Abstract
The basic helix–loop–helix (bHLH).PAS dimeric transcription factors have crucial roles in development, stress response, oxygen homeostasis and neurogenesis. Their target gene specificity depends in part on partner protein choices, where dimerization with common partner Aryl hydrocarbon receptor nuclear translocator (Arnt) is an essential step towards forming active, DNA binding complexes. Using a new bacterial two-hybrid system that selects for loss of protein interactions, we have identified 22 amino acids in the N-terminal PAS domain of Arnt that are involved in heterodimerization with aryl hydrocarbon receptor (AhR). Of these, Arnt E163 and Arnt S190 were selective for the AhR/Arnt interaction, since mutations at these positions had little effect on Arnt dimerization with other bHLH.PAS partners, while substitution of Arnt D217 affected the interaction with both AhR and hypoxia inducible factor-1α but not with single minded 1 and 2 or neuronal PAS4. Arnt uses the same face of the N-terminal PAS domain for homo- and heterodimerization and mutational analysis of AhR demonstrated that the equivalent region is used by AhR when dimerizing with Arnt. These interfaces differ from the PAS β-scaffold surfaces used for dimerization between the C-terminal PAS domains of hypoxia inducible factor-2α and Arnt, commonly used for PAS domain interactions.
INTRODUCTION
The basic helix–loop–helix (bHLH)/Per-Arnt-Sim homology (PAS) transcription factor (TF) family consists of at least 19 structurally related DNA binding proteins in mammals (1). bHLH.PAS TFs are dimeric, with networks centred around two hub proteins: aryl hydrocarbon receptor nuclear translocator (Arnt) and brain and muscle Arnt-like (BMAL). The BMAL cluster is a relatively small network that regulates circadian rhythm and includes BMAL 1 an 2, Clock 1 and 2 and PERIOD (PER) proteins (Supplementary Figure S1). The more extensive Arnt cluster functions in sensing environmental cues such as xenobiotics [aryl hydrocarbon receptor (AhR)] and low oxygen tension [hypoxia inducible factor-αs (HIF-αs)], as well as participating in a broad range of biological processes, including liver and vascular development (AhR and HIF-α, respectively) neurogenesis [single minded proteins (Sim1&2)], synaptic plasticity [neuronal PAS domain protein 4 (NPAS4)] and the progression of many cancers (1–4). Arnt, or the closely related homologue Arnt2, are the obligate partners for all members in the Arnt cluster, where dimerization is required to form active, DNA binding complexes to initiate transcription (4). The dimers recognize asymmetric E-box-like response elements in the regulatory regions of target genes and DNA binding specificity is directed by Arnt’s protein partner. Arnt can also homodimerize and bind the canonical CACGTG E-box sequence in vitro and in vivo (5–7), and although the physiological significance is still unclear, the Arnt homodimer appears to be involved in regulation of CYP2a5 in liver (5).
All bHLH.PAS proteins share similar domain architecture, with a highly conserved N-terminal bHLH motif, adjacent PAS domains, and loosely conserved C-terminal transactivation or transrepression regions (4). The bHLH domain is a well characterized DNA binding and dimerization domain. Strong dimer formation often requires collaboration between bHLH and other regions such as PAS or leucine zipper domains. The PAS domain is a widespread protein interaction and signal transduction motif (8). Most bHLH.PAS TFs have two tandem PAS domains, designated PAS A (N-terminal) and PAS B (4) and both PAS domains contribute to dimer formation and biological activity of transcription complexes (9–11). The N-terminal PAS.A domains have significant roles in dimerization, controlling dimerization specificity (12), stability (11,13) and strengthening the DNA binding (13), and there are several instances where the PAS.A domain alone is sufficient for functional dimerization. Deletion of PAS.B, the ligand binding domain, in AhR produces a constitutively active receptor more potent than the intact protein (14). The AhR Repressor, which lacks PAS.B, competes efficiently with AhR for Arnt binding, to negatively regulate AhR activity (15). Similarly, Inhibitory PAS protein (IPAS), a splicing variant of HIF-3α having only a partial PAS.B domain, negatively modulates HIF-αs activity by dimerizing with HIF-α to prevent formation of active HIF-α/Arnt (16).
As PAS.A is important for directing homo- and heterodimerization within the Arnt cluster, we sought to identify dimerization interfaces in Arnt PAS.A, and to determine whether a common interface is used for all Arnt hub PAS.A interactions, or if the partner proteins use different dimerization interfaces. Both mechanisms are plausible, as several distinct interaction surfaces have been identified for PAS domains, involving the N-terminal α-helical cap, the central β-sheets or the α-helix connecting the N- and C-terminal β-sheets (9,17–21). For other dimeric TFs, such as the related bHLH Leucine Zipper proteins and the nuclear hormone receptors, the same interface is involved in both homo- and hetero-dimerization, with key residues in the interface defining dimer specificity (22,23). Alternatively, proteasome inhibitor PI31 uses different interfaces of a single domain for homo- and heterodimerization. Homodimerization is mediated by α-helical interactions between the N-terminal domain while the β-sheet on the opposite face of this domain is involved in PI31/SCFFBxo7 E3 ubiquitin ligase heterodimerization (24).
Structural analysis coupled with site-directed mutagenesis is the typical approach to investigate such questions mechanistically. The most homologous solved structure for Arnt PAS.A is PER, the negative regulator of circadian rhythm that competes with Clock for BMAL binding. PER has both PAS.A and PAS.B domains and can form a homodimer, but lacks the bHLH domain and does not bind DNA. It is likely that the PER homodimer structure is not representative of bHLH.PAS TF interactions, since the homodimer is antiparallel, where PAS.A of one monomer interacts with PAS.B of the second monomer (25), rather than PAS.A with PAS.A and PAS.B with PAS.B as in bHLH.PAS TFs. The structure of the isolated PAS.B domains of the HIF2α/Arnt dimer has also been solved and shows antiparallel dimerization through the β-sheet surface (9,26). Together, these structures reveal that PAS.A domains differ quite markedly from PAS.B, in having up to three insertion loops between β-stands in the domain fold (25).
In the absence of experimentally determined structures, we have developed a novel reverse bacterial two hybrid system (RevB2H) to investigate Arnt dimerization and define the dimerization interface in the poorly characterized PAS.A region. This genetic approach provided an unbiased and versatile method which identified 22 amino acid residues in Arnt PAS.A and 10 residues in AhR PAS.A critical for AhR/Arnt dimerization. Residues E163 and S190 of Arnt are selective for the AhR/Arnt interaction, as mutations at these positions have little or no effect on other Arnt partner proteins, while substitution at Arnt D217 affected the interaction with both AhR and HIF-1α but not with Sim1, Sim2s or NPAS4.
MATERIALS AND METHODS
RevB2H screening
System validations were performed by PCR-based genotyping and computer-aided colony size comparisons. For PCR genotyping, a mixture of two oligonucleotide pairs (pTRGsense and pTRGantisense, pBTsense and pBTantisense; Supplementary Data) priming from flanking sequences were used, giving a different size product for each encoded protein. Colony size was determined after 40 h incubation at 34°C, when plates were scanned and the frequency distribution of colonies with diameter >0.2 mm was scored in the images, using ImageJ 1.42q software (http://rsbweb.nih.gov/ij/features.html).
Plasmid construction and error-prone PCR generation of mutagenic libraries are described in Supplementary Data. For the Arnt PAS.A mutagenesis screen, pBT_Arnt362_Host_LacZα mutagenic libraries were electroporated into Escherichia coli KS1033 (27) carrying pZS4lacOR2-62_186cI_CTD selection and pZE1_RNAP_AhR278 expression plasmids, then incubated on 100 μg/ml Ampicillin, 30 μg/ml Chloramphenicol, 50 μg/ml Spectomycin, 1 mM IPTG and 50 μg/ml X-gal agar at ≤34°C for 40 h. Recovered plasmid DNA was digested with EcoRI to linearize pZS4lacOR2-62_186cI CTD and pZE1_RNAP_AhR278. Remaining intact pBT_Arnt362_Host_lacZα carrying putative Arnt loss-of-interaction mutations was transformed into either E. coli KS1 (28) harbouring pZE1_RNAP_AhR278 expression plasmid for β-gal validation or XL1-Blue for plasmid propagation and sequencing. AhR PAS.A was screened by electroporating pZE1_RNAP_AhR278_lacZα mutagenic libraries into KS1033 carrying pZS4lacOR2-62_186cI_CTD and pBT_Arnt362_Host and grown as above. Recovered plasmid DNA was digested with XhoI to linearize pZS4lacOR2-62_186cI_CTD and pBT_Arnt362_Host. Intact pZE1_RNAP_AhR278_lacZα carrying putative AhR loss-of-interaction mutations was transformed into KS1 carrying pBT_Arnt362_Host expression plasmid for β-gal validation. Liquid β-gal assays were performed in E. coli KS1 in 96-well plates using 150 µM IPTG as described previously (29). The wt λCI_Arnt362_Host/RNAPα_AhR278 and λCI_Arnt362/RNAPα_AhR278_Host interactions gave 112.8 ± 9.0 and 107.3 ± 4.8 units of β-gal activity (mean ± SEM, n = 9), respectively.
Protein purification and circular dichroism
Expression of 6-histidine tagged proteins was carried out as described (13). Bacterial cells, 60 OD600nm units, were resuspended in 1 ml 100 mM HEPES (pH 7.5), 25 U of Benzonase (Novagen), lysed by sonication and purified by Ni-affinity chromatography using Maxwell® 16 automatic purification Instrument (Promega). CD spectra were recorded in 10 mM sodium phosphate, pH 8.0, at 20°C on a Jasco J-815 spectrometer, from 260 to 185 nm in 0.2 nm steps, with 1 s response time and 1 nm bandwidth, recording seven accumulations per spectrum with buffer correction. Data were normalized for differences in protein concentration using ellipticity at 207 nm to allow comparison with wt (30); concentrations (mg/ml) were wt: 0.3, E163K: 0.3, V179A: 0.07, D217G: 0.06, M267K: 0.05.
Secondary-structure prediction and homology modelling
The positions of secondary-structure elements were derived from structure-based alignments (25) and secondary-structure predictions for the Arnt sequence using Psipred (http://bioinf.cs.ucl.ac.uk/psipred/) and PROF prediction (http://www.aber.ac.uk/∼phiwww/prof/). Homology models of Arnt and AhR PAS.A domain were generated using SwissModel (http://swissmodel.expasy.org/workspace/) with dPER 1WA9 (25) as template. Images were produced using UCSF Chimera (http://www.cgl.ucsf.edu/chimera/) from the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco (NIH P41 RR-01081).
Immunoblotting and antibodies
Cell lysates were prepared in 20 mM HEPES, pH 8.0, 420 mM NaCl, 0.5% Igepal, 25% glycerol, 0.2 mM EDTA, 1.5 mM MgCl2, 1 mM DTT and protease inhibitors and 20 μg, was separated on 10% acrylamide SDS–PAGE before transfer to nitrocellulose. Reporter gene assay extracts from transfection wells were normalized against renilla luciferase levels. Proteins were detected using monoclonal antibodies anti-Myc (4A6, Upstate), anti-α-tubulin (MCA78G, Serotec), anti-AhR (RPT1, Abcom), or anti-Arnt (MA1-515, Affinity BioReagents), horseradish peroxidase-conjugated secondary antibody and visualized by chemiluminescence. Fluorescence-based western blotting of bacterial extracts was performed as described (31) using affinity purified anti-λCI and Cy5-labelled secondary antibody.
Ni-affinity pull-down
Cytosolic proteins were prepared from cultured mouse Hepa C4 cells by lysis in six volumes of hypotonic buffer [1.5 mM MgCl2, 10 mM KCl, 10% glycerol and protease inhibitor cocktail (Sigma) in 10 mM Tris–HCl, pH 8.0] with three freeze-thaw cycles, and centrifugation at 14 000g, 30 min, 4°C. Protein, 80 μg, was treated in vitro with vehicle (0.1% DMSO), 50 nM TCDD or 50 μM YH439, incubated with Ni-NTA resin (Qiagen), washed three times with 250 mM NaCl, 20 mM Imidiazole, 0.1% Igepal and protease inhibitor cocktail (Sigma) in 20 mM HEPES, pH 8.0. Bound protein was recovered by boiling in SDS loading buffer, before western blotting.
Transient transfections and luciferase assays
HEK293T were maintained, and dual luciferase assays performed, as described previously (32). Cells were transiently cotransfected with 200 ng firefly luciferase reporter plasmid [pML-6c-wt, pML-6c-X or pML-6c-AM (33)] 50 ng each of puro6_HisMyc_Arnt (wt or mutant) and puro6-based partner protein expression vectors, and 10 ng of phRL-CMV Renilla luciferase reporter plasmid (Promega) for 24 h. For the XRE reporter, cells were treated after 8 h with vehicle (0.1% DMSO) or 10 µM YH439 for 16 h. Relative luciferase activity was normalized to levels with empty vector to give fold activation over basal levels from endogenous protein. For Arnt homodimer, cells were transiently cotransfected with 200 ng 4(CACGTG)TKMPluc (6) or TKMPluc, 50 ng of puro6_HisMyc_Arnt (wt or mutants) and 0.5 ng of phRL-CMV and incubated for 48 h.
Lentiviral production and infection
Mouse hepatatoma C4 cell lines (34) were cultured in α-MEM containing 10% fetal calf serum, Penicillin/Streptomycin (1% v/v) at 37°C, 5% CO2. The production of lentiviral particles and integration of HisMyc_Arnt wt or mutant expression vectors to generate mouse hepatocyte stable expression lines has been described (32).
RNA extraction, cDNA synthesis and quantitative real time PCR
Cultured Hepa C4 cell lines expressing wt, E163K, S190P or D217G HisMyc_Arnt from integrated lentiviral vectors were seeded at 1.2 × 106 cells/dish in 10 cm dishes and after 48 h, treated with vehicle (0.1% DMSO), 10 µM YH439 or 100 µM 2,2′-dipyridyl (DP) for 16 h. RNA was purified and reverse transcribed and mCYP1A1 and mVEGF expression levels measured by quantitative real time PCR (qRT–PCR) as described previously (32), with all reactions performed in triplicate. The expression level of mouse housekeeping gene hypoxanthine guanine phosphoribosyl transferase (mHPRT), which did not vary with treatments, was used as internal control. The amplification efficiencies for mCYP1A1, mVEGF and mHPRT were 91.1, 93.0 and 88.5%, with amplicons of 165, 101 and 85 bp, respectively.
RESULTS
Reverse bacterial two hybrid system
Our RevB2H was derived from the conventional bacterial two hybrid method (28) by replacing the HIS3-aadA reporter gene cassette with the 186cI CTD gene (Figure 1). This gene encodes a form of the bacteriophage 186 cI repressor protein that lacks the N-terminal DNA binding motif and thus acts as a dominant negative towards full-length 186 cI. Interaction between the DNA bound λCI-bait and the RNAPα-prey fusion proteins activates production of the 186cI CTD protein, which in an appropriate host strain, KS1033, results in cell death. Escherichia coli strain KS1033 is a derivative of XL1-Blue having stable integration of a 186(cIts int−) lysogen (27) and produces a temperature sensitive form of 186cI protein (186cIts). At permissive temperatures (≤34°C), expression of prophage-encoded 186cIts suppresses the lytic cycle of the 186 prophage, permitting cell growth. When interaction between the λCI-bait and RNAPα-prey fusion proteins increases the expression of 186cI CTD, 186cIts is sequestered away from DNA, resulting in prophage mediated cell death or significantly inhibited cell growth. A mutation within the proteins fused to λCI or RNAPα that disrupts their interaction results in reduced expression of 186cI CTD and the host encoded 186cIts is then sufficient for cell survival (Figure 1a).
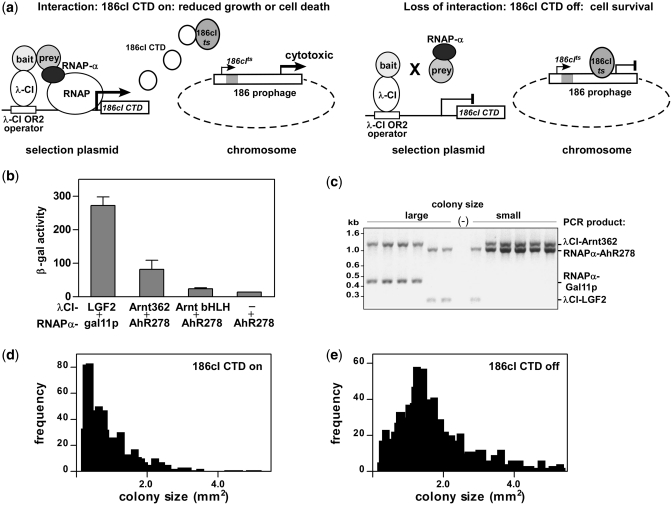
Figure 1.
Characterizing the reverse bacterial two hybrid system (RevB2H). (a) Mechanism: interaction between bait fused to bacteriophage λ-CI repressor (λCI-bait) and prey fused to RNA polymerase α-subunit (RNAPα-prey) (28) drives expression of the C-terminal domain of bacteriophage 186cI repressor [186cI-CTD (27)] from the plac λ-CI OR2-62 promoter (28) on the lacOR2-62_186cI-CTD selection plasmid. 186cI-CTD sequesters chromosomally encoded 186cI repressor (186cIts), from its binding site in the int- 186 prophage. This activates aberrant lytic gene transcription from the prophage, which results in significantly inhibited growth or cell death, depending on the strength of the bait–prey interaction. Mutations in the bait or prey fusion proteins that disrupt interaction reduce 186cI-CTD expression, allowing 186cIts to maintain repression of prophage lytic transcription, permitting normal cell growth. (b) Strength of the indicated λCI-bait and RNAPα-prey fusion proteins interaction measured as β-gal activity from the lacOR2-62_lacZ reporter gene in KS1. Data are mean ± SD, n = 3. (c) Selectivity for interaction: four plasmids encoding two pairs of interacting bait and prey proteins [λCI_Arnt362 and RNAPα_AhR278, λCI_LGF2 and RNAPα_gal11p (35)] were co-transformed into KS1033 carrying the lacOR2-62_186cI-CTD selection plasmid, with induction of fusion protein expression. Plasmid incompatibility permits establishment of only one of the two available λCI and RNAPα plasmids. Genotyping revealed the presence of non-interacting and interacting pairs in large and small colonies, respectively. (d) and (e) Effect of 186cI-CTD production on colony size: induction of protein expression in KS1033 harbouring the lacOR2-62_186cI-CTD selection and λCI_Arnt362 expression plasmids either with (d) or without (e) transformation of the RNAP_AhR278_lacZα mutagenic library.
Initial bait and prey fusions proteins were made where λCI was fused to human Arnt bHLH.PAS.A residues 1–362 (λCI_Arnt362) and RNAPα was fused to mouse AhR bHLH.PAS.A residues 1–278 (RNAPα_AhR278), under the control of isopropyl β-d-1-thiogalactopyranoside (IPTG) inducible lacUV5 promoters. We tested the strength of interaction of these fusion proteins by measuring β-galactosidase (β-gal) activity in a standard forward two-hybrid system using E. coli strain KS1, which carries the lacOR2-62_lacZ reporter cassette on its F’ plasmid (28). The system gave lower β-gal activity than known strong interactors λCI_LGF2 and RNAPα_gal11p (35). Removal of the Arnt PAS.A domain in the λCI_ Arnt bHLH fusion reduced activity to almost background levels seen with λCI alone (Figure 1b). To test the selectivity of the RevB2H system, we cotransformed a mixture of four plasmids encoding two pairs of interacting bait and prey proteins, λCI_Arnt362 and RNAPα_AhR278, λCI_LGF2 and RNAPα_gal11p into KS1033 cells carrying the lacOR2-62_186cI CTD selection plasmid. Genotyping of different sized colonies showed that six out of six large colonies harboured a mismatched interacting pair, while five out of six small colonies carried the λCI_Arnt362 and RNAPα_AhR278 interacting pair (Figure 1c), indicating that the RevB2H system could readily discriminate between interacting and non-interacting protein pairs. No colonies tested harboured both the strongly interacting λCI_LGF2 and RNAPα_gal11p expression plasmids, presumably due to the high 186cI CTD expression sequestering all available 186cIts resulting in cell death, consistent with the relative strength of the interactions measured as β-gal activity (Figure 1b). Expression of both λCI_Arnt362 and RNAPα_AhR278 in the presence of the lacOR2-62_186cI CTD selection plasmid gave predominantly small colonies (Figure 1d), whereas when RNAPα-AhR278 was absent, colony size had a normal distribution (Figure 1e).
RevB2H identified amino acid substitutions in Arnt PAS.A altering AhR/Arnt heterodimerization
A mutagenic library, over the region encoding Arnt PAS.A, residues 152–362, was generated using error-prone PCR (described in Supplementary Data) which we estimated to contain an average of one amino acid mutation per construct. This library was subcloned into the λCI_Arnt362 expression vector, replacing the wt Arnt PAS.A coding region. To improve the screening efficiency, we incorporated lacZα complementation by fusing the lacZα polypeptide (amino acids 1–39 of LacZ) to the C-terminus of Arnt. As the host strain carries the ΔLacZ(M15) allele, which produces a mutant form of β-gal lacking amino acids 11–41, expression of LacZα in the λCI_Arnt362_LacZα fusion protein restores the functionality of β-gal (36). This allows rapid discrimination of nonsense and frame-shift mutations within the N-terminal Arnt362 portion of the fusion protein, since by disrupting the expression of the C-terminal LacZα peptide, these will give rise to white colonies when plated on medium containing X-gal. Cells carrying wt or missense mutations will appear blue due to the restored catalytic activity of β-gal. Alpha complementation also provides a screen for unwanted mutations which directly or indirectly reduce expression levels.
The Arnt PAS.A mutagenic library was transformed into KS1033 harbouring the RNAPα_AhR278 expression and lacOR2-62_186cI CTD selection plasmids and plated on media containing IPTG and X-gal. Colonies that developed showed a marked size distribution, and large colonies were either blue or white while small colonies were predominantly blue. A total of approximately 8000 colonies were screened, and 192 large blue colonies selected for further analysis. The λCI_Arnt362_lacZα expression plasmids harbouring potential loss-of-interaction mutations were isolated, and transformed into E. coli KS1 (28). The forward two-hybrid provided a rapid, quantitative method to validate the initial screening result, allowing the strength of interaction between AhR278 and putative Arnt PAS.A mutants to be measured directly as a function of β-gal activity. Isolates having <80% β-gal activity relative to wt Arnt PAS.A were sequenced (Supplementary Table S1). In this way, we identified 26 single amino acid mutations in Arnt PAS.A that potentially reduced AhR278/Arnt362 heterodimerization. Two of these mutations, T208A and H214R, appeared repeatedly, and mutation of C308 to both tyrosine and arginine was found. As the RevB2H system allows survival due to decreased levels of 186cI CTD gene expression, false positives may emerge due to spontaneous mutations elsewhere that reduce plasmid copy number or reduce fusion protein expression in some way. To eliminate such effects, the coding sequence of each Arnt PAS.A mutation was cloned into fresh λCI_Arnt362 expression vector, and re-analysed for β-gal activity (Figure 2a). At this stage, we compared activity to wt Arnt362 (i.e. Arnt bHLH.PAS.A), Arnt bHLH alone without the adjacent PAS.A domain, and Arnt362 having the G341D substitution, a mutation within Arnt PAS.A identified in the Arnt deficient mouse hepatoma cell line, Hepa C4, which reduces dimerization with AhR and HIF (37). These gave, respectively, 22 and 47% of wt activity in this assay. Twenty-two out of the 26 Arnt PAS.A mutations isolated from the RevB2H screening showed statistically significant reduction in β-gal production, consistent with having disrupted AhR278/Arnt362 heterodimerization.
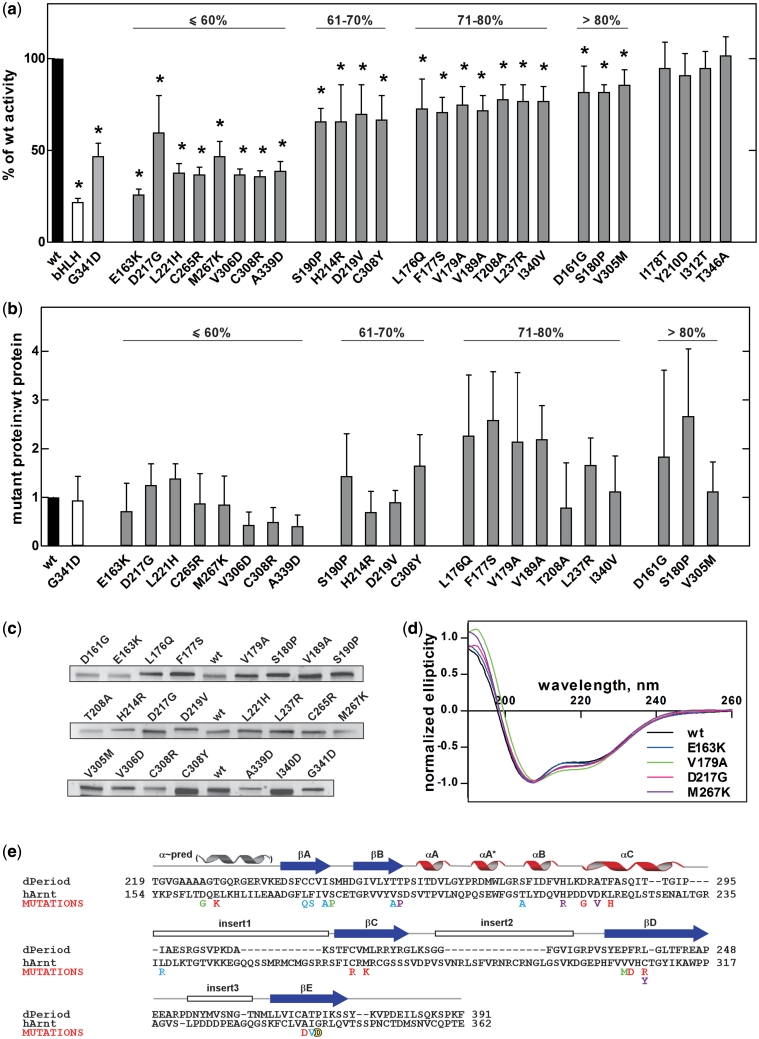
Figure 2.
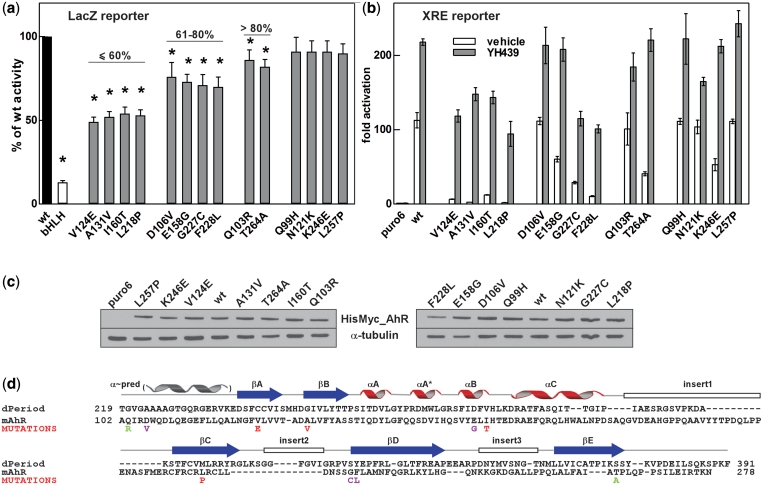
Single amino acid mutations in Arnt PAS.A domain with disrupted AhR/Arnt heterodimerization isolated using RevB2H. (a) The strength of interaction between RNAPα_AhR278 and the potential λCI_Arnt362 loss-of-interaction mutants selected as large blue colonies from RevB2H was measured in a standard forward bacterial two hybrid system with the lacZ reporter gene (28). Data are mean with 95% confidence intervals, shown as a percentage of β-gal activity obtained with wt λCI_Arnt362 protein, n = 9. bHLH: λCI_Arnt bHLH domain only; *P < 0.05 (one tailed unpaired Student’s t-test). (b) Expression of mutant λCI_Arnt362 relative to wt; mean ± SEM. from three independent bacterial extracts. Percent symbol denotes grouping by β-gal activity relative to wt from (a). (c) Shows a representative western blot. (d) CD spectra of wt H6_Arnt362 and selected loss-of-interaction mutants, normalized to allow comparison with wt (30). CD spectra of all mutants are shown in Supplementary Figure S2. (e) The dimerization deficient mutations identified from RevB2H mapped onto the sequence of Arnt PAS.A, aligned with dPer PAS.A. dPer PAS.A is representative of PAS.A domains, which have one to three insert loops within the PAS domain fold (25). The positions of secondary-structure elements, including the predicted N-terminal α-helix are indicated above. The mutations are coloured according to the strength of disruption observed in the β-gal assay; red: <60%, magenta: 61–70%, blue: 71–80% and green: >80% of wt activity, yellow: Hepa c4 mutation G341D (37).
Although we had incorporated alpha complementation in our RevB2H screen to eliminate severe λCI_Arnt362 expression and misfolding mutations, the observed decrease in β-gal activity might in principle reflect small reductions in levels of fusion protein or mutations resulting in protein misfolding and degradation, rather than the desired compromised AhR278/Arnt362 heterodimerization. To address this concern, the expression levels of all 22 positive λCI_Arnt362 PAS.A mutants were assayed by quantitative fluorescence-based western blotting using affinity purified λCI antibodies (Figure 2b and c). While expression was quite variable with mutations, protein levels relative to wt did not correlate with reduced β-gal activity except for V306D, C308R and A339D, suggesting that the lower activity seen for the remaining mutations was not likely to be predominantly the result of protein instability. In fact, as many of the mutant proteins showing reduced β-gal production expressed at higher levels than wt Arnt, there may be greater effects on dimerization than are reported by the assay.
The overall structure of the Arnt PAS.A putative dimerization mutants was further characterized using circular dichroism (CD) spectroscopy. Figure 2d and Supplementary Figure S2 show the CD spectra of nickel affinity purified 6-Histidine tagged-Arnt362 (bHLH.PAS.A) wt and mutant proteins. Although there were minor differences in the CD spectra of mutant proteins, as well as differences in yield from the purification, these did not correlate with the reduction in β-gal activity using the LacZ reporter gene (Figure 2a). Therefore we conclude that our data are consistent with the mutations in Arnt PAS.A isolated using the RevB2H system having disrupted dimerizaton with AhR, rather than the reduced β-gal activity arising primarily from gross misfolding or instability, and demonstrate the utility of the additional alpha complementation screening approach. Thus, from this analysis, we did not discard any mutants as having reduced β-gal activity arising simply from gross structural changes.
Arnt PAS.A mutation sites were clustered in key structural elements and conserved throughout evolution
The mutations identified in Arnt PAS.A having deficient dimerization with AhR mapped across the entire PAS.A region and clustered in predicted key structural elements (Figure 2e). Only one mutation, L237R, was found in a predicted insert loop region. Furthermore, most of the mutations found in the N-terminal β-strands (βA and βB) had relatively mild effects, whereas mutations within the α-helical connector or C-terminal β-strands (βC, βD and βE) tended to have more severe effects on AhR/Arnt heterodimerization (cf Figure 2a). Interestingly, amino acids D161 and E163 found N-terminal of the structured PAS domain, in a sequence predicted to form an α-helix (Figure 2e), also influenced dimerization, with the E163K substitution having the largest single effect on dimerization in the β-gal activity assay. Cross-species alignment of Arnt PAS.A domains from fly to fish and mammals showed the mutation sites are highly conserved throughout evolution, implying a significant role of these amino acid residues in function (Supplementary Figure S3).
Arnt PAS.A mutations identified using the RevB2H reduced AhR/Arnt transactivation activity and dimerization in mammalian cells
As the RevB2H screen was performed in bacterial cells with truncated proteins containing only the bHLH and PAS.A domains, next we tested the effect of these mutations in the context of full-length proteins in mammalian cells in culture. In reporter gene assays using an artificial XRE-driven luciferase reporter construct pML_6c_X (33), most of the Arnt PAS.A mutants identified from the RevB2H screen reduced transactivation by the AhR/Arnt heterodimer, both without activating ligand and upon treatment with the ligand YH439 (Figure 3a and Supplementary Figure S4). In general, there was a clear correlation between the effects on β-gal induction using truncated proteins in bacteria and the function of full-length proteins in mammalian cells. Significantly, mutants giving <60% of wt β-gal activity, i.e. E163K, L221H, C265R, M267K, V306D, C308R and A339D, were also severely compromised with full-length proteins in mammalian cells, both without and in response to AhR ligand (compare Figures 2a and 3a). These reductions in reporter gene activity were unlikely due to poor expression, as levels of exogenous wt and mutants Arnt proteins were mostly similar (Figure 3b). Expression of three mutant proteins (i.e. D219V, A339D and S190P) was detectably lower than wt Arnt. Arnt D219V mutant was almost indistinguishable from wt in terms of reporter gene activation, while A339D and S190P mutants were >100-fold less active than wt protein in the absence of ligand, which does not correlate with the small observed reduction in protein expression.
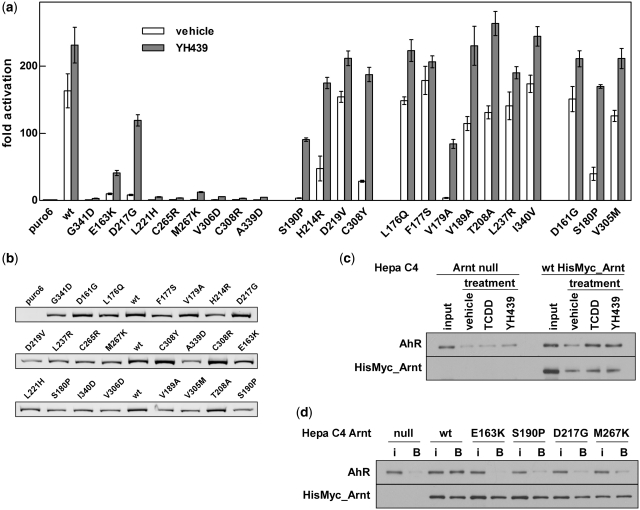
Figure 3.
Arnt PAS.A mutations identified from RevB2H disrupt full-length AhR/Arnt heterodimerization in mammalian cells. (a) Activation of the pML-6c-X XRE-driven reporter gene in 293T cells by HisMyc-tagged AhR and full-length wt or mutant HisMyc-tagged Arnt in response to vehicle or the ligand YH439, shown as fold activation over basal levels (puro6) from endogenous protein. Data are mean ± SD of transfections performed in triplicate and representative of two independent experiments. (b) Expression of HisMyc_Arnt wt and mutants in the transfected cells used for the reporter gene assay in (a), detected with anti-Arnt monoclonal antibody and Cy5 labelled anti-mouse secondary antibody. (c) Cytosolic extracts from parental Arnt-deficient mouse Hepa C4 cells (Arnt null) and the derived line constitutively expressing HisMyc-tagged wt Arnt, from an integrated lentiviral vector, were treated in vitro with vehicle, TCDD or YH439. Protein captured by Ni resin was analysed by western blot, using anti-Myc antibody for HisMyc_Arnt or anti-AhR antibody to detect co-immobilized AhR. (d) Mutations in Arnt reduced co-capture of AhR in Ni affinity pull-downs. Cytosolic extracts from the Arnt null and derived cell lines constitutively expressing wt or the indicated mutant HisMyc_Arnt from integrated lentiviral vectors were in vitro transformed with YH439. Captured HisMyc_Arnt and AhR were detected by western blot using anti-Myc and anti-AhR antibodies. i, 5% of input; B, fraction bound to Ni resin. Data are representative of three independent experiments.
In general, the difference in the reporter gene activation between wt and mutant Arnt was greater in the absence of ligand. Activation of the XRE-driven reporter gene in the absence of ligand is commonly observed with exogenous expression of AhR/Arnt (32,38) and most of the mutations showed a reduction in this activity, consistent with reduced dimerization through PAS.A. For the less severe mutations, this was largely overcome in the presence of ligand, presumably due to the addition of the PAS.B dimerization interface (39).
Both the β-gal and luciferase reporter gene assays were indirect measures of dimerization, with the reasonable assumption that dimerization is required for reporter gene activation. To demonstrate directly that the Arnt PAS.A mutants identified using the RevB2H indeed have disrupted AhR/Arnt heterodimerization, we stably expressed a subset of Arnt mutants as full-length proteins with a HisMyc tag in the Arnt-deficient mouse hepatoma Hepa C4 cell line (34), using lentiviral mediated infection. HisMyc-tagged wt and mutant E163K, S190P, D217G and M267K Arnt proteins from cell extracts treated with YH439 in vitro were successfully captured by Ni resin to a similar extent. However, there was a marked difference in AhR co-immobilization, with only wt Arnt, but not the mutant Arnt proteins, enabling significant capture of AhR (Figure 3d). AhR ligand YH439 promoted capture of AhR/HisMyc_Arnt wt heterodimer from cell extracts to the same extent as the prototypical AhR ligand, 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD; Figure 3c) and similar results were obtained with TCDD (data not shown). Thus, these mutations in Arnt PAS.A disrupted AhR/Arnt heterodimerization in the full-length protein in response to ligand.
Arnt PAS.A mutations selectively alter dimerization with partner proteins Sim1, Sim2s, NPAS4, HIF-1α and Arnt itself
Since Arnt is the common dimerization partner for all other proteins in the Arnt cluster (Supplementary Figure S1), we looked at the effect of Arnt PAS.A mutants on dimerization with bHLH.PAS proteins other than AhR. Sim1/Arnt heterodimerization was assayed using a six CNS midline enhancer (CME) driven luciferase reporter gene construct derived from the Drosophila Sim target gene toll 6 (33) (Supplementary Figure S4). Interestingly, the E163K, V179A, S190P and D217G mutations, which significantly disrupted AhR/Arnt heterodimerization, had little or no effect on Sim1/Arnt interaction in this assay (Figure 4a). In contrast, mutations such as L221H, C265R, V306A, C308R and A339D showed reduced reporter gene expression for both Sim1/Arnt and AhR/Arnt (c.f. Figure 3a). E163K, V179A, S190P and D217G Arnt also had little effect on reporter gene activation when partnered with Sim2s or NPAS4 (Figure 4b and c) whereas M267K reduced activation for each of AhR, Sim1, Sim2s and NPAS4.
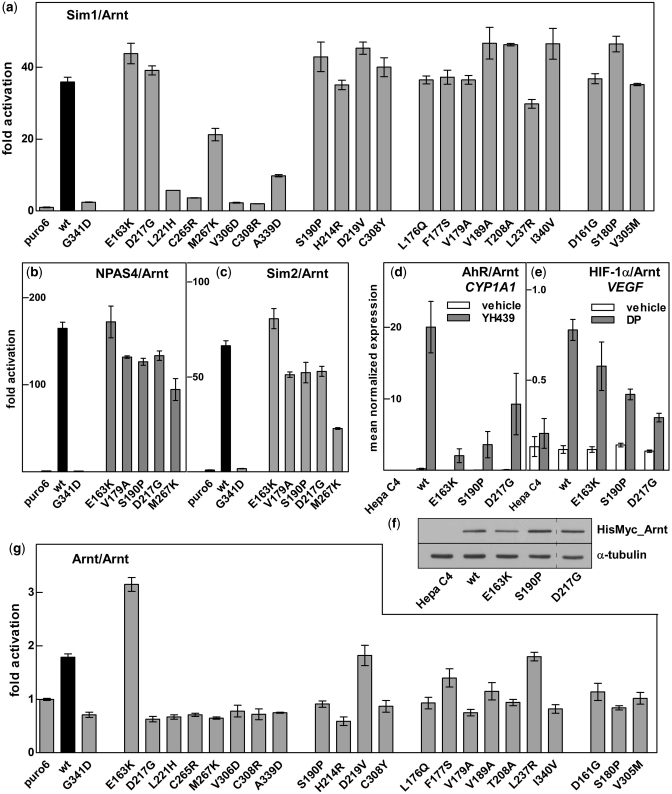
Figure 4.
Arnt mutations differentially alter dimerization with other partner proteins. Activation of the CME driven reporter (pML-6c-wt) in 293T cells by wt or mutant HisMyc_Arnt together with hSim1_Myc (a), NPAS4_Myc (b) or hSim2s_Myc (c) shown as fold activation over basal levels (puro6). Data are mean ± SD of transfections performed in triplicate and are representative of two independent experiments. Hepa C4 cell lines expressing wt, E163K, S190P or D217G HisMyc_Arnt were treated with vehicle, YH439 or 2,2′-dipyridyl (DP) and assayed for CYP1A1 (d) or VEGF (e) mRNA induction by qRT-PCR. Data are mean ± SEM, n = 3. (f) Whole cell extracts from the Hepa C4 Arnt deficient and derived cell lines constitutively expressing wt, E163K, S190P or D217G HisMyc_Arnt were separated by SDS–PAGE, followed by western blotting for Myc tagged Arnt and α-tubulin loading control. Dashed line indicates removal of an empty lane from the image. (g) Activation of the E-box driven 4(CACGTG)TKMPluc reporter gene in 293T cells by wt or mutant HisMyc_Arnt. Data are mean ± SD of transfections performed in triplicate and are representative of two independent experiments.
Next, we used our Hepa C4 lines expressing wt or mutant HisMyc-Arnt to investigate activation of prototypical endogenous target genes cytochrome P450 1A1 (CYP1A1) or vascular endothelial growth factor (VEGF) by AhR/Arnt or HIF1-α/Arnt heterodimers, respectively. The Arnt E163K, S190P and D217G mutations impaired AhR ligand-dependent CYP1A1 activation in a manner consistent with activity measured in the XRE reporter gene assays (Figure 4d). In contrast, the response of the HIF1-α/Arnt target gene VEGF in the presence of the hypoxia mimetic 2,2′-dipyridyl showed that the E163K and S190P Arnt mutant proteins were able to dimerize with HIF-1α to induce VEGF expression to greater levels relative to wt Arnt than with AhR on CYP1A1. The D217G mutation significantly reduced both AhR and HIF-1α activity in this assay (Figure 4e). These differences were not due to protein levels (Figure 4f). Our data indicate that Arnt probably uses the same interface to interact with each of its partner proteins, with particular residues having partner-specific effects.
The effect of mutations in the Arnt PAS.A domain on Arnt homodimerization was investigated by looking at the ability of the full-length mutant proteins to activate transcription in mammalian cells in culture using a reporter gene driven by the canonical E-box sequence (6). Since exogenous mutant Arnt protein levels were much greater than endogenous wt Arnt (Figure 3b), competing homodimer interactions with endogenous protein have no significant effect in this experiment. Overall, the pattern was similar to that seen for heterodimerization with partner proteins AhR or Sim1, suggesting the use of the same interface for homo- and heterodimerization. Surprisingly, the E163K mutation, which appeared specific for the AhR/Arnt interaction, increased activity of the Arnt homodimer by ∼2-fold relative to wt (Figure 4g). This difference was even more dramatic at sub-saturating concentration, where wt Arnt gave no reporter gene induction while E163K Arnt showed ∼2-fold induction, comparable to the maximum for wt Arnt homodimer, suggesting the E163K mutation resulted in stronger homodimer formation (Supplementary Figure S5).
AhR mutant RevB2H screen
Having identified mutations in Arnt affecting dimer formation, we applied the RevB2H screen to the AhR PAS.A domain to isolate mutations in AhR which disrupted dimerization with Arnt. The wt AhR PAS.A coding region (amino acids 88–278) in the RNAPα_AhR278 expression vector was replaced with a mutagenic library generated using error-prone PCR and fused to the lacZα 1–39 polypeptide at the C-terminus to allow screening by alpha complementation. The resulting AhR library was transformed into KS1033 harbouring the λCI_Arnt362 expression and lacOR2-62_186cI CTD selection plasmids and grown in selective conditions. As in the Arnt experiment, the transformants arising showed a size distribution that differed distinctly from those without library transformation (Figure 1d and e). Again, large colonies which developed under selective conditions were either blue or white and small colonies predominantly blue. Of approximately 4000 transformants, RNAPα_AhR278 expression plasmids harbouring potential loss-of-interaction mutations were isolated from 120 large blue colonies and assayed for β-gal activity in the lacOR2-62_lacZ reporter strain, E. coli KS1, with co-transformation of λCI_Arnt362 expression plasmid. Putative AhR PAS.A mutants having <90% of wt activity were sequenced (Supplementary Table S1). At this stage, we found 14 single amino acid mutations in AhR PAS.A that potentially reduced AhR278/Arnt362 heterodimerization. After cloning the coding sequence of each AhR PAS.A mutation into fresh RNAPα_AhR278 expression vector without the LacZα fusion, mutants were re-analysed for β-gal activity. From this we identified 10 AhR PAS.A mutations with a statistically significant reduction in β-gal activity, consistent with having disrupted dimerization with Arnt (Figure 5a). These were again spread across the PAS.A region, clustered in the same structural elements as we saw for the Arnt dimerization mutations (Figure 5d) and largely conserved across species (Supplementary Figure S6). There were also several residues in the region N-terminal of the structured PAS domain that appeared to influence dimerization with Arnt, as seen for Arnt.
Figure 5.
Mutations in AhR PAS.A that alter dimerization with Arnt. (a) The strength of interaction between λCI_Arnt362 and the potential RNAPα_AhR278 loss-of-interaction mutants selected as large blue colonies from RevB2H was measured in the forward bacterial two hybrid system with the lacZ reporter gene (28). Data are mean with 95% confidence intervals, shown as a percentage of β-gal activity obtained with wt RNAPα_AhR278. bHLH: RNAPα_AhR bHLH domain only; *P < 0.05 (one tailed unpaired Student’s t-test). (b) AhR PAS.A mutations identified from RevB2H disrupted full-length AhR/Arnt function. Activation of the XRE-driven reporter gene in 293T cells by wt or mutant HisMyc_AhR and HisMyc_Arnt in response to vehicle or the ligand YH439, shown as fold activation over basal levels (puro6) from endogenous protein. Data are mean ± SD of transfections performed in triplicate and representative of two independent experiments. (c) Expression of wt and mutant HisMyc_AhR in the transfected cells used for the reporter gene assay in (b), detected by western blotting for Myc-tagged AhR and α-tubulin loading control. (d) AhR mutations deficient in dimerization with Arnt mapped onto the PAS.A sequence, illustrated with dPer PAS.A. See Figure 2e legend for details. Colouring of mutations according to the strength of disruption observed in the β-gal assay (a): red: <60%, magenta: 61–80%, green: >80% of wt activity.
On the XRE-driven luciferase reporter gene, the most severe AhR PAS.A mutations, V124E, A131V, I160T, L218P, G227C and F228L, reduced the ability of the AhR/Arnt heterodimer to activate transcription in full-length AhR in mammalian cells in culture, both in the basal activation in the absence of stimulus and in response to YH439 (Figure 5b). These observed differences were not due to protein expression levels (Figure 5c). Interestingly, E158G and T264A, having somewhat compromised dimerization with Arnt in the LacZ bacterial reporter gene assay using truncated bHLH.PAS.A proteins, showed reduced basal activity but close to wt activation in response to exogenous ligand, suggesting that addition of the PAS.B dimerization interface in the ligand-bound form can compensate for mild defects in PAS.A dimerization.
DISCUSSION
Protein–protein interaction is constant, dynamic and governs every aspect of cellular processes. This is clearly exemplified in budding yeast Saccharomyces cerevisiae, which is estimated to contain more than 30 000 protein–protein interactions for its approximately 6000 identified genes (40,41). Thus, on average, every yeast protein interacts with at least five others, demonstrating the essential role of protein networking in biology. A mechanistic understanding of protein interactions is important in health and disease, since many diseases are interaction related. Predictions suggest that at least 1200 of experimentally verified disease-related mutations present in Online Mendelian Inheritance in Man (OMIM) and UniPort databases can be attributed to protein interaction (42). A recent study of pair-wise TF interactions, for more than 1000 TFs in human and mouse, demonstrates how a better understanding of combinatorial TF interactions will greatly advance our current understanding and prediction accuracy on tissue differentiation and specification (43).
A number of techniques have been developed to identify protein interactions, including TAP tag purification (44), and yeast, bacterial and mammalian two hybrid approaches (28,45). More recently, studies of protein interaction networks are providing a global picture of the protein interactome landscape (41,43). Methods such as yeast split-hybrid (46), one- plus two-hybrid (47), and absence of interference (48), have been developed to delineate interaction interfaces and define amino acids essential to interaction. These yeast based methods can be time consuming and difficult for plasmid retrieval. Here, we describe a simple, robust bacterial system, RevB2H, to our knowledge the first reported bacterial system designed to detect loss of protein interactions. We have successfully incorporated alpha complementation, fusing lacZα to the C-terminus of the mutagenic library, reducing false positives and simplifying the subsequent validation procedure. As a result, nonsense and frame-shift mutations were readily filtered out (Supplementary Table S1). In addition, as the C-terminally encoded lacZα requires at least some folding of the partner protein to function, this approach helped to eliminate severe expression and mis-folding mutants. In fact, we found that the CD spectra of the Arnt mutant proteins were largely consistent with there being no gross structural perturbation. Some differences in the proportions of secondary structural elements, which may be indicative of changes in the local environment, were evident in the CD spectra for a number of the mutants. It should be noted that the protein concentration in the CD experiments varied with mutations, since severe mutations generally resulted in lower yield of purified protein. This can be attributed to reduced Arnt homodimerization (Figure 4g) consistent with previous work indicating that Arnt bHLH.PAS A protein purifies as a homodimer (13). Finally, the RevB2H mutations were readily transferable to full-length proteins, where effects on activity in mammalian cells correlated well with that in the bacterial β-gal assays using truncated proteins, encompassing only the N-terminal bHLH-PAS.A region. This indicates that the RevB2H system is robust and presents an easy alternative to yeast systems for systematic study of mammalian protein interactions.
Based on known PAS domain structures and interactions, the mechanism and specificity of dimerization mediated by Arnt PAS.A could potentially involve either the same or independent surfaces of the domain for homo- and hetero- dimerization (9,17–21). Our data favours the former, since most of the Arnt mutations we identified had similar effects on both homo- and hetero-dimerization with different partner proteins. Key residues discriminated between partner proteins, showing partner-specific effects. In particular, Arnt E163K, which had no effect on Sim1/Arnt, Sim2/Arnt, and NPAS4/Arnt dimerization, and only a mild effect on HIF1α/Arnt dimerization, completely abolished AhR/Arnt interaction, and yet strengthened Arnt homodimerization. Arnt S190 was also selective for the AhR/Arnt interaction, with the S190P mutation having little effect on other Arnt partner proteins, while substitution of D217 affected both AhR and HIF-1α but not Sim1, Sim2s or NPAS4.
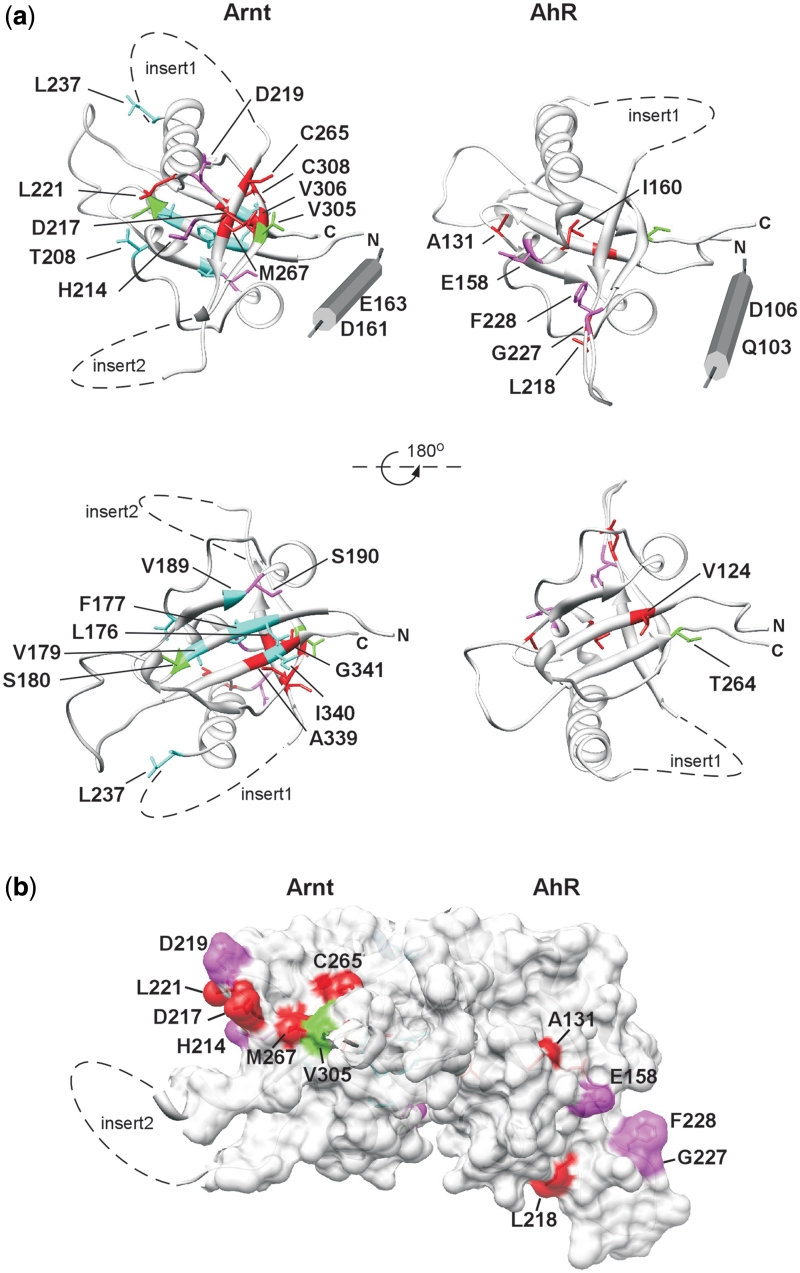
We generated homology models of the Arnt and AhR PAS.A domains, using the dPER crystal structure (25) as template, and mapped the residues that we found to be important for dimerization onto these models (Figure 6a). When the modelled structures were superimposed on the crystal structure of the HIF-2α/Arnt PAS.B dimer, where interaction is through the β-sheet surfaces (49), it was clear that the most severe of our identified mutations in both Arnt and AhR cluster on faces away from the HIF-2α/Arnt PAS.B β-scaffold dimerization interface (Figure 6b). This suggests the PAS.A interactions use a face of the domain, encompassing α-C, β-C and β-D, that is different from the HIF-2α/Arnt PAS.B interface, as well as other examples of PAS domain interactions (9,17–21). Further, AhR and Arnt appear to interact through equivalent regions of PAS.A (Figure 6a). Partner specific Arnt D217 is centrally located in the putative dimerization interface in this model, while the S190P substitution on the opposite face may alter interaction with AhR through effects on packing. Arnt E163, where substitution with lysine resulted in the most severe and discriminating alteration to dimerization, lies outside the PAS fold, in a predicted α-helical extension. This may be functionally equivalent to the N-terminal cap found in many PAS domains, which forms an interaction interface in some cases (20,21). While the packing of this predicted helix in the Arnt structure is unknown, it could be positioned to bring E163K close to the cluster of significant residues on Arnt (red and magenta in Figures 2 and 6) to form the dimerization interface. Two residues in the predicted N-terminal α-helix in AhR similarly appear important for dimerization with Arnt, albeit with less drastic effects than Arnt E163K, consistent with the suggestion that these helices form part of the AhR/Arnt PAS.A dimerization interface. A number of the less severe Arnt mutations that reduced interaction with all partner proteins map on the β-sheet surface in the model (lower view in Figure 6a), thus may disrupt interaction indirectly as a result of relatively minor perturbation of the β-scaffold. Alternatively, the β-sheet surface may contribute in some manner to the protein–protein interactions that regulate dimerization.
Figure 6.
Mutations altering dimerization of Arnt and AhR PAS.A domains define a putative dimerization interface. (a) Residues found to be important for Arnt and AhR dimerization are illustrated on homology models of Arnt and AhR PAS.A domain. The mutations are coloured according to the severity of the defect in the lacZ reporter assay. For Arnt red: <60%, magenta: 61–70%, blue: 71–80% and green: >80% of wt activity (Figure 2a) and for AhR red: <60%, magenta: 61–80%, green: >80% of wt activity (Figure 5a). (b) Superimposition of PAS.A models of Arnt and AhR onto the structure 3F1P of HIF-2α/Arnt PAS.B (49) suggests dimerization occurs through different interfaces for PAS A and PAS B domains.
The mechanism of a common interface within which particular residues control specificity occurs with other dimeric TFs. The bHLH.ZIP TF Max can homodimerize and heterodimerize with bHLH.ZIP proteins, Myc and Mad. Structural studies show that in the Max/Max homodimer, Q91 of one Max monomer interacts with N92 of the other, stabilizing the dimer interface. The presence of charged amino acid residues at corresponding positions in Myc (R423 and R424) and Mad (E125) further strengthens dimerization. As a result, Max preferentially forms heterodimers with Myc and Mad over homodimerization (23). Similarly, single amino acid changes in mouse retinoid X receptor (RXR) alters RXR dimer specificity, such that mRXR Y402A displays significantly weakened dimerization with retinoid acid receptor (RAR), peroxisome proliferator activating receptor (PPAR) and vitamin D receptor (VDR), but a marked increase in homodimerization (22). This mode of dimerization regulation may be favoured during protein evolution, as computational studies suggest that many dimeric TFs arise from gene duplication, which initially results in homodimer formation. Subsequent diversification of one of the genes then accommodates more specialized function (50,51). However, during this process, the mode of interaction is likely to be preserved, with key residues defining specificity.
Arnt can form homodimers which bind to the canonical E-box DNA sequence in vitro and in vivo (5–7). However, the physiological significance of Arnt homodimerization is still unclear, partially due to the weak dimerization affinity between Arnt monomers. As a result, it is hard to investigate effects mediated by Arnt homodimers separately from Arnt heterodimers. Here, we reported a mutant form of Arnt which showed a marked increase in Arnt homodimerization, at least in reporter gene assays, which may be useful for studying the physiological role of Arnt homodimers. In addition, sequence analysis show that E163 is also conserved between Arnt and BMAL 1 and 2. Thus, it will be interesting to investigate the effect of the same mutation on BMAL, since like Arnt, BMAL can homodimerize and heterodimerize with proteins such as Clock (Supplementary Figure S1), both of which are essential proteins in regulation of circadian rhythm.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Australian Research Council (grant numbers DP0559011 to A.C.-S. and M.L.W., DP0665185 to K.E.S.); Australian Government National Health and Medical Research Council (grant number 627140). Funding for open access charge: Waived by Oxford University Press.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank J. Pelletier for pML-6c vectors and the 2009 Third year Molecular and Structural Biology class at the University of Adelaide for performing part of the initial Arnt PAS.A mutagenesis screen as a supervised practical project.
REFERENCES
- 1.McIntosh BE, Hogenesch JB, Bradfield CA. Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu. Rev. Physiol. 2010;72:625–645. doi: 10.1146/annurev-physiol-021909-135922. [DOI] [PubMed] [Google Scholar]
- 2.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
- 3.DeYoung MP, Tress M, Narayanan R. Identification of Down's syndrome critical locus gene SIM2-s as a drug therapy target for solid tumors. Proc. Natl Acad. Sci. USA. 2003;100:4760–4765. doi: 10.1073/pnas.0831000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol. 2004;36:189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- 5.Arpiainen S, Lamsa V, Pelkonen O, Yim SH, Gonzalez FJ, Hakkola J. Aryl hydrocarbon receptor nuclear translocator and upstream stimulatory factor regulate Cytochrome P450 2a5 transcription through a common E-box site. J. Mol. Biol. 2007;369:640–652. doi: 10.1016/j.jmb.2007.03.075. [DOI] [PubMed] [Google Scholar]
- 6.Kewley RJ, Whitelaw ML. Phosphorylation inhibits DNA-binding of alternatively spliced aryl hydrocarbon receptor nuclear translocator. Biochem. Biophys. Res. Commun. 2005;338:660–667. doi: 10.1016/j.bbrc.2005.08.073. [DOI] [PubMed] [Google Scholar]
- 7.Antonsson C, Arulampalam V, Whitelaw ML, Pettersson S, Poellinger L. Constitutive function of the basic helix-loop-helix/PAS factor Arnt. Regulation of target promoters via the E box motif. J. Biol. Chem. 1995;270:13968–13972. doi: 10.1074/jbc.270.23.13968. [DOI] [PubMed] [Google Scholar]
- 8.Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erbel PJ, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix–loop–helix-PAS transcription factor hypoxia-inducible factor. Proc. Natl Acad. Sci. USA. 2003;100:15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, Zhang L, Erbel PJ, Gardner KH, Ding K, Garcia JA, Bruick RK. Functions of the Per/ARNT/Sim domains of the hypoxia-inducible factor. J. Biol. Chem. 2005;280:36047–36054. doi: 10.1074/jbc.M501755200. [DOI] [PubMed] [Google Scholar]
- 11.Lindebro MC, Poellinger L, Whitelaw ML. Protein-protein interaction via PAS domains: role of the PAS domain in positive and negative regulation of the bHLH/PAS dioxin receptor-Arnt transcription factor complex. Embo J. 1995;14:3528–3539. doi: 10.1002/j.1460-2075.1995.tb07359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pongratz I, Antonsson C, Whitelaw ML, Poellinger L. Role of the PAS domain in regulation of dimerization and DNA binding specificity of the dioxin receptor. Mol. Cell. Biol. 1998;18:4079–4088. doi: 10.1128/mcb.18.7.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman-Smith A, Lutwyche JK, Whitelaw ML. Contribution of the Per/Arnt/Sim (PAS) domains to DNA binding by the basic helix-loop-helix PAS transcriptional regulators. J. Biol. Chem. 2004;279:5353–5362. doi: 10.1074/jbc.M310041200. [DOI] [PubMed] [Google Scholar]
- 14.McGuire J, Okamoto K, Whitelaw ML, Tanaka H, Poellinger L. Definition of a dioxin receptor mutant that is a constitutive activator of transcription: delineation of overlapping repression and ligand binding functions within the PAS domain. J. Biol. Chem. 2001;276:41841–41849. doi: 10.1074/jbc.M105607200. [DOI] [PubMed] [Google Scholar]
- 15.Kikuchi Y, Ohsawa S, Mimura J, Ema M, Takasaki C, Sogawa K, Fujii-Kuriyama Y. Heterodimers of bHLH-PAS protein fragments derived from AhR, AhRR, and Arnt prepared by co-expression in Escherichia coli: characterization of their DNA binding activity and preparation of a DNA complex. J. Biochem. 2003;134:83–90. doi: 10.1093/jb/mvg115. [DOI] [PubMed] [Google Scholar]
- 16.Makino Y, Kanopka A, Wilson WJ, Tanaka H, Poellinger L. Inhibitory PAS domain protein (IPAS) is a hypoxia-inducible splicing variant of the hypoxia-inducible factor-3alpha locus. J. Biol. Chem. 2002;277:32405–32408. doi: 10.1074/jbc.C200328200. [DOI] [PubMed] [Google Scholar]
- 17.Amezcua CA, Harper SM, Rutter J, Gardner KH. Structure and interactions of PAS kinase N-terminal PAS domain: model for intramolecular kinase regulation. Structure. 2002;10:1349–1361. doi: 10.1016/s0969-2126(02)00857-2. [DOI] [PubMed] [Google Scholar]
- 18.Lee J, Tomchick DR, Brautigam CA, Machius M, Kort R, Hellingwerf KJ, Gardner KH. Changes at the KinA PAS-A dimerization interface influence histidine kinase function. Biochemistry. 2008;47:4051–4064. doi: 10.1021/bi7021156. [DOI] [PubMed] [Google Scholar]
- 19.Buttani V, Losi A, Eggert T, Krauss U, Jaeger KE, Cao Z, Gartner W. Conformational analysis of the blue-light sensing protein YtvA reveals a competitive interface for LOV-LOV dimerization and interdomain interactions. Photochem. Photobiol. Sci. 2007;6:41–49. doi: 10.1039/b610375h. [DOI] [PubMed] [Google Scholar]
- 20.Ma X, Sayed N, Baskaran P, Beuve A, van den Akker F. PAS-mediated dimerization of soluble guanylyl cyclase revealed by signal transduction histidine kinase domain crystal structure. J. Biol. Chem. 2008;283:1167–1178. doi: 10.1074/jbc.M706218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR. Conformational switching in the fungal light sensor Vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vivat-Hannah V, Bourguet W, Gottardis M, Gronemeyer H. Separation of retinoid X receptor homo- and heterodimerization functions. Mol. Cell. Biol. 2003;23:7678–7688. doi: 10.1128/MCB.23.21.7678-7688.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112:193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 24.Kirk R, Laman H, Knowles PP, Murray-Rust J, Lomonosov M, Meziane el K, McDonald NQ. Structure of a conserved dimerization domain within the F-box protein Fbxo7 and the PI31 proteasome inhibitor. J. Biol. Chem. 2008;283:22325–22335. doi: 10.1074/jbc.M709900200. [DOI] [PubMed] [Google Scholar]
- 25.Yildiz O, Doi M, Yujnovsky I, Cardone L, Berndt A, Hennig S, Schulze S, Urbanke C, Sassone-Corsi P, Wolf E. Crystal structure and interactions of the PAS repeat region of the Drosophila clock protein PERIOD. Mol. Cell. 2005;17:69–82. doi: 10.1016/j.molcel.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 26.Card PB, Erbel PJ, Gardner KH. Structural basis of ARNT PAS-B dimerization: use of a common beta-sheet interface for hetero- and homodimerization. J. Mol. Biol. 2005;353:664–677. doi: 10.1016/j.jmb.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 27.Pinkett HW, Shearwin KE, Stayrook S, Dodd IB, Burr T, Hochschild A, Egan JB, Lewis M. The structural basis of cooperative regulation at an alternate genetic switch. Mol. Cell. 2006;21:605–615. doi: 10.1016/j.molcel.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 28.Dove SL, Joung JK, Hochschild A. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature. 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 29.Palmer AC, Ahlgren-Berg A, Egan JB, Dodd IB, Shearwin KE. Potent transcriptional interference by pausing of RNA polymerases over a downstream promoter. Mol. Cell. 2009;34:545–555. doi: 10.1016/j.molcel.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raussens V, Ruysschaert JM, Goormaghtigh E. Protein concentration is not an absolute prerequisite for the determination of secondary structure from circular dichroism spectra: a new scaling method. Anal. Biochem. 2003;319:114–121. doi: 10.1016/s0003-2697(03)00285-9. [DOI] [PubMed] [Google Scholar]
- 31.Schubert RA, Dodd IB, Egan JB, Shearwin KE. Cro's role in the CI Cro bistable switch is critical for {lambda}'s transition from lysogeny to lytic development. Genes Dev. 2007;21:2461–2472. doi: 10.1101/gad.1584907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whelan F, Hao N, Furness SG, Whitelaw ML, Chapman-Smith A. Amino acid substitutions in the aryl hydrocarbon receptor ligand binding domain reveal YH439 as an atypical AhR activator. Mol. Pharmacol. 2010;77:1037–1046. doi: 10.1124/mol.109.062927. [DOI] [PubMed] [Google Scholar]
- 33.Moffett P, Pelletier J. Different transcriptional properties of mSim-1 and mSim-2. FEBS Lett. 2000;466:80–86. doi: 10.1016/s0014-5793(99)01750-0. [DOI] [PubMed] [Google Scholar]
- 34.Sun W, Zhang J, Hankinson O. A mutation in the aryl hydrocarbon receptor (AHR) in a cultured mammalian cell line identifies a novel region of AHR that affects DNA binding. J. Biol. Chem. 1997;272:31845–31854. doi: 10.1074/jbc.272.50.31845. [DOI] [PubMed] [Google Scholar]
- 35.Dove SL, Hochschild A. Conversion of the omega subunit of Escherichia coli RNA polymerase into a transcriptional activator or an activation target. Genes Dev. 1998;12:745–754. doi: 10.1101/gad.12.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ullmann A, Jacob F, Monod J. Characterization by in vitro complementation of a peptide corresponding to an operator-proximal segment of the beta-galactosidase structural gene of Escherichia coli. J. Mol. Biol. 1967;24:339–343. doi: 10.1016/0022-2836(67)90341-5. [DOI] [PubMed] [Google Scholar]
- 37.Numayama-Tsuruta K, Kobayashi A, Sogawa K, Fujii-Kuriyama Y. A point mutation responsible for defective function of the aryl-hydrocarbon-receptor nuclear translocator in mutant Hepa-1c1c7 cells. Eur. J. Biochem. 1997;246:486–495. doi: 10.1111/j.1432-1033.1997.00486.x. [DOI] [PubMed] [Google Scholar]
- 38.Murray IA, Reen RK, Leathery N, Ramadoss P, Bonati L, Gonzalez FJ, Peters JM, Perdew GH. Evidence that ligand binding is a key determinant of Ah receptor-mediated transcriptional activity. Arch. Biochem. Biophys. 2005;442:59–71. doi: 10.1016/j.abb.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 39.Soshilov A, Denison MS. Role of the Per/Arnt/Sim domains in ligand-dependent transformation of the aryl hydrocarbon receptor. J. Biol. Chem. 2008;283:32995–33005. doi: 10.1074/jbc.M802414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 41.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuster-Bockler B, Bateman A. Protein interactions in human genetic diseases. Genome Biol. 2008;9:R9. doi: 10.1186/gb-2008-9-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 45.Lievens S, Lemmens I, Tavernier J. Mammalian two-hybrids come of age. Trends Biochem. Sci. 2009;34:579–588. doi: 10.1016/j.tibs.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shih HM, Goldman PS, DeMaggio AJ, Hollenberg SM, Goodman RH, Hoekstra MF. A positive genetic selection for disrupting protein-protein interactions: identification of CREB mutations that prevent association with the coactivator CBP. Proc. Natl Acad. Sci. USA. 1996;93:13896–13901. doi: 10.1073/pnas.93.24.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim JY, Park OG, Lee JW, Lee YC. One- plus two-hybrid system, a novel yeast genetic selection for specific missense mutations disrupting protein/protein interactions. Mol. Cell Proteomics. 2007;6:1727–1740. doi: 10.1074/mcp.M700079-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Dhayalan A, Jurkowski TP, Laser H, Reinhardt R, Jia D, Cheng X, Jeltsch A. Mapping of protein-protein interaction sites by the ‘absence of interference’ approach. J. Mol. Biol. 2008;376:1091–1099. doi: 10.1016/j.jmb.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheuermann TH, Tomchick DR, Machius M, Guo Y, Bruick RK, Gardner KH. Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor. Proc. Natl Acad. Sci. USA. 2009;106:450–455. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amoutzias GD, Robertson DL, Oliver SG, Bornberg-Bauer E. Convergent evolution of gene networks by single-gene duplications in higher eukaryotes. EMBO Rep. 2004;5:274–279. doi: 10.1038/sj.embor.7400096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pereira-Leal JB, Levy ED, Kamp C, Teichmann SA. Evolution of protein complexes by duplication of homomeric interactions. Genome Biol. 2007;8:R51. doi: 10.1186/gb-2007-8-4-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.