Abstract
hSSB1 is a recently discovered single-stranded DNA binding protein that is essential for efficient repair of DNA double-strand breaks (DSBs) by the homologous recombination pathway. hSSB1 is required for the efficient recruitment of the MRN complex to sites of DSBs and for the efficient initiation of ATM dependent signalling. Here we explore the interplay between hSSB1 and MRN. We demonstrate that hSSB1 binds directly to NBS1, a component of the MRN complex, in a DNA damage independent manner. Consistent with the direct interaction, we observe that hSSB1 greatly stimulates the endo-nuclease activity of the MRN complex, a process that requires the C-terminal tail of hSSB1. Interestingly, analysis of two point mutations in NBS1, associated with Nijmegen breakage syndrome, revealed weaker binding to hSSB1, suggesting a possible disease mechanism.
INTRODUCTION
Cells frequently encounter DNA damage; with DNA double-strand breaks (DSBs) being among the most cytotoxic of these lesions. Chromosomal instability may occur even from a single DSB, if it is repaired incorrectly, and this may ultimately lead to cell death. It is essential that DSBs in human cells are detected, signalled and repaired efficiently in order to prevent the accumulation of damage, which can lead to chromosomal instability or malignant transformation. DSBs may be induced by a number of factors including ionizing radiation (IR), reactive chemical species and via normal cellular processes such as DNA replication. Once a DSB is detected, DNA repair proteins are recruited to the site of the DSB and a multi-faceted DSB pathway is activated. This complex signalling network includes altered transcriptional and translational regulation and the induction of DSB repair and cell cycle arrest via the activation of checkpoints. While non-homologous end joining (NHEJ) may be used to repair DSBs in any phase of the cell cycle, homologous recombination (HR) may be used to repair DSBs that specifically occur in the S or G2 phases of the cell cycle (1–3).
One of the first events in the process of HR is the recruitment of the MRN repair complex to the DSB site. Once located at the DSB, MRN activates the ATM kinase and tethers the DNA ends (4). When ATM is activated it in turn initiates signalling cascades, which leads to the resection of the DSBs to produce single-stranded DNA (ssDNA) (5). The resulting ssDNA acts as a substrate for Rad51-mediated strand exchange (4,6). Recent studies have shown that MRN also has a role in classical and alternative NHEJ (7,8).
The ssDNA binding (SSB) protein family have a fundamental role in the repair of DNA damage in all three domains of life. The simple SSBs and the replication protein A (RPA)-like SSBs form the sub-groups of this family of proteins (9). RPA is the most widely studied member of the SSB family in humans and is believed to be a pivotal factor of both DNA replication and DNA repair pathways (10–12). However, human RPA has a complex oligomeric structure not seen in the simple bacterial SSBs (13). The simple SSBs were believed to be restricted to the bacterial and archaeal domains of life, however, recently we have identified two new members of the SSB family in humans: hSSB1 and hSSB2 (14). hSSB1 and hSSB2 are structurally much more closely related to the bacterial and archaeal SSBs than to RPA (9). Both hSSBs consist of a ssDNA oligonucleotide binding (OB) fold, a divergent spacer domain, followed by a conserved C-terminal tail predicted to be required for protein–protein interactions (14). The crenarchaeal SSB, from Sulfolobus solfataricus, also has a flexible spacer followed by basic and acidic regions near the C-terminus; this region plays no part in DNA binding but is known to modulate protein–protein interactions (15).
We have previously shown that hSSB1 is required for the efficient signalling of DSBs following exposure to IR (14). Recently, we have also shown that hSSB1 is rapidly recruited to sites of DSBs and its presence is required for the efficient recruitment of the MRN complex and subsequent downstream partners (16). Furthermore, hSSB1 deficient cells fail to generate the ssDNA tracts associated with the nuclease activity of MRN and CtIP (16). We, and others, have also demonstrated that hSSB1 is a component of a complex containing IntS3 (17,18).
In this study, we demonstrate that hSSB1 forms a DNA damage-independent complex with Mre11:Rad50:NBS1 (MRN) which is distinct from the hSSB1:IntS3 complex. We show that hSSB1 plays an essential role in the recruitment and function of MRN at sites of DSBs. The MRN complex is believed to be the primary sensor of DSBs and promotes the activation of ATM kinase, which in turn initiates downstream DSB signalling. MRN also functions in the resection of the DSB, a process required for ATR signalling and preparation of DNA for HR in particular Rad51 mediated strand invasion (6,19,20). Our data now demonstrate how hSSB1 functions to directly recruit the MRN complex to DSBs, a process that also stimulates MRN nuclease activity. These data implicate hSSB1 as a pivotal member of the DNA damage response, cementing its role in maintaining genomic stability in metazoans.
MATERIALS AND METHODS
Cell lines, plasmids and siRNA
HeLa and HEK293T cells were maintained in DMEM supplemented with 10% fetal bovine serum (Gibco). Transfection of plasmids was performed using Lipofectamine 2000 (Invitrogen) as per manufacturer’s instructions. Full-length hSSB1 and truncations were cloned into bacterial expression vectors encoding a His-tag (pET28c). GFP-hSSB1 tail (153–211amino acids) was expressed from pEGFP-C1. FLAG tagged Mre11 in pFastBac1 (TP813) and NBS1 in pFastBac1 (TP28) were kindly supplied by Tanya Paull. The HisRad50 was excised from the pVL1392 vector (TP11) (gift from Tanya Paull) using EcoR1 restriction enzyme and then cloned directly into an EcoR1 digested pFastBac1 vector. Clones of the correct orientation were selected following a diagnostic digest with EcoRV. Virion phiX174 was supplied by New England Biolabs.
Antibodies
The following antibodies were used in the study: Rad50 (Calbiochem), Mre11, NBS1 (Sigma), IntS3 (Bethyl laboratories) and Alexa secondary antibodies (Invitrogen). Sheep antiserum to hSSB1 has been described previously (14).
Immunoprecipitation experiments
Co-immunoprecipitations were performed using the ice-cold NP40 buffer; 20 mM HEPES pH 8, 150 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, 0.2% NP40, 0.5 mM DTT, 5% glycerol, 1 mM NaF, 1 mM NaVO4, protease inhibitor cocktail (Sigma). Assays were performed at 5°C for 2 h using the antibodies as indicated in the ‘Results’ section. Antibodies were captured using magnetic protein G agarose beads (Invitrogen), washed three times in NP40 buffer before being analysed by immunoblot.
hSSB1 pull-down assays
Recombinant hSSB1 was bound to cyanogen bromide agarose as per manufacturer’s instructions (GE Healthcare) with bound protein being quantified by coomassie blue staining. hSSB1 pull-downs were performed in NP40 buffer (described above for immunoprecipitations) and the washed beads were analysed by immunoblot.
DNA pull-down assays
Annealed double-stranded oligonucleotides with six bp overhangs were generated from the following two sequences: oligo 1, 5′-GATCCACTGGGTTAACGCCGGACATTGCCCGGAT; oligo 2, 5′-TCCATGATCCGGGCAATGTCCGGCGTTAACCCAGTGGATC. Oligo 1 was modified with a 5′ biotin label. Oligonucleotides were annealed and bound to streptavidin agarose prior to assay. Each assay consisted of 10 ng oligo bound to beads with 20 ng (or 80 ng) of purified MRN, and a final concentration of 2 µM hSSB1, RPA or hSSB1-T1 (amino acids 1–114). Binding was performed for 15 min at room temperature in DNA pull-down buffer: 20 mM HEPES pH 8, 150 mM KCl, 5 mM MgCl2, 5% glycerol, 0.05% NP40, prior to analysis on a NUPAGE 4–12% SDS gel.
Preparation of nuclear extract
Nuclear extract was prepared as described previously (21).
Protein purification
Recombinant hSSB1 was expressed in Escherichia coli Rosetta 2 strain and purified by a three-step purification method. Clarified soluble E. coli cell lysate treated with DNaseI (10 µg/ml final concentration) was applied to a Hi-trap metal chelating column loaded with Nickel in a high-salt buffer containing 20 mM Tris pH 8.0, 500 mM NaCl and 30 mM imidazole at a flow rate of 1 ml/min. hSSB1 was eluted using a gradient of 0–100% 500 mM imidazole (1 ml/min) on a BioLogic System (GE). Protein fractions were collected from the middle of the broad protein peak and diluted 1:10 in heparin buffer consisting of 20 mM Tris pH 7.6, 0.5 mM EDTA, 1 mM DTT. This allowed the salt concentration to be reduced to 50 mM NaCl when protein was loaded onto heparin column. Protein was eluted with a 0–100% gradient of 1 M NaCl. Protein was concentrated, then subjected to size-exclusion chromatography on a Superdex75 column (in 20 mM Tris pH 7.6, 150 mM NaCl, 1 mM DTT) at a flow rate of 1 ml/min to purify protein on the basis of size. Protein eluted as one sharp peak. Purified hSSB1 was electrophoresed through a NuPAGE protein gel and stained with Sypro Ruby (GE Healthcare) to further determine the purity of the preparation. GST fusion proteins were purified as described previously (22).
Nuclear protein fractionation
Pre-filtered nuclear extract (3 mg) prepared from HEK293T, as described previously (21), cells was passed through a Hi-load Superdex 16/60 size-exclusion column (GE Healthcare) at a flow rate of 1 ml/min in buffer D. Samples were collected during the entire run.
MRN nuclease activity
Nuclease assays were performed essentially as previously described (23). Assays were performed for 40 min (to demonstrate MRN endonuclease activity) or 5 min (assays with addition of hSSB1) as indicated. All assays were performed in the presence of 5 mM MnCl2 unless otherwise indicated.
RESULTS
hSSB1 interacts with the MRN complex and is required for MRN recruitment to DSBs
We first sought to identify functional protein complexes containing hSSB1. An N-terminal GST-hSSB1 fusion protein was covalently linked to a cyanogen bromide sepharose column as the bait. Nuclear extract (10 mg) was passed over the hSSB1 affinity column, containing 10 µg/ml ethidium bromide to dissociate protein–DNA interactions. The column was washed extensively and interacting proteins eluted with 1 M NaCl. Eluted proteins were then lyophilized along with the eluate from a GST-only control column and both samples analysed by tandem mass spectrometry (MS/MS). We identified the IntS3 complex, which has previously been reported to interact with hSSB1 (17,18,24). We also identified strong sequence coverage for Rad50, a component of the MRN complex (Supplementary Figure S1). To confirm this interaction we performed reciprocal co-immunoprecipitation experiments with hSSB1 and Mre11 from nuclear extracts. This revealed that hSSB1 interacts with Mre11 and NBS1 in a DNA damage-independent manner (±IR) (Figure 1a and b), suggesting that hSSB1 is in complex with MRN.
Figure 1.
hSSB1 interacts with the MRN complex. (a and b) Co-immunoprecipitation of hSSB1 with NBS1 and or Mre11 using antibodies as indicated or NS (nonspecific serum). Immunoprecipitates obtained from cells with or without exposure to IR (6 Gy) were immunoblotted with indicated antibodies.
We next fractionated nuclear extract from HEK293T cells stably expressing Flag-tagged hSSB1 using size-exclusion chromatography. This method allows for the detection of stable protein complexes in solution. hSSB1 and Mre11 co-elute in fraction 36 (smaller complex) before IR (Figure 2a and b). Interestingly following IR (6 Gy, 1 h recovery), Mre11 shifts to be predominantly in fraction 34 (larger complex). IntS3 also elutes in these same fractions, raising the possibility of a common complex. To investigate this further we performed co-immunoprecipitation experiments using Fraction 34 (IR) and found that although hSSB1 precipitated with MRN components and IntS3; Mre11 failed to precipitate IntS3 and IntS3 failed to co-precipitate Mre11 (Figure 2c and d). This suggests that the hSSB1:MRN complex is distinct from the hSSB1:IntS3 complex. We performed extensive immunoprecipitation experiments but were unable to detect any interaction between MRN and IntS3 from nuclear or whole cell lysates under our experimental conditions (data not shown). Interestingly, the interaction between MRN and IntS3 observed by Huang et al. (24) required the over-expression of all components of MRN and IntS3 in insect or HEK 293 cells, suggesting the interaction may represent a small pool, or specific conditions which are not easily observed with endogenous protein in asynchronous cells.
Figure 2.
hSSB1 exists in two independent complexes with either MRN or IntS3. (a) hSSB1 and Mre11 co-elute from a Hiload Superdex 16/60 column (GE Healthcare). Fractionation of nuclear extract (prepared as described in materials and methods from HEK293T cells) over a Superdex 16/60 column following IR (6 Gy) or mock treatment (-IR) as indicated. (b) Densitometry of fractions from (a) normalized to exposure times were performed using the Image Gauge software (Fuji). Molecular weight markers are indicated below the graph. (c) Immunoprecipitation of hSSB1 from fraction 34 (6 Gy IR) showing co-immunoprecipitation of MRN complex and IntS3. (d) Immunoblot of IntS3 and Mre11 immunoprecipitates demonstrating that hSSB1 co-immunoprecipitates with Mre11 independently of IntS3 and vice-versa.
hSSB1 interacts directly with the MRN complex via its C-terminal tail
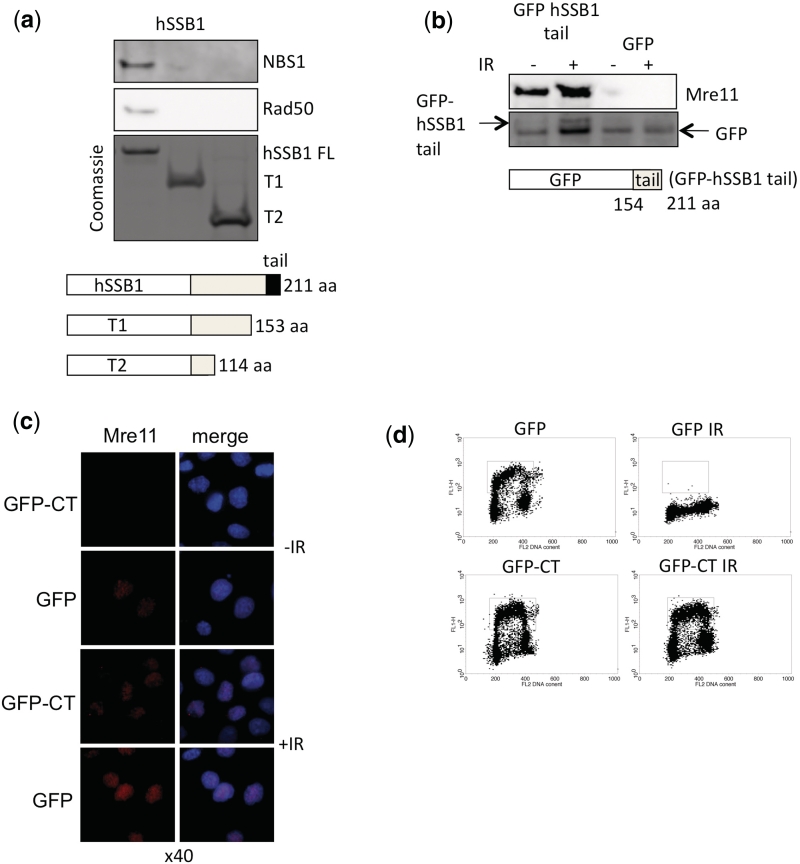
We next sought to determine if hSSB1 interacted directly with the MRN complex. Purified MRN was incubated with recombinant hSSB1 (Supplementary Figure S2a–c), and truncations of hSSB1 that were chemically crosslinked to sepharose beads. These data confirmed the interaction was direct and like other SSB interactions (9,22), was mediated through the C-terminal tail of hSSB1, since deletion of this region in hSSB1 abrogated the interaction (Figure 3a). To further confirm this interaction we ectopically expressed the hSSB1 C-terminal tail (amino acids 154–211) as an N-terminal GFP fusion in HeLa cells. Co-immunoprecipitation using GFP antibodies from nuclear extract, demonstrated that the hSSB1 tail was sufficient to mediate this MRN interaction (Figure 3b). Since the GFP-hSSB1 tail binds MRN we reasoned that this would compete with endogenous hSSB1 binding to MRN, thus preventing MRN recruitment to sites of DSBs. To confirm this, we irradiated cells (2 Gy) and looked for chromatin bound Mre11. In GFP positive cells we were able to observe normal Mre11 foci formation, however, in cells expressing the GFP-hSSB1 tail we were unable to observe normal Mre11 foci formation (Figure 3c). Furthermore, transient over-expression of the hSSB1-tail region attenuated the ATM dependent, IR-induced G1/S checkpoint thus mimicking the effect of hSSB1-knockdown on ATM activity, as reported previously (14) (Figure 3d). Taken together these data suggest that hSSB1 functions directly to recruit the MRN complex to DNA DSBs.
Figure 3.
Mapping of region in hSSB1 required to pull-down MRN complex. (a) Pull-down reactions were performed by incubating full-length hSSB1 or C-terminal truncations of hSSB1 (T1 1–153, T2 1–114) bound to sepharose beads with purified recombinant MRN (20 ng) complex. After washing, beads were subjected to immunoblotting with indicated antibodies. Coomassie stained gel shows the relative input of hSSB1 protein. (b) hSSB1 tail (amino acids 153–211) is sufficient for interaction with Mre11 in nuclear extracts. Cells were transfected with GFP-tagged hSSB1-tail or GFP vector alone and anti-GFP immunoprecipitates were immunoblotted with indicated antibodies. (c) Overexpression of GFP-hSSB1-tail (MRN interacting domain) results in impaired Mre11 foci formation after IR. HeLa cells were transfected with GFP or GFP-hSSB1-tail. Forty-eight hours after transfection cells were treated with IR (6 Gy, 30 min) and immunostained with indicated antibodies. Overexpression of GFP-hSSB1 tail (MRN interacting domain) results in impaired Mre11 foci formation after IR. HeLa cells were transfected with GFP or GFP-hSSB1-tail. Forty-eight hours after transfection cells were treated with IR (6 Gy, 30 min) and immunostained with indicated antibodies. (d) Overexpression of GFP-hSSB1 tail (MRN interacting domain) abrogates the IR induced G1/S checkpoint. HeLa cells were transfected with GFP or GFP-hSSB1-tail (GFP-CT). Forty-eight hours after transfection cells were treated with IR (6 Gy, 30 min) and allowed to recover for 16 h after which cells were pulse labeled with with BrdUrd (30 min, 10 μg/ml). Cells were subsequently stained with anti-BrdUrd followed by Alexa 488 secondary antibodies and propidium iodide before being analysed by flow cytometry. The box illustrating BrdUrd positive cells indicates that GFP-transfected cells effectively arrest at the G1/S boundary compared to GFP-CT transfected cells, which continue to enter S-phase.
hSSB1 interacts with the N-terminus of NBS1
To determine which component of the MRN complex interacts with hSSB1, GST fusion constructs spanning the length of Rad50, Mre11 and NBS1 were used to map the interaction with hSSB1. Pull-down assays using sepharose-linked hSSB1 revealed that the N-terminal domain of NBS1 interacted with hSSB1 (Figure 4a, Supplementary Figure S3). We further confirmed that this interaction was mediated through the C-terminus of hSSB1 (Figure 4b). Furthermore, hSSB1 failed to interact with full-length Mre11, RAD50 or their deletion mutants (data not shown).
Figure 4.
Mapping of region within NBS1 required for interaction with hSSB1. (a and b) hSSB1 c-terminal tail interacts directly with the N-terminal fragment of NBS1. hSSB1 pull-down experiments were performed with purified recombinant hSSB1 bound to sepharose and purified GST-tagged NBS1 fragments (NBS1A 1-221, NBS1B 199-473 and NBS1C 454-754).
Nijmegen breakage syndrome-associated NBS1 mutants fail to efficiently interact with hSSB1
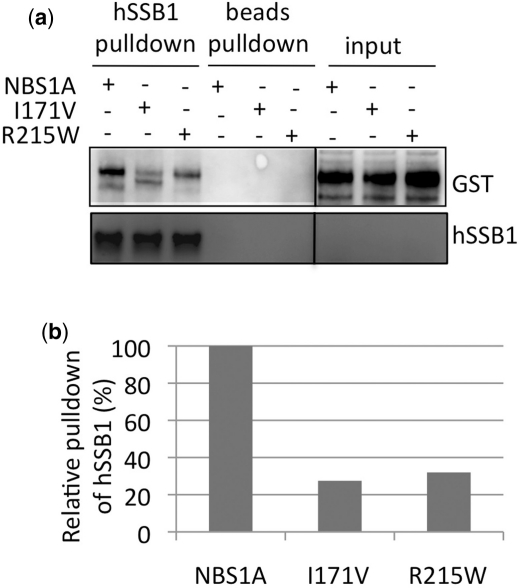
Nijmegen breakage syndrome is a disease associated with chromosomal instability. Two point mutations associated with a subset of this disease (I171V and R215W) located within hSSB1 interaction domain were examined for their ability to interact efficiently with hSSB1. Interestingly, neither mutant efficiently interacted with hSSB1 (Figure 5a and b, Supplementary Figure S4). This may be due to the mutations having a specific effect on the binding site for hSSB1 or that altered folding or misfolding of these mutants inhibits the hSSB1 interaction. However, a loss in the efficiency of hSSB1 binding to NBS1 would explain the chromosomal instability associated with these mutations.
Figure 5.
Nijmegen breakage syndrome mutants fail to interact efficiently with hSSB1. (a) Pull-down reactions were performed using hSSB1 bound to sepharose incubated with NBS1A, or NBS1A with the mutations I171V or R215W in the BRCT domain and immuno-blotted with indicated antibodies. (b) Quantification of data presented in (a), densitometry was performed using Image Gauge software (Fuji).
hSSB1 stimulates the endonuclease activity of the MRN complex by facilitating its recruitment to DSBs with short ss DNA overhangs
To determine if hSSB1 functions to directly recruit MRN to DSBs, we performed DNA pull-down assays. Since most endogenously generated DSBs (e.g. collapsed replication forks) possess short ssDNA overhangs, which have been shown to enhance ATM activation (25), we utilized a double-stranded DNA (dsDNA) substrate with a short 6 bp overhang. While MRN itself bound weakly in the experimental conditions used, the addition of hSSB1 resulted in a significant increase in MRN binding (Figure 6a), suggesting that hSSB1 was functioning to stimulate MRN recruitment to the DNA substrate. In contrast, recombinant RPA was unable to significantly stimulate binding of the MRN complex directly (Figure 6a, Supplementary Figure S5). Interestingly, although RPA could not bind this DNA substrate alone, addition of MRN allowed binding to occur. This is likely due to the end processing activity of MRN in these assays, generating ssDNA to which RPA could bind. We also observed that DNA binding of MRN was further enhanced when both hSSB1 and RPA were added. One possible explanation for this is that once hSSB1 stimulates binding, RPA may function to stabilize the bound MRN. To confirm that hSSB1 directly recruited MRN to the DNA substrate we next examined whether truncated hSSB1 (T1), (which binds ssDNA with a similar affinity to full-length hSSB1 (KD ≃15 nM), but is unable to interact with MRN could stimulate MRN binding. hSSB1-T1 failed to enhance MRN binding under normal assay conditions, however, with higher concentrations of MRN (where MRN binding could normally be observed in the absence of full-length hSSB1) hSSB1-T1 inhibited MRN binding (Figure 6b). This inhibition in the presence of hSSB1-T1 is likely due to competition for the DNA substrate. In contrast, the full-length hSSB1 functioned to further enhance MRN binding under the same conditions.
Figure 6.
hSSB1 directly facilitates MRN DNA binding and enhances the nuclease activity of the MRN complex. (a) Immunoblot of DNA pull-down assays using Rad50 (as a marker of the MRN complex), RPA and hSSB1 as indicated. Proteins, MRN 20 ng, 2 µM hSSB1 or 2 µM RPA, as indicated were precipitated with a biotinylated-double-stranded DNA oligo with a 6 bp overhang bound to streptavidin agarose beads. (b) Immunoblot of DNA pull-down assays using Rad50 as a marker of the MRN complex. Proteins, MRN 80 ng, 2 µM hSSB1 or 2 µM hSSB1-T1, as indicated were precipitated with a biotinylated-double-stranded DNA oligo with a 6 bp overhang bound to streptavidin agarose beads. (c) hSSB1 stimulates MRN nuclease activity. MRN nuclease activity assays contained phiX DNA, 20 ng MRN, 2 µM hSSB1, 2 µM hSSB1 truncated hSSB1 (1–153 amino acids, T1) or 2 µM RPA as indicated. Assays were incubated for 5 min before being resolved on a 1% agarose gel and stained with ethidium bromide.
Since both hSSB1 and MRN are needed for effective DNA resection, and hSSB1 is directly required for recruitment of MRN to DSBs, we analysed the effect of hSSB1 on the nuclease activity associated with MRN. While MRN has both exo- and endonuclease activities, its ssDNA endonuclease activity is important for DSB resection (26). We tested this activity using a closed circular ss phiX174 DNA as a substrate (23). As shown previously (23), MRN exhibited nuclease activity in the presence of manganese but not magnesium chloride (Supplementary Figure S6). The addition of hSSB1 to the assay drastically increased the MRN nuclease activity while the addition of RPA had no effect on MRN activity (Figure 6c, Supplementary Figure S7). To discount co-purification of any potential nuclease activity with recombinant hSSB1 we incubated the same substrate with hSSB1 for 1 h. Under our experimental conditions we were unable to observe nuclease activity (Supplementary Figure S8). Addition of a truncated hSSB1 missing the carboxy terminal tail (T1:1–153 amino acids), which is unable to interact with the MRN complex, failed to activate the MRN nuclease activity (Figure 6c). These results indicate that hSSB1 interacts directly with the MRN complex and enhances its nuclease activity.
DISCUSSION
We have previously shown that hSSB1 is rapidly recruited to sites of DSBs (16); this process is required for the efficient recruitment of MRN and activation of ATM. More recently, we have shown that hSSB1 is rapidly recruited to sites of DSBs and is required for generation/stability of ssDNA at the sites of damage (16). We now demonstrate that hSSB1 forms a complex with MRN, and that this complex is required for the recruitment of MRN to DSBs with short ssDNA overhangs. Interestingly RPA cannot substitute for hSSB1 in these assays, providing further evidence that these two members of the SSB family have distinct roles in the repair of DSBs in humans.
We have demonstrated that hSSB1 directly interacts with NBS1 via its C-terminal tail. Interestingly, hSSB1 interacts with the N-terminal domain of NBS1, a region known to also interact with MDC1 (27). The N-terminal domain of NBS1 contains both the FHA and BRCT domains; strikingly, mutations in this region severely impair HR, a process also dependent on hSSB1 (14,28). We also demonstrate, using size-exclusion fractionation and co-immunoprecipitation, that hSSB1 exists in two distinct complexes with IntS3 and MRN. This is in contrast with Huang et al. (24) who suggested that over-expression of NBS1 and IntS3 in insect and HEK 293 cells allows an IntS3: NBS1 complex to form. After repeated attempts, we have been unable to observe an interaction between endogenous IntS3 and MRN components in cell lysates. This may suggest that over-expression of both components forces the interaction kinetics to allow a rare, but potentially real, event to be observed. We have also shown that by inhibiting the hSSB1:MRN interaction using dominant negative hSSB1 proteins, MRN recruitment to DSBs is abolished, resulting in a failure to activate DSB induced cell cycle checkpoints. Finally, we have demonstrated that hSSB1 can facilitate binding of MRN to DSBs with short overhangs (6 bp), and that hSSB1 stimulates Mre11 nuclease activity. These data provide a plausible mechanism through which hSSB1 could facilitate MRN dependent DSB resection.
It has been well established that depletion of hSSB1 results in dramatic reduction in ATM activation (14,17). Our data, taken together with previous studies on MRN dependent ATM activation and DNA end resection, now provides a model for the role of hSSB1 in mediating initial steps of DNA end resection that are essential for HR. MRN is proposed to tether together broken DNA ends and promote ATM activation via mediating the recruitment of ATM to sites of DNA damage (6,19,20). ATM in turn stimulates nuclease activity of Mre11 in response to DSBs and Mre11 in cooperation with CtIP carries out early limited resection at the sites of DSBs (5,23,26). Our data suggests that hSSB1 is a crucial component of MRN mediated resection. We find that hSSB1 exists in a stable DNA damage-independent complex with MRN. In response to DNA damage, hSSB1 recruits the MRN complex to DSBs with short ssDNA overhangs, or to naturally breathing DNA ends at DSBs. hSSB1-mediated recruitment of the MRN complex to DSBs then facilitates ATM activation and DNA end resection. This together with our recent data demonstrating rapid recruitment of hSSB1 to sites of DSBs (16) suggests that free hSSB1 may bind to DSBs causing localized duplex melting and opening of the DNA end, as has been shown previously for S. solfataricus SSB (29). This in turn may provide the substrate required for hSSB1: MRN binding. Therefore, it is possible that the recruitment of hSSB1 to the initial DSB functions to protect the DSB from incorrect processing by nucleases. It may also then function to present the DSB to the hSSB1: MRN complex. In addition, we have demonstrated that hSSB1-dependent MRN recruitment stimulates/facilitates Mre11 endo-nuclease activity. This would then lead to MRN-dependent generation of ssDNA, thereby providing a substrate for RPA to bind, allowing subsequent Rad51 loading and sister chromatid strand invasion. This model is consistent with the data presented and previous findings in which we have demonstrated a loss of RPA and Rad51 foci formation and sister chromatid exchange upon hSSB1 depletion (14,16).
The development of many anti-cancer drugs is now focusing on the inhibition of DNA repair processes. Therefore, further studies into the mechanism of the hSSB1:MRN interaction, particularly as it acts at the earliest stages of the DNA damage response, could provide valuable information to aid drug development.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancer Council Queensland Project Grant (to D.J.R.); Program Grant from National Health and Medical Research Council of Australia (to K.K.K.). Funding for open access charge: Queensland Institute of Medical Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank all colleagues in the Khanna laboratory for discussion and Stephen Miles for technical assistance.
REFERENCES
- 1.Lieber MR. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 2.Saleh-Gohari N, Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saintigny Y, Delacote F, Boucher D, Averbeck D, Lopez BS. XRCC4 in G1 suppresses homologous recombination in S/G2, in G1 checkpoint-defective cells. Oncogene. 2007;26:2769–2780. doi: 10.1038/sj.onc.1210075. [DOI] [PubMed] [Google Scholar]
- 4.D’Amours D, Jackson SP. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 5.Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V. Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. Embo J. 2008;27:1953–1962. doi: 10.1038/emboj.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 7.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 8.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richard DJ, Bolderson E, Khanna KK. Multiple human single-stranded DNA binding proteins function in genome maintenance: structural, biochemical and functional analysis. Crit. Rev. Biochem. Mol. Biol. 2009;14:1–19. doi: 10.1080/10409230902849180. [DOI] [PubMed] [Google Scholar]
- 10.Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 11.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 12.Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J. Cell Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 14.Richard DJ, Bolderson E, Cubeddu L, Wadsworth RI, Savage K, Sharma GG, Nicolette ML, Tsvetanov S, McIlwraith MJ, Pandita RK, et al. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature. 2008;453:677–681. doi: 10.1038/nature06883. [DOI] [PubMed] [Google Scholar]
- 15.Wadsworth RI, White MF. Identification and properties of the crenarchaeal single-stranded DNA binding protein from Sulfolobus solfataricus. Nucleic Acids Res. 2001;29:914–920. doi: 10.1093/nar/29.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richard DJ, Kienan S, Emma B, Liza C, Sairei SO, Mihaela G, David JC, Malcolm FW, Kerry R, Kevin MP. hSSB1 rapidly binds at the sites of DNA double-strand breaks and is required for the efficient recruitment of the MRN complex. Nucleic Acids Res. 2010;2010 doi: 10.1093/nar/gkq1098. [doi:10.1093/nar/gkq1098; Epub ahead of print 3 November 2010] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skaar JR, Richard DJ, Saraf A, Toschi A, Bolderson E, Florens L, Washburn MP, Khanna KK, Pagano M. INTS3 controls the hSSB1-mediated DNA damage response. J. Cell Biol. 2009;187:25–32. doi: 10.1083/jcb.200907026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Bolderson E, Kumar R, Muniandy PA, Xue Y, Richard D, Seidman M, Pandita TK, Khanna KK, Wang W. hSSB1 and hSSB2 form similar multi-protein complexes that participate in DNA damage response. J. Biol. Chem. 2009;284:23525–23531. doi: 10.1074/jbc.C109.039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupre A, Boyer-Chatenet L, Gautier J. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat. Struct. Mol. Biol. 2006;13:451–457. doi: 10.1038/nsmb1090. [DOI] [PubMed] [Google Scholar]
- 20.Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 21.Richard DJ, Schumacher V, Royer-Pokora B, Roberts SG. Par4 is a coactivator for a splice isoform-specific transcriptional activation domain in WT1. Genes Dev. 2001;15:328–339. doi: 10.1101/gad.185901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richard DJ, Bell SD, White MF. Physical and functional interaction of the archaeal single-stranded DNA-binding protein SSB with RNA polymerase. Nucleic Acids Res. 2004;32:1065–1074. doi: 10.1093/nar/gkh259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J, Gong Z, Ghosal G, Chen J. SOSS complexes participate in the maintenance of genomic stability. Mol. Cell. 2009;35:384–393. doi: 10.1016/j.molcel.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiotani B, Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol. Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu L, Luo K, Lou Z, Chen J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc. Natl Acad. Sci. USA. 2008;105:11200–11205. doi: 10.1073/pnas.0802885105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakamoto S, Iijima K, Mochizuki D, Nakamura K, Teshigawara K, Kobayashi J, Matsuura S, Tauchi H, Komatsu K. Homologous recombination repair is regulated by domains at the N- and C-terminus of NBS1 and is dissociated with ATM functions. Oncogene. 2007;26:6002–6009. doi: 10.1038/sj.onc.1210428. [DOI] [PubMed] [Google Scholar]
- 29.Cubeddu L, White MF. DNA damage detection by an archaeal single-stranded DNA-binding protein. J. Mol. Biol. 2005;353:507–516. doi: 10.1016/j.jmb.2005.08.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.