280 nm (slit width 4.6 nm); tryptophan emission was measured using a 335 nm cut-off filter. Experiments were performed using 10 or 2 mm path lengths when higher sensitivity in absorption mode or lower inner-filter effect in the fluorescence regime, respectively, was required. Typically, 3–7 traces were acquired and averaged from reactions containing 3 µM MTase, 2.5 µM DNA and 0–200 µM AdoMet or AdoHcy in Reaction buffer (10 mM Tris–HCl pH 7.4, 50 mM NaCl, 0.5 mM EDTA, 2 mM 2-mercaptoethanol) at 25°C. Normalized absorbance and fluorescence curves were fitted into three-exponential functions with two shared rate constants as described above. Multicurve analysis was performed using Origin 8.1 SS2 software (OriginLab).
RESULTS
Direct observation of cytosine flipping and covalent bond formation
Binding of M.HhaI to its GCGC target in duplex DNA induces flipping of the target cytosine base (underlined) out of the DNA helix with little disturbance to the rest of the duplex. Since the target cytosine is unstacked completely, and the two adjacent guanines are unstacked on one face (2), we presumed that these changes might give a detectable hyperchromic change for direct observation of the flipping motion. Subsequent covalent catalysis by M.HhaI involves the formation of a covalent bond from the Cys81 residue to the C6 atom of the flipped out target cytosine (Figure 1B). The attack at C6 disrupts the aromatic system in the cytosine ring and dramatically alters the spectral properties of the chromophore (λmax shifts from 270 nm in cytosine to 225–240 nm in 5,6-dihydrocytosines) (18–20). We therefore examined if the UV absorbance changes deriving from the target cytosine can be detected in a 25-mer duplex substrate using stopped flow techniques. Remarkably, a clearly defined A280 signal comprising a 25–30 mAU increase followed by a 18–20 mAU decrease (in the background of total absorbance of ∼1 AU) was obtained upon fast mixing of a 2.5 µM cognate hemimethylated 25-mer DNA duplex with an excess of M.HhaI (3 µM) and AdoHcy (100 µM) (Figure 2A) in a 10 mm cuvette. Control experiments with DNA duplexes containing an abasic residue replacing the target cytosine, a non-flippable 2-aminopurine:thymine base pair (8), a non-cognate target site all showed none or minor changes in absorbance (amplitudes of 3–6 mAU) (Figure 2A). Nor was flipping signal observed with the Q237A mutant of M.HhaI, which is strongly impaired in target base flipping (8) (Supplementary Figure S1A). A wavelength scan of the absorbance signal in the cognate system revealed a maximal change at ∼280–290 nm (Supplementary Figure S2). Finally, our observation of a similar absorbance trace (Supplementary Figure S1A) in the W41F mutant excludes the possibility that the observed signal originates from the unique tryptophan residue (W41) in the protein. The observed upward amplitude (Figure 2A, Table 2) represents a 150% hyperchromic change based on the absorbance of one 2′-deoxycytidine residue (derived from a molar extinction coefficient in neutral milieu of ε280 = 7200 M−1 cm−1). Taking into account that a much smaller absorbance change is observed upon flipping of an abasic target residue, one can conclude that the ascending transients in the absorbance traces largely represent the outgoing conformational transitions of the target nucleotide.
Figure 2.
Stopped-flow absorption traces during M.HhaI–DNA interaction. Reactions contained 3 µM MTase, 2.5 µM DNA and 100 µM AdoHcy (final concentrations). (A) WT M.HhaI was rapidly mixed with different DNA substrates in the presence of AdoHcy: blue—GCGC/GMGC (cognate target site), yellow—GsGC/GMGC (abasic target site), green—TCGA (non-cognate target site), purple—GPGC/GMTC (non-flippable target site); black—the catalytic C81S mutant rapidly mixed with GCGC/GMGC in the presence of 100 µM AdoHcy. (B) Reaction profiles at different mixing orders: blue—MTase was rapidly mixed with GCGC/GMGC in the presence of AdoHcy, red—MTase was first premixed with GCGC/GMGC and then mixed with AdoHcy, grey—MTase rapidly mixed with GCGC/GMGC in the absence of cofactor. Parameters derived from fitting experimental traces to exponential equations are shown in Table 2. Fits are shown as black thin lines.
Table 2.
Kinetics of target base flipping and covalent bonding upon M.HhaI interaction with cognate DNA
| Reaction | HhaI >< DNA |
HhaI + AdoHcy >< DNA + AdoHcy |
HhaI + DNA >< AdoHcy |
HhaI + AdoMet >< DNA + AdoMet |
||||
|---|---|---|---|---|---|---|---|---|
| Parameters | Rate, s−1 | Amplitude, mAU | Rate, s−1 | Amplitude, mAU | Rate, s−1 | Amplitude, mAU | Rate, s−1 | Amplitude, mAU |
| Flip-out | 27.4 ± 0.3 | 8.3 ± 0.04 | 19.1 ± 0.1 | 28 ± 0.1 | 19.7 ± 0.9 | 7.6 ± 0.5 | 120 ± 3 | 7.1 ± 0.1 |
| – | – | 2.5 ± 0.2 | 2.5 ± 0.1 | 5.5 ± 0.4 | 7.1 ± 0.4 | 14.4 ± 0.2 | 8.5 ± 0.1 | |
| C6-bond | 0.23 ± 0.06 | −0.7 ± 0.07 | 0.71 ± 0.003 | −23 ± 0.1 | 0.60 ± 0.01 | −16 ± 0.1 | 0.26 ± 0.003 | −12.8 ± 0.4 |
| Flip-back | – | – | – | – | – | – | 0.16 ± 0.004 | 5.6 ± 0.4 |
Apparent kinetic parameters derived from fitting absorption traces to multi-exponential functions for reactions involving 3 µM M.HhaI, 2.5 µM GCGC/GMGC duplex and 100 µM cofactor as shown in Figures 2 and 5. Premixed components in two chambers (separated by ><) were rapidly mixed in a stopped-flow cell to observe reaction-associated changes in absorbance. Values are reported with standard errors of the fit.
The observed downward amplitude of 16–20 mAU agrees well with the loss of one cytosine chromophore; an equally high although significantly slower absorbance change was also observed (Supplementary Figure S3) upon formation of an irreversible covalent M.HhaI–DNA complex involving 5-fluorocytosine (21). To confirm the nature of the descending absorbance change, we performed a control experiment with a catalytic mutant of M.HhaI, C81S, which lacks the catalytic thiol group required for the covalent bonding to the target cytosine (22). As expected, the downward signal was absent in the mutant system (Figure 2A). These findings clearly indicate that the descending transient is a reporter of the covalent bond formation in the M.HhaI–DNA complex. Altogether, our set up thus permits a direct real-time observation of the target cytosine flipping and subsequent covalent bond formation by the HhaI methyltransferase in a chemically unperturbed system.
In the absence of cofactor, interaction with DNA gives similar transients but several-fold reduced amplitudes as compared to the ternary complex (Figure 2B). The remaining signal is regained upon addition of AdoHcy to the preformed M.HhaI–DNA complex. This suggests that in the binary complex, a fast equilibrium is achieved in which about one-third of the target cytosine occurs in the flipped-out covalently bound state. This observation is consistent with the previously described dynamic nature of the complex (4,5) and catalysis of fast C5-hydrogen exchange in the absence of cofactor (3).
Simultaneous sensing of the catalytic loop motion with built-in fluorophores
In addition to base flipping, M.HhaI itself undergoes major conformational changes during the catalytic cycle (2). A large motion of the catalytic loop (residues 81–100), engulfs the bound DNA and the catalytic nucleophile Cys81 is brought in contact with the flipped out cytosine. To follow the motions of the catalytic loop during catalysis, we have redesigned the fluorophore locations in the HhaI methyltransferase by site directed mutagenesis of two positions. The unique intrinsic Trp residue (W41) in the cofactor binding pocket has been replaced by a Phe, whereas the bulky Ile86 residue in the catalytic loop was replaced with a Trp. The double W41F/I86W mutant showed similar kinetic parameters with the previously described W41F variant (Supplementary Table S1) (11); it differed from the WT M.HhaI mainly in the lower affinity towards the cofactor AdoMet or its product AdoHcy. A similar behavior has recently been noted for another two engineered single-tryptophan variants of M.HhaI, W41F/K91W and W41F/E94W (13,14).
Stopped flow experiments with the W41F/I86W mutant M.HhaI showed a strong decrease in the Trp fluorescence (excitation at 280 nm, emission at >335 nm) intensity upon formation of the ternary complex with the cognate DNA duplex and AdoHcy (Figure 3B). Null or small fluorescence changes were detected when either the DNA or cofator was omitted from the reaction, indicating that the signal is dependent on the formation of the closed ternary complex (4,5,14). The fluorescence signal was also absent in the catalytically active W41F mutant, which contains no tryptophan residues (Supplementary Figure S1). In the presence of the AdoMet cofactor, the fluorescence decrease was followed by an ascending fluorescence signal (see below and Figure 5B), consistent with the reversal of the conformational change after the methylation reaction. Thus we conclude that the observed quenching of the tryptophan fluorescence derives from the closure of the catalytic loop whereby the Trp86 residue approaches the major groove of the bound DNA duplex; similarly with the previously reported K91W and E94W variants (13,14), the engineered Trp86 residue is thus capable of reporting on conformational transitions of the catalytic loop through changes in fluorescence intensity. Although this engineered system differs slightly form the WT enzyme, it permits direct comparison of the base flipping and the loop motion. Incidentally, both the target cytosine and the Trp residue in the catalytic loop can be analyzed at the same excitation wavelength (typically 280 nm), and therefore both conformational changes can be simultaneously followed in real time as absorbance and fluorescence signals (emission at a 180° and 90° angle from the incident beam, respectively) in each individual stopped-flow measurement.
Figure 3.
Flipping of the target cytosine is tightly coupled to the catalytic loop motion in W41F/I86W mutant. W41F/I86W (3 µM) was mixed with 100 µM AdoHcy (gray) or 2.5 µM GCGC/GMGC (black); 3 µM W41F/I86W was mixed with 2.5 µM GCGC/GMGC in the presence of 100 µM AdoHcy (blue). (A) Absorption traces, (B) fluorescence traces, (C) superimposition of normalized fluorescence (red) and absorbance (blue) traces obtained from the same shot; the polarity of the fluorescence signal is inverted for clarity. (D) Reaction time courses at different AdoHcy concentration. W41F/I86W (3 µM) was first premixed with 2.5 µM GCGC/GMGC and mixed with AdoHcy (15, 30, 60, 100 and 200 µM, traces from right to left). Normalized absorption (blue) and fluorescence (red) traces are shown together with combined fits into a three-exponential model with two shared rate constants.
Figure 5.
Optical and covalent changes observed during catalytic turnover of M.HhaI. WT (A) or W41F/I86W (B) variant of M.HhaI (3 µM) was mixed with 2.5 µM cognate DNA duplex in the presence of 100 µM AdoMet. Stopped-flow absorbance traces (blue) showing flipping of the target cytosine and formation of the covalent complex, and fluorescence traces showing movements of the catalytic loop (red). The catalytic transfer of methyl groups (black) was measured under single turnover conditions using 3H-AdoMet. Absorption traces are shown in actual scale, fluorescence and single turnover traces are normalized to unity. Parameters derived from fitting experimental traces to exponential equations are shown in Tables 2 and 3 and in Supplementary Table S1.
In reaction with cognate DNA and 100 µM AdoHcy, the absorbance time course of the W41F/I86W mutant (Figure 3A) was similar to that of WT M.HhaI. The data fitted well into a three-exponential function yielding a slightly lower rates of base flipping (9 versus 19 s−1 for WT M.HhaI) whereas the rate of covalent bond formation was nearly identical with that of WT M.HhaI (0.64 s−1). The fluorescence intensity also followed a three-exponential decay with similar apparent rates. Since the W41F/I86W mutant is somewhat impaired in cofactor binding, we investigated the possibility that the slower base flipping rate is caused by incomplete saturation of the W41F/I86W–DNA complex with AdoHcy. In contrast to the WT enzyme (Supplementary Figure S4), the reaction profile showed a marked dependence on AdoHcy concentration up to the highest attainable values (15–200 µM) (Figure 3D). Interestingly, multicurve analysis of the absorbance and fluorescence signals obtained at a series of AdoHcy concentrations revealed that both processes can be fitted into a three exponential function with two identical rate constants (as shown in Table 3). Superimposition of normalized fluorescence and absorbance traces obtained from the same shots strongly suggests that flipping of the target cytosine and closure of the catalytic loop proceed simultaneously (Figure 3C). The third transient in the fluorescence profiles showed substantially smaller amplitude as compared with the preceding two steps, and it was not fully synchronous with the covalent bond formation in the absorbance profile. Since this minor signal varied with DNA and M.HhaI preparations, its nature has not been investigated further.
Table 3.
Kinetics of cytosine flipping and catalytic loop motion in Trp-engineered M.HhaI
| Reaction | Cognate dead-end (Figure 3): Target base pair = C:G; cofactor = AdoHcy |
Mismatch dead-end (Figure 4): Target base pair = C:T; cofactor = AdoHcy |
Cognate full cycle (Figure 5B): Target base pair = C:G; cofactor = AdoMet |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Goodness of fit (χ2 × 105) | 12.5 |
8.65 |
7.32 |
||||||
| Parameters | Rate, s−1 | Amplitude |
Rate, s−1 | Amplitude |
Rate, s−1 | Amplitude |
|||
| Abs. | Fl. | Abs. | Fl. | Abs | Fl. | ||||
| Step 1 | 19.0 ± 1.1 | 0.26 ± 0.03 | −0.32 ± 0.03 | 63 ± 2 | 0.53 ± 0.02 | −0.56 ± 0.02 | 180 ± 5 | 0.23 ± 0.004 | −0.13 ± 0.004 |
| Step 2 | 6.9 ± 0.2 | 0.81 ± 0.03 | −0.57 ± 0.03 | 19.6 ± 0.6 | 0.44 ± 0.02 | −0.38 ± 0.02 | 4.68 ± 0.03 | 0.46 ± 0.002 | −0.59 ± 0.002 |
| Step 3 | 0.64 ± 0.01/ 0.76 ± 0.05a | −0.74 ± 0.005 | −0.10 ± 0.004a | 0.60 ± 0.004/ 0.68 ± 0.06 a | 0.70 ± 0.001 | − 0.05 ± 0.001a | 0.36 ± 0.01 | −0.23 ± 0.01 | −0.44 ± 0.02 |
| Step 4 | – | – | – | – | – | – | 0.12 ± 0.002 | 0.16 ± 0.01 | 0.95 ± 0.02 |
Apparent kinetic parameters derived from simultaneous fitting of normalized absorption and fluorescence traces into multi-exponential functions for reactions involving 3 µM M.HhaI (W41F/I86W), 2.5 µM DNA and 100 µM cofactor. Values are reported with standard errors of the fit. Steps 1–2 correspond to target base flipping and catalytic loop closure; step 3 - covalent bond formation; step 4 - covalent bond breakage, base flip-back and loop opening.
aUnassigned change in fluorescence intensity.
Interaction with mismatched DNA substrates
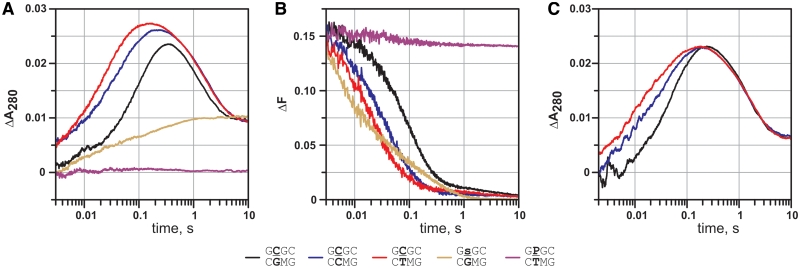
To better understand the relationship between the base flipping and the catalytic loop motion, we investigated interactions of the W41F/I86W mutant with mispaired substrates in the presence of AdoHcy. It has been reported that M.HhaI and other DNA MTases bind strongly DNA substrates containing mismatched bases at the target position and the increased affinity of M.HhaI towards mismatched DNA was correlated to the decreased stability of the base pair to be disrupted (4,8). Using different partner bases we examined how the motions of the catalytic loop and the target cytosine are influenced by the nature of the target base pair. We found that the rate of cytosine flipping as well as the rate of the closure of the catalytic loop increased as the stability of the target base pair decreased (Figure 4A and B). As with the cognate substrate, the base flipping and loop closure could be fitted with identical rate constants (Table 3), suggesting a synchronized nature of both processes. An increased rate in the reaction with mismatched substrates was also observed in the case of WT M.HhaI, when base flipping and covalent complex formation was monitored (Figure 4C, Supplementary Table S2). In addition to the dominant flipping rate observed with correctly paired substrate (19 s−1), a faster step was detectable with mismatched target base pairs (130 s−1 for C:T and 154 s−1 for C:C base pairs, respectively). These results agree well with a model in which a mismatched base pair promotes the accumulation of the target cytosine in a flipped-out state which facilitates the transition of the complex into the fully closed conformation.
Figure 4.
Interaction of M.HhaI with mismatched DNA substrates. (A and B) W41F/I86W (3 µM) was mixed with 2.5 µM DNA in the presence of 100 µM AdoHcy. (A, C) Stopped-flow absorption traces reporting flipping of the target cytosine and formation of the Michael adduct. (B) Stopped-flow fluorescence traces reporting the closure of the catalytic loop. (C) WT M.HhaI (3 µM) was mixed with 2.5 µM DNA in the presence of 30 µM AdoHcy. Black—GCGC/GMGC (target base pair C:G), blue—GCGC/GMCC (target base pair C:C), red—GCGC/GMTC (target base pair C:T), yellow—GsGC/GMGC (abasic target site), purple—GPGC/GMTC (non-flippable target site).
The fastest closure of the catalytic loop was observed upon interaction with a DNA duplex containing an abasic target site (initial rate >200 s−1, Figure 4B). The rate of the loop closure with this substrate was essentially cofactor independent, consistent with the previously noted ability of M.HhaI to form highly compact binary complexes with abasic DNA substrates (6). This suggests that the motion of the catalytic loop from an open to a closed conformation is potentially faster than is observed with natural DNA substrates, and is largely limited by the target nucleotide flipping.
Ternary complexes with AdoMet: the complete reaction cycle
In the presence of AdoMet, the reaction can proceed through the full catalytic cycle yielding the ultimate reaction products: methylated DNA and AdoHcy (Figure 1C). Three initial transients corresponding to target base flipping and covalent cytosine activation, can be readily discerned in the absorbance and fluorescence profiles of the reaction, since they show similar signals and apparent rates as observed in the corresponding AdoHcy profiles (Figure 5B and Table 3). In addition, an ascending transient of ∼0.1 s−1 is observed in both the fluorescence and absorbance traces. Obviously, the reverse fluorescence change of a matching amplitude can be safely assigned to the opening of the catalytic loop after the catalytic reaction. However, the interpretation of a much smaller increase in absorbance is not as straightforward, mostly because the methyltransfer step itself does not give a detectable optical change. Although the C5-methylation of cytosine results in a substantial bathochromic shift (Figure 1B), it remains obscure until the subsequent β-elimination step occurs in which the transient covalent bond is broken and the aromatic system of the cytosine ring is restored. However, subsequent back-flipping of the methylated target base in the presence of high concentrations of AdoMet (14) leads to a downward signal due to the hypochromic effect, which nearly cancels out the upward absorbance changes. Altogether, a single step with a small, AdoMet concentration-dependent (Supplementary Figure S4B) increase in absorbance is observed at 280 nm. Since the observed transient closely follows the chemically determined methyltransfer step (Supplementary Table S1) and is coincident with the major upward transient in the fluorescence trace (reopening of the catalytic loop) in the Trp-engineered M.HhaI and (Figure 5A and Table 3), it appears that the above three steps occur nearly simultaneously and cannot be resolved by the present data. Therefore, combining absorption, fluorescence and chemical methylation data, the four transients can be reliably characterized in the engineered M.HhaI variant.
The WT enzyme shows a very similar absorption profile with four-exponential kinetics (Figure 5A and Table 2). The slowest transient again closely resembles the rate of methyltransfer determined under single-turnover conditions in a rapid-quench experiment (Supplementary Table S1). Although the amplitude of the slowest transient is quite low at 280 nm, traces collected at 290–300 nm (Supplementary Figure S2B) give somewhat higher upward signals (due to a higher spectral difference between C and 5mC), permitting a fairly reliable kinetic assignment of this final step.
DISCUSSION
Here we provide the first direct real-time observation of DNA base flipping in an unperturbed enzymatic system at physiological conditions. Since the approach is based on the general phenomenon of hyperchromicity, it is thus applicable to many other base-flipping systems. Although such subtle absorbance changes may not always permit a unequivocal discrimination between base flipping and other distortions of DNA or RNA duplexes that involve unstacking of nucleobases [such as DNA kinks or RNA strand looping (23)], there is no inherent limitation with respect to the nature of the target base analyzed.
To our knowledge, the hyperchromic effect of a single nucleotide flipping in DNA has not been experimentally characterized before. It’s magnitude (150% at 280 nm) turned out to be 2- to 3-fold higher than the average contribution of a disrupted base pair (60–80% at 260 nm), suggesting that a single base gap in the double helical stack may impact its electronic structure to a larger extent than can be anticipated from the nearest-neighbor effects alone (24,25). As the signal largely originates from the loss of stacking interactions between the target cytosine and the adjacent guanines, it is important to note that in general these effects are largest when A or G are involved as neighbors (26,27) and that C:G base pairs have a maximum hypochromicity at 280 nm (28). In addition, the absorbance of the flipped-out cytosine can be influenced by the protein environment since it is known that the N3 protonation of the cytosine leads to a 9 nm red shift and a 1.5-fold increase in molar absorbance [εmax = 9000 M−1 cm−1 at λmax = 271 nm under physiological conditions in solution, εmax = 13 200 M−1 cm−1 at λmax = 280 nm at pH <3.0 (29)]. Crystal structures, biochemical and computational studies of M.HhaI–DNA complexes indeed suggest extended protonation and/or hydrogen bonding of the N3 and at O2 positions of the cytosine ring in the catalytic site (2).
The observed base flipping in the ternary complex involving M.HhaI, cognate DNA and cofactor generally occurs at a rate of 10–20 s−1. Previous studies of N6-adenine MTases EcoRI, EcoRV and Ecodam (30,31) as well as the HhaI (12) MTase using 2-aminopurine fluorescence have not provided reliable estimates for the outgoing motion of the natural target nucleotide. The base flipping is synchronous with the motion of the catalytic loop in the M.HhaI mutant (Figures 3 and 5B), and is very likely to be the same in the WT enzyme. Indirectly, similar estimates for the rate of loop closure were derived from the 2-aminopurine flipping experiments (12) as well as observed using other M.HhaI catalytic loop mutants (14). The concerted flip-lock motion is consistent with the notion that efficient extrahelical trapping of the flipped out base occurs only within a fully assembled catalytic site (4,5). Although the intrinsic rate of the loop closure can be higher with abasic DNA substrates, the assembly of the closed conformer is dependent on the strength of the target base pair. It is thus possible that the accumulation of a (partially) unpaired base-flipping intermediate would give rise to a faster base flipping phase, thereby explaining biphasic kinetics of the flip-lock motion. On the other hand, the biphasic behavior may also derive from an intermediate stable conformer in the trajectory of the catalytic loop or the target cytosine. Notably, crystal structures of the open and closed protein show extensive internal rearrangements in the 20-residue loop during the locking motion (2).
The established tight coupling between the target base flipping and the catalytic loop closure is consistent with two alternative models for the M.HhaI-induced base flipping. The first model suggests that the target base flipping occurs via the minor groove of the DNA, which then induces the catalytic loop closure (5). This hypothesis is largely based on steric considerations (2), and solvent accessibility of the target nucleotide replaced with mismatched bases such 2-aminopurine or thymine (12,32). Alternatively, a molecular dynamics study suggested that the closure of the catalytic loop upon the bound DNA would induce target base flipping via the major groove inside the protein (33); this point of view gained some indirect support from crystallographic studies (10). Although the Ser87 residue of M.HhaI implicated in the base-pair opening turned out to be dispensable for both base flipping and catalytic activity (8), this counter-intuitive (due to steric constrains to base flipping inside the closed ternary complexes) model cannot be decisively excluded or confirmed by the present data either.
Our first direct observation of the covalent bond formation to the target cytosine in solution provides evidence on several important aspects of the reaction mechanism. First, the observed rate of the covalent bonding in the presence of AdoHcy or AdoMet (0.3–0.7 s−1) is considerably slower than the cytosine flipping and locking on the ternary complex. This suggests that the formation of the covalent activated intermediate in the cognate reaction complex is largely dependent on the intrinsic rate of nucleophilic addition of Cys81, rather than the rate of cytosine locking in the catalytic site. However, the cytosine locking may be an important mechanistic factor that contributes to the catalytic rate of the MTase upon interaction with near-cognate or non-specific DNA substrates (9,14).
In the presence of product AdoHcy, the reaction reaches a dead-end equilibrium state. The extent of the covalent bond that is formed under these conditions has not been known. Numerous X-ray structures of the ternary M.HhaI complex with AdoHcy show that the bond length between the catalytic sulfur and the C6 atom of the cytosine is around 2.6–2.8 Å (34). This represents a weighed average between the length of a covalent C-S bond (1.8 Å) observed in a irreversible covalent complex involving 5-fluorocytosine (2) and a van der Waals distance (3.6 Å) (34), suggesting a partial (∼50%) formation of the covalent bond in the crystals. We find that the downhill amplitude of the absorbance signal in the presence of AdoHcy, which reflects the covalent bond formation (Figure 2A and Table 2), approximately matches the absorbance value of one cytosine chromophore at physiological conditions. Taking into account that the spectral properties of the bound cytosine may be affected (prior to the covalent bond formation) by the interactions with residues in the active site (see ‘Discussion’ section above), the extent of covalent bonding to the target base in the ternary complex in solution can be estimated to be in the range of 70–100%.
In the presence of AdoMet, the covalent complex undergoes subsequent transfer of the methyl groups from the cofactor. We observe that the formation of the covalent bond temporally precedes the methyl transfer step, i.e. the two events appear as distinct kinetic steps in the reaction cycle (Figure 5; Tables 2 and 3). These findings agree with a recent quantum-mechanical (QM) study (35) and disprove a QM calculations-derived concerted mechanism with simultaneous catalytic activation and methyltransfer steps (36).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Gražvydas Lukinavičius for M.HhaI C81S preparations and Elmar Weinhold for stimulating discussions.
FUNDING
Ministry of Education and Science of Lithuania; Howard Hughes Medical Institute (International research scholarship grant 55000317). Funding for open access charge: European Community's Seventh Framework Programme (FP7-REGPOT-2009-1 project 245721 MoBiLi).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Saenger W. Principles of Nucleic Acid Structure. New York: Springer-Verlag; 1984. [Google Scholar]
- 2.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 3.Wu JC, Santi DV. Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem. 1987;262:4778–4786. [PubMed] [Google Scholar]
- 4.Klimasauskas S, Roberts RJ. M.HhaI binds tightly to substrates containing mismatches at the target base. Nucleic Acids Res. 1995;23:1388–1395. doi: 10.1093/nar/23.8.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klimasauskas S, Szyperski T, Serva S, Wüthrich K. Dynamic modes of the flipped-out cytosine during HhaI methyltransferase-DNA interactions in solution. EMBO J. 1998;17:317–324. doi: 10.1093/emboj/17.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang P, Brank AS, Banavali NK, Nicklaus MC, Marquez VE, Christman JK, MacKerell AD. Use of oligodeoxyribonucleotides with conformationally constrained abasic sugar targets to probe the mechanism of base flipping by HhaI DNA (cytosine C5)-methyltransferase. J. Am. Chem. Soc. 2000;122:12422–12434. [Google Scholar]
- 7.Vilkaitis G, Merkiene E, Serva S, Weinhold E, Klimasauskas S. The mechanism of DNA cytosine-5 methylation. Kinetic and mutational dissection of HhaI methyltransferase. J. Biol. Chem. 2001;276:20924–20934. doi: 10.1074/jbc.M101429200. [DOI] [PubMed] [Google Scholar]
- 8.Daujotyte D, Serva S, Vilkaitis G, Merkiene E, Venclovas C, Klimasauskas S. HhaI DNA methyltransferase uses the protruding Gln237 for active flipping of its target cytosine. Structure. 2004;12:1047–1055. doi: 10.1016/j.str.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Svedruzic ZM, Reich NO. The mechanism of target base attack in DNA cytosine carbon 5 methylation. Biochemistry. 2004;43:11460–11473. doi: 10.1021/bi0496743. [DOI] [PubMed] [Google Scholar]
- 10.Horton JR, Ratner G, Banavali NK, Huang N, Choi Y, Maier MA, Marquez VE, MacKerell AD, Cheng X. Caught in the act: visualization of an intermediate in the DNA base-flipping pathway induced by HhaI methyltransferase. Nucleic Acids Res. 2004;32:3877–3886. doi: 10.1093/nar/gkh701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merkiene E, Klimasauskas S. Probing a rate-limiting step by mutational perturbation of AdoMet binding in the HhaI methyltransferase. Nucleic Acids Res. 2005;33:307–315. doi: 10.1093/nar/gki175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vilkaitis G, Dong A, Weinhold E, Cheng X, Klimasauskas S. Functional roles of the conserved threonine 250 in the target recognition domain of HhaI DNA methyltransferase. J. Biol. Chem. 2000;275:38722–38730. doi: 10.1074/jbc.m005278200. [DOI] [PubMed] [Google Scholar]
- 13.Estabrook RA, Reich N. Observing an induced-fit mechanism during sequence-specific DNA methylation. J. Biol. Chem. 2006;281:37205–37214. doi: 10.1074/jbc.M607538200. [DOI] [PubMed] [Google Scholar]
- 14.Estabrook RA, Nguyen TT, Fera N, Reich NO. Coupling sequence-specific recognition to DNA modification. J. Biol. Chem. 2009;284:22690–22696. doi: 10.1074/jbc.M109.015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daujotyte D, Vilkaitis G, Manelyte L, Skalicky J, Szyperski T, Klimasauskas S. Solubility engineering of the HhaI methyltransferase. Protein Eng. 2003;16:295–301. doi: 10.1093/proeng/gzg034. [DOI] [PubMed] [Google Scholar]
- 16.Dila D, Sutherland E, Moran L, Slatko B, Raleigh EA. Genetic and sequence organization of the mcrBC locus of Escherichia coli K-12. J. Bacteriol. 1990;172:4888–4900. doi: 10.1128/jb.172.9.4888-4900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 18.Skaric V, Gaspert B, Hohniec M, Ladan G. Some dihydro-cytidines and -isocytidines. J. Chem. Soc. Perkin. 1974;I:267–271. doi: 10.1039/p19740000267. [DOI] [PubMed] [Google Scholar]
- 19.Sverdlov ED, Monastyrskaya GS, Guskova LI, Levitan TL, Sheichenko VI, Budowsky EI. Modification of cytidine residues with a bisulfite–O-methylhydroxylamine mixture. Biochim. Biophys. Acta. 1974;340:153–165. [PubMed] [Google Scholar]
- 20.Ulanov BP, Matorina TI, Emanuel NM. Double modification of cytidine residues in DNA. Mol. Biol. 1976;10:1211–1220. [PubMed] [Google Scholar]
- 21.Osterman DG, DePillis GD, Wu JC, Matsuda A, Santi DV. 5-Fluorocytosine in DNA is a mechanism-based inhibitor of HhaI methylase. Biochemistry. 1988;27:5204–5210. doi: 10.1021/bi00414a039. [DOI] [PubMed] [Google Scholar]
- 22.Mi S, Roberts RJ. The DNA binding affinity of HhaI methylase is increased by a single amino acid substitution in the catalytic center. Nucleic Acids Res. 1993;21:2459–2464. doi: 10.1093/nar/21.10.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Möller A, Wild U, Riesner D, Gassen HG. Evidence from ultraviolet absorbance measurements for a codon-induced conformational change in lysine tRNA from Escherichia coli. Proc. Natl Acad. Sci. USA. 1979;76:3266–32701. doi: 10.1073/pnas.76.7.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajski SR, Kumar S, Roberts RJ, Barton JK. Protein-modulated DNA electron transfer. J. Am. Chem. Soc. 1999;121:5615–5616. [Google Scholar]
- 25.Kelley SO, Barton JK. Electron transfer between bases in double helical DNA. Science. 1999;283:375–381. doi: 10.1126/science.283.5400.375. [DOI] [PubMed] [Google Scholar]
- 26.Cantor CR, Warshaw MM, Shapiro H. Oligonucleotide interactions. III. Circular dichroism studies of the conformation of deoxyoligonucleolides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 27.Cavaluzzi MJ, Borer PN. Revised UV extinction coefficients for nucleoside-5'-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004;32:e13. doi: 10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puglisi JD, Tinoco I. Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 29.Fox JJ, Shugar D. Spectrophotometric studies of nucleic acid derivatives and related compounds as a function of pH. II. Natural and synthetic pyrimidine nucleosides. Biochim. Biophys. Acta. 1952;9:369–384. doi: 10.1016/0006-3002(52)90181-9. [DOI] [PubMed] [Google Scholar]
- 30.Reich NO, Coffin SR. M·HhaI and M·EcoRI: paradigms for understanding the conformational mechanisms of DNA methyltransferases. In: Grosjean H, editor. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Vol. 6. Austin TX, USA: Landes Bioscience; 2009. pp. 47–57. [Google Scholar]
- 31.Jeltsch A, Jurkowski TP. Mechanism and evolution of DNA recognition by DNA-(adenine N6)-methyltransferases from the EcoDam family. In: Grosjean H, editor. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Vol. 7. Austin TX, USA: Landes Bioscience; 2009. pp. 98–108. [Google Scholar]
- 32.Serva S, Weinhold E, Roberts RJ, Klimasauskas S. Chemical display of thymine residues flipped out by DNA methyltransferases. Nucleic Acids Res. 1998;26:3473–3479. doi: 10.1093/nar/26.15.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang N, Banavali NK, MacKerell AD. Protein-facilitated base flipping in DNA by cytosine-5-methyltransferase. Proc. Natl Acad. Sci. USA. 2003;100:68–73. doi: 10.1073/pnas.0135427100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar S, Horton JR, Jones GD, Walker RT, Roberts RJ, Cheng X. DNA containing 4'-thio-2'-deoxycytidine inhibits methylation by HhaI methyltransferase. Nucleic Acids Res. 1997;25:2773–2783. doi: 10.1093/nar/25.14.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zangi R, Arrieta A, Cossio FP. Mechanism of DNA methylation: the double role of DNA as a substrate and as a cofactor. J. Mol. Biol. 2010;400:632–644. doi: 10.1016/j.jmb.2010.05.021. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, Bruice T. The mechanism of M.HhaI DNA C5 cytosine methyltransferase enzyme: a quantum mechanics/molecular mechanics approach. Proc. Natl Acad. Sci. USA. 2006;103:6148–6153. doi: 10.1073/pnas.0601587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.