Abstract
Although accumulating evidence has provided insight into the various functions of long-non-coding RNAs (lncRNAs), the exact functions of the majority of such transcripts are still unknown. Here, we report the first computational annotation of lncRNA functions based on public microarray expression profiles. A coding–non-coding gene co-expression (CNC) network was constructed from re-annotated Affymetrix Mouse Genome Array data. Probable functions for altogether 340 lncRNAs were predicted based on topological or other network characteristics, such as module sharing, association with network hubs and combinations of co-expression and genomic adjacency. The functions annotated to the lncRNAs mainly involve organ or tissue development (e.g. neuron, eye and muscle development), cellular transport (e.g. neuronal transport and sodium ion, acid or lipid transport) or metabolic processes (e.g. involving macromolecules, phosphocreatine and tyrosine).
INTRODUCTION
Large-scale analyses of full-length cDNA sequences have detected large numbers of long-non-coding RNAs (lncRNAs) in human (1), mouse (2) and fly (3). These lncRNAs have been shown to play key roles in imprinting control, cell differentiation, immune responses, human diseases, tumorigenesis and other biological processes (4–6). In particular, the regulatory roles of lncRNAs in the expression, activity and localization of protein coding genes have attracted much attention (5). For example, the lncRNA MEG3 activates the expression of Tp53 and enhances its binding affinity to the promoter of its target gene, Gdf15, implying a role for MEG3 in regulating the expression and transcriptional activation of Tp53 (7). Although an increasing number of lncRNAs are being characterized, the functions of most lncRNA genes are still unknown. Generally, lncRNAs are as poorly conserved as the introns of coding genes and less conserved than the 5′- or 3′-untranslated regions (UTRs) of mRNAs (8). However, lack of conservation does not necessarily mean lack of function, as demonstrated by the very poorly conserved lncRNA Xist transcript, which plays a critical role in regulation of imprinted and random X inactivation (9). The low-conservation level of lncRNAs suggests they evolve more quickly than protein-coding genes, rendering functional prediction by genomic comparison very difficult. Besides, functional prediction of lncRNAs is also hampered by the lack of collateral information such as molecular interaction data and expression profiles. It has been proposed that the functional properties of lncRNAs are mainly related to their secondary structures (10). However, our ability to decipher RNA function based on the secondary-structure information is still rudimentary, and only a few reports on the functional validation of lncRNAs have been published (10–12). Guttman et al. (12) used chromatin-state maps to identify a large number of long-intervening ncRNAs, and developed an approach for functional assignment of these based on coding–non-coding gene co-expression relationships extracted from custom-designed tiling array data. In spite of much effort, the number of lncRNAs with known functions still remains scarce, and efficient prediction of lncRNA functions is still a considerable challenge. The fact that ncRNAs have regulatory roles in a wide range of processes have led to the realization that question of ncRNA functions cannot be ignored (4), and excavating the hidden layer of lncRNA function is necessary in order to obtain a comprehensive understanding of the operational mechanisms of the mammal.
The rapid update of genomic information over the past years has drawn some attention to the accuracy of microarray probe annotation and mapping (13–15). For example, on the Affymetrix GeneChip U95A, ∼11% of the probes are non-specific and 9% of the probes are mismatched to the genome (14). Many EST sequences that previously were assumed to be mRNA fragments have turned out to be the fragments of lncRNAs, and a number of microarray probes which were designed based on EST have been verified to match lncRNAs perfectly. For example, by re-annotating the ABA probes, Mercer et al. (11) identified 849 ncRNAs that were expressed in the adult mouse brain. Similarly, through re-annotation of the probes in the GNF Gene Expression Atlas data, Pang et al. (10) found over 1000 ncRNAs that were expressed in human and mouse CD8+ T cells. These reports suggest that much latent information on ncRNAs can be obtained from other high-throughput microarrays. By examining the Affymetrix arrays, we identified similar inaccuracies in probe annotation, consequently designed a strict computational pipeline to re-annotate the probes corresponding to both coding and non-coding genes in the Affymetrix Mouse Genome 430 2.0 Array (Mouse 430 2.0 array). We created a new chip-description-file (CDF) named the ‘CNC-Mouse4302cdf’ to replace the old CDF file ‘Mouse4302cdf’, and demonstrated its accuracy and consistency by several methods.
Biological processes and cellular regulation networks are very complex, involving interactions of various molecules such as proteins, RNAs and DNAs (16). Co-expression networks, in which a node represents a molecule and an edge an expressional correlation, have previously been used to identify cellular modules and predict the functions of unknown protein coding genes (16–18). However, owing to the vast amount of ‘noise’ in microarray data, a co-expression network should be constructed using multiple microarray data sets, since genes with similar expression patterns under multiple, but resembling experimental conditions have a higher probability of sharing similar functions (19) or being involved in related biological pathways (20). Microarray-based co-expression networks have generally been constructed with proteins or protein coding genes, as probes targeting non-coding transcripts have been either lacking or not considered. Here, we re-annotated the expression profiles of both coding and non-coding genes in a widely used commercial array, and constructed a coding–non-coding gene co-expression (CNC) network which included both coding and non-coding genes. By this approach, we predicted the functions of more than 300 mouse lncRNAs from the FANTOM3 project, thereby increasing our understanding of lncRNAs as well as of biological networks. We propose that this method can be used as a novel technical platform to predict the functions of lncRNAs in other organisms.
MATERIALS AND METHODS
Probe re-annotation pipeline
The probes sequences provided by Affymetrix (http://www.affymetrix.com) were aligned to non-coding transcript sequences from the FANTOM3 project (21) and to the coding transcript sequences from the RefSeq database (22), respectively, using BLASTn. The alignment results were filtered by the following steps:
Only probes perfectly matched to a transcript were retained, resulting in two sets of probes targeting protein coding and non-coding transcripts, respectively.
Probes targeting non-coding transcripts that also perfectly matched coding cDNA sequences in the FANTOM3 project were removed.
All transcripts corresponding to retained probes were mapped to the genome and annotated at the gene level.
Genes matched by less than three probes were discarded.
Non-coding genes whose genomic regions could not be transformed from the 5 to 9 mm versions of the mouse genome were discarded.
Non-coding genes with a Codon Substitute Frequency (CSF, see below) score no less than 300 were removed.
A new CDF package (called CNC-Mouse4302cdf corresponding to the original CDF package Mouse4302cdf) covering the re-annotated probe–gene relationships was created by using the makecdfenv R package (makecdfenv: CDF Environment Maker. R package version 1160 2006 http://www.bioconductor.org/packages/2.5/bioc/html/makecdfenv.html).
The pipeline for re-annotation of the Affymetrix Mouse 430 2.0 array probes is illustrated in Supplementary Figure S2.
Calculation of the codon substitution frequency score
To filter out potentially unrecognized coding genes among the annotated non-coding loci, we used the CSF method proposed by Lin and colleagues (23). First, two codon substitution matrices (CSM) corresponding to coding and non-coding genes, respectively, were created based on an estimate of the frequencies at which all pairs of codons are substituted between genes in target species and informants [see ref. (23) for details]. Coding exon sequence alignment data for 30 species including the mouse were downloaded from the UCSC genome browser (build 9 mm, http://hgdownload.cse.ucsc.edu/goldenPath/mm9/multiz30way/) (24). The coding CSM training data was alignments of Refseq exons, excluding exons targeted by probes of the Mouse 430 2.0 array, while the non-coding CSM training data was alignments of non-coding sequences with the same length distribution as the coding training sequences. The non-coding training sequences were randomly selected from intergenic sequences that had not been annotated as repeat regions by UCSC (24). Based on the above non-coding and coding training alignment data, we created non-coding and coding CSMs [CSMN and CSMC, respectively; see ref. (23) for details]. The CSF method assigns to a codon substitution (a, b) a score  As there are multiple informant species in the alignment data, we calculated a CSF matrix for each informant species. The final CSF score of a sequence was determined by the score of each codon substitution (a, b) in the sequence.
As there are multiple informant species in the alignment data, we calculated a CSF matrix for each informant species. The final CSF score of a sequence was determined by the score of each codon substitution (a, b) in the sequence.
For each non-coding gene targeted by the Mouse 430 2.0 array, we computed CSF scores by summing up all the 30 codon substitution frequency scores across a sliding windows of 90 bp in each informant species. We then scanned all the six possible open reading frames in each window, and finally selected the maximum CSF score for the non-coding gene. Coding genes were treated likewise. Based on the CSF score distribution of coding and non-coding genes targeted by the probes of Mouse 430 2.0 Array, we removed non-coding genes with a CSF score under a threshold of 300 (Supplementary Figure S3).
Preparation of expression data
Thirty-four Mouse 430 2.0 microarray data sets were obtained from the Gene Expression Omnibus (GEO) database (25). Preprocessing of the data consisted of Robust Multichip Average (RMA) background correction, constant normalization and expression summarization as described by Irizarry et al. (26). Genes were regarded as expressed under an experimental condition only if they were detected in ≥50% of all the replicated samples according to MAS5CALLS (27). Genes were considered for further analysis only if they were expressed in at least one experimental condition. The above processing was implemented using the affy package of the R Bioconductor software (28). The signal intensity in the gene expression matrix was log2-transformed and standardized so that each gene within each column had a median value of 0 and a variance of 1.
Comparison of the coding gene expression as measured by Mouse4302cdf and CNC-Mouse4302cdf
Mouse4302cdf is the original CDF package of the Mouse 430 2.0 array data, while our new re-annotated CDF package was named CNC-Mouse4302cdf. The expression profile data of GSE1986 (17 normal tissues) and GSE9954 (22 normal tissues) were used to compare the two CDF packages. By applying the Mouse4302cdf and the CNC-Mouse4302cdf, expression signal intensities of the original and re-annotated probes were calculated as given in the section of ‘Preparation of expression data’ (the ‘expression summarization’ step excepted). Pearson correlation coefficients (Pccs) for the expression values of every two probes within the same coding probe set were calculated. Then the average and the variance of coefficient of the Pccs for each probe set were calculated to represent the probe expression consistency of the probe sets.
Comparison of the Affymetrix Mouse Genome 430 2.0 Array and the RIKEN cDNA array
The original RIKEN cDNA array, consisting of expression profiles of FANTOM3 transcripts across 20 tissues (RIKEN 60 K microarray set), were downloaded from the FANTOM project web site (http://read.gsc.riken.jp/fantom2/) (29). This data set was compared with two re-annotated data sets (GSE1986 and GSE9954). As the RIKEN cDNA data relates expression levels to transcripts while re-annotated Mouse 430 2.0 data relates expression levels to genes, only genes that have a single transcript were included in the comparison. Genes with one or more NA values and genes with expression variance in the bottom 25percentile in each data set were removed. Expression matrices of non-coding genes from the RIKEN cDNA and the re-annotated Mouse 430 2.0 data were generated. For each data set, the expression values were ranked for each tissue, and Spearman correlation coefficients for the same non-coding genes in the two data sets were calculated. As a control, non-coding genes were paired randomly and Spearman correlation coefficients were computed. The control step was repeated 1000 times.
Construction of the co-expression network
Thirty-four data sets each including nine or more experiments were used to construct the coding–non-coding gene co-expression network. For each data set, the data processing was as follows:
Genes with expressional variance ranked in the top 75 percentile of each data set were retained.
A set of Pcc P-values for each gene pair was estimated through Fisher's asymptotic test implemented in the WGCNA library of R (30), and adjusted with the Bonferroni multiple test correction implemented in the multtest package of R (multtest: Resampling based multiple hypothesis testing, 2005. R package version: 2.2.0.).
Only gene pairs with a P-value of 0.01 or less and with a Pcc value ranked in the top or bottom 0.05 percentile for each gene were regarded as co-expressed in the given data set.
Finally, each gene pair was assigned a parameter according to the number of data sets in which the gene pair was co-expressed in the same ‘direction’ (i.e. positively or negatively). Only gene pairs co-expressed in the same direction in three or more data set were included in the co-expression network.
Random network
In the CNC network, we identified the edges as either coding–coding, coding–non-coding and non-coding–non-coding. To obtain a random network with a similar distribution of edges, we randomly selected two connected gene pairs (e.g. A–B and C–D), and exchanged two nodes (e.g. B and D) if these two links satisfied the below two conditions: (i) all four nodes are different, and (ii) the new links generated after the node exchange do not exist in the network before the exchange. If the above conditions are satisfied, the links A–B and C–D are exchanged for links A–D and C–B. As the numbers of the three types of connections are different, the exchange steps were repeated 1 000 000, 100 000 and 50 000 times for coding–coding, coding–non-coding and non-coding–non-coding links, respectively.
The hub-based method
The network hub-based method is the most direct method for functional prediction. It determines the function of a protein based on the enrichment of functional annotations of genes in its immediate neighborhood. In the CNC network, only non-coding genes with 10 or more immediate coding neighbors with gene ontology (GO) biological process (BP) annotations were considered. Coding genes with GO BP annotations and 10 or more known coding neighbors were used as a test set for evaluating prediction performance. For each gene in the test set, GO enrichment analysis was performed using the g:profiler web server (31). The P-value of the functional enrichment (PV) and the number of coding neighboring genes annotated with the enriched GO BP term (GN) were used as parameters in the function prediction of non-coding genes. The precision and specificity defined below were used to evaluate the prediction performance.
Precision and specificity of the prediction performance
All enriched GO BP terms were reduced to MGI GO Slim BP terms (excluding the ‘other biological processes’ term). For each gene in the test set, we counted the number of known MGI GO–Slim–BP terms (denoted as Nki), the number of predicted MGI GO–Slim–BP terms, (denoted as Npi) and the number of MGI GO–Slim–BP terms occurring as both known and predicted terms (noted as Noi). The precision of the predictive performance can be defined as,
and the specificity as,
RESULTS
Re-annotation of the microarray probes
The Mouse 430 2.0 array is composed of probes targeting more than 39 000 transcripts, and has been widely used by biological researchers. Of the 242 known mouse ncRNAs from the RNAdb (32), we found that 78 lncRNAs have at least one perfectly matched probe (Supplementary Table S1), and that 73 lncRNAs have >3 probes (Supplementary Figure S1A). For example, the Air RNA (RNAdb ID: LIT1838), which is transcribed in the antisense orientation to the imprinted Igf2r locus, has 96 probes, and the Jpx RNA (RNAdb ID: LIT1008), which is located in the ChrX inactivation center, has 22 probes (Supplementary Figure S1B). Since genome annotation has progressed considerably, a strict computational pipeline was established to re-annotate the 496 468 probes of the Mouse 430 2.0 array (Figure 1A and Supplementary Figure S2). According to our results, there were 67 089 probes (13.5%) that were perfectly matched to the FANTOM3 non-coding RNAs but not to any Refseq mouse coding transcript, and 248 116 probes (50.0%) that were perfectly matched to Refseq coding transcripts, but not to any non-coding RNA. The remaining were composed of 39 775 probes (8.0%) which perfectly matched both Refseq coding transcripts and FANTOM3 lncRNAs, and 141 488 probes (28.5%) that did not match any transcripts, and these were all discarded. In order to avoid ambiguities, we also removed the 8655 probes that perfectly matched FANTOM3 coding transcripts, and mapped the remaining probes to their corresponding genomic loci. The Entrez GeneID was used to represent a coding gene, while the FANTOM transcriptional framework (TK) ID (21) was used to represent a non-coding gene. To further reduce the noise, probes that matched to more than one gene were removed, and to increase the accuracy, genes that were matched by less than three probes were discarded, leaving 14 861 coding genes and 5169 non-coding genes. To obtain an even more reliable set of non-coding genes, we removed non-coding genes with a Codon Substitution Frequency (CSF, ‘Materials and Methods’ section) score <300 (Supplementary Figure S3), as well as those lncRNA loci whose genomic region could not be transformed from the mm5 to mm9 version of the mouse genome sequence. Finally, 14 861 coding genes and 4571 lncRNA genes were retained and assembled into a new chip-description-file (CNC-Mouse4302cdf). On average, coding and non-coding genes were targeted by 14.9 and 11.2 probes, respectively (Supplementary Figure S4). Of the 14 861 coding genes, 12 250 genes (82.4%) were annotated with at least one GO term and 9846 genes (66.3%) had at least one GO BP term.
Figure 1.
Re-annotation of Affymetrix Mouse Genome 430 2.0 Array probes. (A) Computational pipeline for re-annotating the probes of the Mouse 430 2.0 array. (B) The relative distribution of the 496 468 original probes of the Affymetrix Mouse Genome 430 2.0 Array.
Probe re-annotation according to the most recent genome annotation should enhance the quality of the microarray data, and to test this we compared the performance of CNC-Mouse430cdf and Mouse430cdf. As expected, after removing the ambiguous probes and accurately mapping the remaining probes, the mean Pcc between every two probes targeting the same coding gene was significantly increased (P < 2.20e-16 by the Kolmogorov–Smirnov test; Figure 2A and Supplementary Figure S5A), while the coefficient variance of the Pccs was reduced (P < 2.2e-16, Kolmogorov–Smirnov test; Figure 2B and Supplementary Figure S5B).
Figure 2.
Specificity and accuracy of the r-Mouse4302cdf file. (A) Mean expressional correlation of probes (GSE9954) targeting the same coding gene before and after re-annotation (P < 2.20e-16 by Kolmogorov–Smirnov Test). (B) Coefficient of variance on the expressional correlation of probes (GSE9954) targeting the same coding gene between before and after re-annotation (P < 2.2e-16, Kolmogorov–Smirnov Test). (C) Expressional correlations of lncRNAs. Dark grey line: distribution of Spearman correlation coefficients for the expression of identical lncRNAs in corresponding tissues in the Riken cDNA array data set and in the re-annotated Mouse 430 2.0 (GSE9954) array data. Light grey lines: distribution of Spearman correlation coefficients for the expression of randomly selected lncRNA pairs, repeated 1000 times. (Mean Spearman correlation coefficient was 0.26, mean P-value of the KS test was 4.39e-08). (D) Expression profiles of lncRNAs TK27265 and TK100617 in the re-annotated Mouse 430 2.0 array data and in the Riken cDNA array data.
We next compared the lncRNA expression levels in the re-annotated Mouse 430 2.0 array data to the original expression profiles of the RIKEN cDNA array (RIKEN 60 K microarray set), which contains 11 084 FANTOM3 non-coding transcripts from 20 tissues (29). The comparison showed that the average correlation coefficient for the same lncRNAs from the two independent studies was significantly higher than for randomly selected lncRNA pairs. For example, in the comparison between expression profiles of the Riken cDNA array and the GSE9954 data, the mean Spearman correlation coefficient and the mean P-value of the KS test were 0.26 and 4.39 × 10−8, respectively (Figure 2C). Similar results were also found for the GSE1986 data (Supplementary Figure S6). We also observed tissue-specific-expression patterns for several lncRNAs in both the re-annotated Mouse 430 2.0 array data and the original RIKEN cDNA expression data. For example, 10 tissue specific lncRNAs were detected by both the RIKEN cDNA array and the GSE9954 data (Supplementary Table S2). Among them, TK27265 and TK100617 were only expressed in testis and brain, respectively, and similar expression patterns for these lncRNAs were also seen in the GSE1986 data (Figure 2D).
Construction of the coding–non-coding gene co-expression network
As of September 2010 there were 1398 data sets in the GEO database, including a total of 18 082 expression profiles arising from the Affymetrix Mouse Genome 430 2.0 Array. Instead of constructing a network based on single data set, we considered a combination of many data sets involving different conditions as a more robust approach (19). This also ensures that the number of samples in each data set is large enough to obtain the required co-expression patterns, and we therefore selected as many relevant microarray data sets as possible. As a result, 34 data sets, each comprising nine or more different experimental conditions or cellular states, were used to construct a ‘two-color’ co-expression network including both coding and non-coding genes. The experimental conditions included a number of biochemical and biophysical conditions, various tissue resources, and diverse biological processes (Supplementary Table S3). For each expression profile, genes with high-expressional variance (top 75 percentile) were selected for identification of co-expressed gene pairs. The P-value of each Pcc was estimated by Fisher's asymptotic distribution, and the set of P-values for each gene were adjusted by the Bonferroni method. We defined a gene pair as co-expressed in a given expression profile only when the adjusted P-value was <0.01 and the Pcc ranked in the top or bottom 0.05% of the Pccs for each gene.
As an additional requirement, we required that an edge between two genes could be included in the CNC network only if the two genes were co-expressed in the same direction (i.e. either positive or negative) in more than a given number of data sets. To determine this minimum number of data sets, we evaluated the networks with different cutoffs of data set number by several network parameters (Supplementary Table S4). The size of the network naturally decreased with a higher cutoff value. Furthermore, GO term overlap analysis showed that the higher the cutoff, the more similar the annotated functions of the connected gene pairs were in the network (Figure 3A). Based on the size and quality of the networks, we selected the network that was constructed with a cutoff of three for further analysis. In this CNC network, there were 1720 non-coding genes and 10 420 coding genes that were linked by 59 591 edges. Nearly 50 000 edges (49 912; 83.75%) connected coding genes, and 4840 edges (8.18%) connected coding and non-coding genes, whereas another 4839 edges (8.17%) linked pairs of non-coding genes (Figure 3B). Further information about the topological structure of CNC network is found in the Supplementary Data.
Figure 3.
The coding–non-coding gene co-expression network. (A) The relationship between the number of data sets in which gene pairs were co-expressed and the similarity of the annotated functions of the connected gene pairs. The figure shows the probability that gene pairs co-expressed in a number of data sets have X or less common GO BP terms. (B) Visualization of the CNC network. Green nodes represent non-coding genes while pink nodes represent coding genes. Several of the largest modules are shown. (C) GO enrichment analysis result of 2858 coding genes co-expressed with at least one lncRNA gene. (D) GO enrichment analysis of 243 coding genes enriched for co-expressed lncRNA genes. (E) GO enrichment analysis result of the 1249 coding genes associated with 189 lncRNAs enriched for co-expressed coding genes.
Of the 10 420 coding genes in the network, 8789 (84.3%) were annotated with at least one GO term, most commonly (7077 genes, or 67.9%) with a GO BP term. The 2585 coding genes that were co-expressed with at least one non-coding gene were enriched in GO annotations concerning cellular component organization, neurotransmitter transport, neurotransmitter secretion and synaptic transmission (Figure 3C). We subsequently identified genes with three or more neighbors (including 7118 coding genes and 1028 non-coding genes) that were preferentially co-expressed with coding genes or non-coding genes (hypergeometric test with a cutoff of 0.05). Of these, 243 coding genes were significantly enriched in non-coding gene partners. With respect to functional annotation, these coding genes were enriched in GO BP terms associated with nervous system processes such as synaptic transmission (P = 1.55 × 10−14), regulation of neurotransmitter levels (P = 3.50 × 10−9) and nervous system development (P = 8.31 × 10−9) (Figure 3D). This finding is consistent with previous research which suggested that lncRNAs are particularly active and play regulatory roles in brain (11). On the other hand, 1054 non-coding genes had at least one co-expressed protein coding partner. Among these, there were 189 non-coding genes with three or more neighbors that were significantly enriched in coding gene partners. These non-coding genes had a total of 1249 coding neighbors, which were enriched in the GO BP terms associated with muscle contraction (P = 2.29 × 10−7) and visual perception (P = 1.09 × 10−6) (Figure 3E). In addition, 676 non-coding genes were significantly enriched in non-coding gene partners.
Prediction of lncRNA function based on co-expression and genomic co-location
The transcriptional patterns of mammalian non-coding genes are very complex (21), with non-coding gene loci located within the intronic regions of coding genes, overlapping coding exons either in sense or antisense orientation, or positioned between two coding genes (6). It has been shown that the transcription of non-coding genes can affect the expression of their flanking coding genes (6). For example, an lncRNA is co-expressed with its bilateral coding genes, Fank1 and Adam12, and its down-regulation reduces the expressions of both coding genes by establishing active chromatin structures (33). In the re-annotated Mouse 430 2.0 array, there are 3618 and 4105 lncRNAs (out of a total of 4571) that are located within 10 and 100 kb, respectively, of any of the 14 861 protein-coding genes, resulting in, respectively, 6155 (<10 kb) and 13 407 (<100 kb) co-located coding–non-coding gene pairs. Among these, only 141 (∼2.3%, 138 lncRNAs) and 148 (∼1.1%, 143 lncRNAs) pairs, respectively, were observed in our co-expression network. This indicates that most lncRNAs are not co-expressed with their nearby coding genes and thus most likely independently transcribed. Besides, if an lncRNA is co-expressed with a nearby coding gene, the two genes are frequently separated by a distance of <10 kb. Here, we defined two genes as a co-expressed and co-located pair if they were co-expressed and spaced by <100 kb. For further analysis, we classified these pairs as ‘internal’, ‘upstream’ or ‘downstream’ according to the position of the lncRNA locus relative to the coding locus. In the CNC network, there were 84 downstream, 55 internal and 9 upstream coding–non-coding gene pairs (involving 143 lncRNAs, Supplementary Table S5), compared to only one co-expressed and co-located coding–non-coding gene pair in the random network. Interestingly, we found that more lncRNAs were co-expressed with their upstream coding genes than with downstream or host genes, which is consistent with the previous finding that the transcription of lncRNA loci is frequently initiated from the 3′-UTR of coding genes (21).
Internal lncRNAs are mainly derived from the introns of coding genes, while some may also fall within the 5′- or 3′-UTR regions. Internal non-coding genes are involved in multiple kinds of biological processes such as regulation of expression at the transcriptional and post-transcriptional level, alternative splicing, subcellular localization and regulation of the host protein activity (34). In the CNC network, the non-coding gene TK226771 was co-expressed with its host gene Plagl1 (Figure 4A), a transcription factor and tumor suppressor gene (35). Plagl1 is located in a candidate imprinting center, a region showing hypermethylation in patients with ovarian cancer and loss of methylation in patients with transient neonatal ‘diabetes mellitus’ (35).
Figure 4.
Genomic contexts of four non-coding genes. (A) Intronic lncRNA TK226771. (B) Upstream lncRNA TK118632. (C) Downstream lncRNA TK79018.
Upstream lncRNAs may overlap the promoter regions of their co-expressed coding genes, and may regulate their expression at the transcriptional or post-transcriptional level (6, 36). For instance, TK118632 was co-expressed with the downstream (∼400 bp) coding gene Lrrc4c, which plays an important role in the regulation of axon guidance and excitatory synaptic formation (37–39) (Figure 4B). Interestingly, TK118632 was also detected in the cerebellum tissue, implicating it also may function in brain (21).
Downstream lncRNAs initiate transcription from the 3′ UTRs or downstream regions of protein-coding genes, and may be involved in the intergenic regulatory interactions (21). For example, TK79018 is located about 900 bp downstream of its co-expressed gene Mef2c, which is a transcription factor playing a key role in cardiac development (Figure 4C).
Several co-expressed and co-located gene pairs have been corroborated by independent research. For example, Ponjavic et al. (40) experimentally characterized six co-expressed and co-located coding–non-coding gene pairs in the embryonic or neonatal mouse brain, and four of these pairs were present in the re-annotated Mouse 430 2.0 array, all showing high Pcc values. The expression of Meis1 and its intronic non-coding gene, TK116311, were highly and significantly correlated in five GSE data sets. Rbms1 and its downstream non-coding gene TK98616 were co-expressed with a Pcc of about 0.8 in four GSE data sets. The expression of the remaining two pairs (TK109313 and Vangl2, TK151497 and Eif2c3) were also highly correlated (Pcc > 0.7) in at least three GSE data sets. The non-coding partner of Slitrk1, also studied by Ponjavic et al. (40), was not targeted by the re-annotated probes, however, Slitrk1 was linked to several other non-coding genes (TK125716, TK76136, TK84100, TK116414, TK168361, TK99201, TK77830 and TK105892) in the CNC network. Taken together, the above observations strongly suggest that the CNC network reflected real relationships between the mouse coding and non-coding genes.
Hub-based prediction of lncRNA functions
The hub-based method assigns functions to un-annotated genes according to the functional enrichment of its neighboring genes. Being the first to apply this method to predict the functions of non-coding genes in a CNC network, we evaluated the accuracy of this method by cross validating it on coding genes with known GO BP terms. In the CNC network, there were 7077 coding genes annotated with at least one GO BP term, among which 1319 had 10 or more coding neighbors with known functions. For each of the 1319 coding genes, we calculated the functional enrichment of their neighbors using the g:Profiler web server with default parameters (31). Of the 1000 genes whose neighbors showed functional enrichment, 595 (59.5%) were annotated with at least one of the enriched GO BP terms. In the random network, there were 1311 annotated coding genes with 10 or more known coding neighbors, however, the neighbors of most of these (1108) showed no functional enrichment, and of the 203 genes whose neighbor showed functional enrichment, only four genes (1.97%) had at least one GO BP term that corresponded to those enriched in the neighbors.
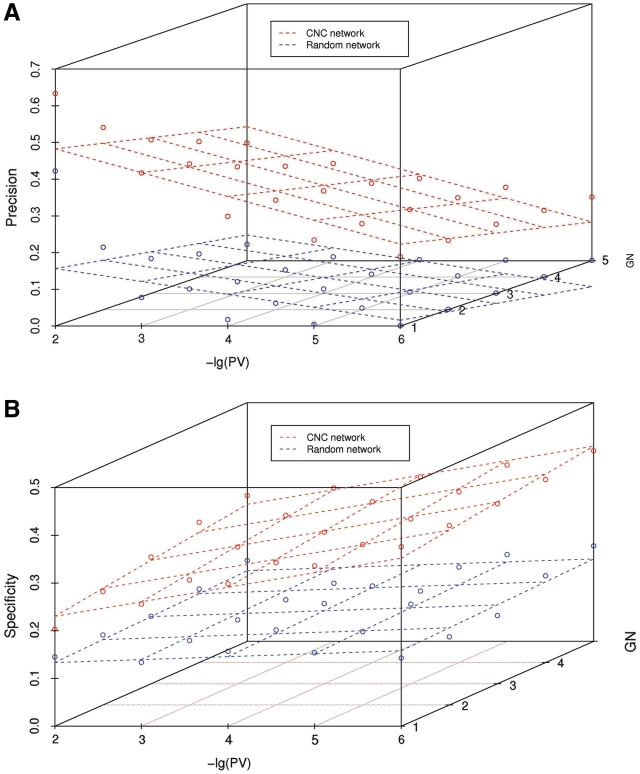
To improve its predictive performance, we adjusted the parameters of the hub-based prediction method. We first defined ‘precision’ and ‘specificity’ standards of the prediction (‘Materials and Methods’ section). Both the P-value of the GO BP term enrichment (PV), and the number of neighboring genes enriched with the GO BP term (GN) influence the precision and specificity values (Figure 5). Requiring a low PV (e.g. 10−6) results in a low precision, irrespective of GN, thus, to obtain a reasonable precision, the PV should not be set too low (Figure 5A). The specificity, on the other hand, is strongly affected by changes in GN, but less so by changes in PV (Figure 5B). We found that a PV ≤ 0.01 and a GN ≥ 5 gave a reasonable trade-off between precision (32.1%) and specificity (30.5%, Supplementary Table S6). In comparison, the same cutoffs applied to the random network produced far lower precision (4.45%) and specificity (16.9%) values.
Figure 5.
Hub-based functional prediction. (A) The relationship among PV, GN and precision in the CNC network. (B) The relationship among the PV, GN and specificity in the CNC network. Random networks are shown for comparison.
Using the above PV and GN cutoffs, we randomly selected 10% of the 1319 coding genes to serve as ‘unknown’, and predicted their functions. After repeating this procedure 100 times, on average 79.3% of the ‘unknown’ genes were ‘assigned’ with at least one GO BP term. Among these, 72.2% were ‘assigned’ with at least one ‘correct’ GO BP term, corresponding to precision and specificity values of 32.3% (variance = 2.52 × 10−3) and 33.4% (variance = 2.23 × 10−3), respectively. In comparison, the precision and specificity values were 4.49 and 19.2%, respectively, in the random network (Supplementary Figure S7).
Applying the same method and cutoffs to the 84 non-coding genes with 10 or more annotated coding neighbors, 70 non-coding genes were annotated with at least one significantly enriched GO BP term (Supplementary Table S7 for details). On average, each non-coding gene was assigned with 13 GO BP terms (including multiple level terms). After reducing these GO BP terms to the MGI GO Slim terms, we found that the predicted lncRNA functions were mainly associated with development processes (32.5%), transport (18.3%), cell–cell signaling (16.7%), metabolism (14.2%) and cell organization and biogenesis (11.5%; Supplementary Figure S8A). Interestingly, the known lncRNA TK170500 (Dlx1as) was assigned functions such as brain development, central nervous system development, neuron differentiation, neurogenesis and other neuron related GO BP terms. This finding is consistent with the report that Dlx1as is expressed in forebrain and in regions associated with neurogenesis in the mouse embryo (41).
Prediction of lncRNA functions by network modules
Genes within a co-expressed module commonly have similar functions, thus, mining modules in a network is an efficient approach for predicting gene functions (42,43). The Markov cluster algorithm (MCL) is an efficient and powerful algorithm, which identifies modules based on the simulation of random walks in a network. With default parameters (inflation value = 1.8), the MCL algorithm found 1695 modules with three or more genes, of which 550 modules were composed of both coding and non-coding genes. Sixty-two of these modules were significantly enriched for at least one GO BP term (P < 10−18, Fisher's one-tailed test; Figure 6 and Supplementary Table S8). We named each module after the most significantly enriched function, and annotated the 218 long-non-coding genes contained in these modules accordingly (Supplementary Table S9). Among the 218 lncRNA genes, there were 54 lncRNA genes whose functions had also been predicted by the hub-based method (above). Moreover, all of the 54 lncRNAs had at least one common GO BP term predicted by both methods, and on average each lncRNA had 10 GO BP terms predicted by both methods. The main functional categories of the lncRNAs were similar to the predictions by the hub-based method (Supplementary Figure S8B). The two modules with the highest number of non-coding gene were the ‘synaptic transmission’ module (47 non-coding genes) and the ‘male gamete generation’ module (20 non-coding genes). This finding is consistent with previous studies, suggesting that non-coding genes be particularly active in the brain or in embryo development (11,40,41). The predicted functions of a number of lncRNAs were consistent with previous reports. For example, TK78533 (AK044422) belongs to a module which was significantly enriched with functions related to neurotransmitter secretion and transport as well as GABA signaling, consistent with the reported involvement of TK78533 in the regulation of neuronal specification and differentiation (44). The same report (44) also suggested a role for the lncRNA TK170605 (AK079380) in oligodendrocyte lineage commitment. In the CNC network, TK170605 was co-expressed with Map6d1, a member of the STOP family that is responsible for the stabilization of neuronal microtubules (45).
Figure 6.
The largest modules of the co-expression network. The color depth signifies the number of data sets in which the gene pairs were co-expressed.
The ‘synaptic transmission’ module
The ‘synaptic transmission’ module comprised 47 non-coding genes and 148 coding genes, of which 106 coding genes had GO BP annotations. This module was enriched in neuronal signal transmission functions, such as synaptic transmission (P = 1.14 × 10−18), transmission of nerve impulse (P = 1.79 × 10−17) and cell–cell signaling (P = 1.21 × 10−15) (Figure 7A), and most genes in the module are expressed in brain or sensory organ tissues (Figure 7B and Supplementary Figure S9), which is consistent with the FANTOM3 project observations that most non-coding transcripts are detected in brain tissues (21). For example, the three different transcripts giving rise to TK99165 in the Mouse 430 2.0 array were detected in separate regions of the brain (21). TK99165 had the largest number of co-expressed partners (44 coding genes and 13 non-coding genes) in the module, and 35 of its coding partners are functionally related to the nervous system or are active in the mammalian brain (e.g. Neuro1, Gabrg2, Snap25, Slc6a1, Cadm2; Supplementary Table S11). Besides, TK99165 is transcribed from the 3′-UTR of Cadm2 (Figure 7C), a member of the Necl protein family which is important in the central and peripheral nervous system (46). Thus, the network topology, expression patterns and genomic locations all suggested neuronal functions for the non-coding loci in the ‘synaptic transmission’ module.
Figure 7.
The ‘synaptic transmission’ module. (A) GO enrichment analysis of coding genes within the module. (B) Expression patterns of coding and non-coding genes within the module (GSE9954 was used). (C) Network visualization of the module. Green circles indicate lncRNAs, yellow circles represent coding genes involved in the neuron active ligand–receptor interaction pathway, while blue circles represent other coding genes. Co-expressed and co-located gene pairs are marked by red rectangles.
Other modules
Confident function predictions could also be made for non-coding genes in other modules. For example, in the module ‘muscle contraction’, the lncRNA TK124882 was linked to its genomic neighboring genes Myh1, Myh2 and Myh4 (Supplementary Figure S10). TK124882 overlaps the last exon and the 3′-UTR of Myh1 gene. According to the FANTOM3 project annotation, TK124882 is a cis-antisense transcript to Myh1 and a trans-antisense transcript to Myh2 and Myh4. The Myh family consists of at least 10 different isoforms expressed in the striated and smooth muscle cells and in certain non-muscle cells. It has been reported that their expression levels are spatially and temporally regulated during mammalian development (47). The results from the hub-based method further support the annotation of TK124882 as involved in muscle contraction and muscle development. (More examples are available in the Supplementary Data.)
DISCUSSION
In this study, a high-quality CNC network was constructed by re-annotating both the coding and non-coding probes of the Affymetrix Mouse Genome 430 2.0 Array, and 340 lncRNAs were functionally annotated based on the network characteristics and genomic locations. We propose that functional annotation based on re-annotated expression profiles could in the future be applicable to thousands of lncRNAs.
Several re-annotations of the Affymetrix Array probes have been reported, but these have mainly been directed at coding genes (13–15). Recently, the expression of numerous lncRNAs in the brain and immune system was analyzed through re-annotation of both coding and non-coding probes of certain customized microarrays (10,11). The fact that pre-existing microarrays have probes perfectly matching known lncRNAs suggests that the re-annotation of other microarrays for coding–non-coding analysis is feasible. The probe re-annotation carried out in this work shows that, in principle, all expression profiles of the Mouse430 2.0 array can be re-used to mine lncRNAs data. The computational pipeline designed by employing the comprehensive and accurate FANTOM3 and Refseq databases may serve as a model for future work. Particularly, as there may be lncRNAs with coding potential present in the FANTOM3 project, we used the CSF score to filter out these. The quality of probe re-annotation was demonstrated by the higher accuracy and specificity obtained in subsequent tests.
Much emphasis was also put on the quality of the network and the accuracy of the function prediction. We required a high number of experimental conditions (≥9) in each data set included in the analysis, and the selection of co-expressed pairs in each data set was based on a stringent statistical method. In addition, the gene pairs in the CNC network must be co-expressed in the same direction (i.e. either positively or negatively) in at least three data sets. Both the P-value on the GO term enrichment and the number of neighboring genes annotated with the enriched GO term were taken into consideration, yielding relatively high precision and specificity values.
In order to obtain an indication of the functional characteristics of as many lncRNAs as possible, we predicted functions using three different methods. This not only had the advantage of increasing the number of lncRNAs for which we obtained a function prediction, but also extended the range of potential functions that could be reliably ascribed to a given lncRNA. In a number of cases, the functional predictions obtained with the three methods were coherent and complementary, further strengthening the validity of the predictions. For example, the lncRNA TK111271 is co-expressed and co-located with Lck, a key signal gene in T-cell development, and was predicted by both the hub-based and the module-based methods to be functionally related to the immune system. The intronic lncRNAs TK99129 and TK105282 were both co-expressed with their host gene Bai3, a brain specific inhibitor of angiogenesis. In accordance with previous research (44), TK99129 was predicted by the module-based method to have functions related to neuron development and differentiation, whereas TK105282 was ascribed with the functions related to synaptic transmission and neurological system processes by the hub-based method.
Of the 340 lncRNAs with predicted functions in this study, 286 were located within 10 kb of any protein-coding gene targeted by the Mouse 430 2.0 array, and 143 lncRNAs were observed to be co-expressed with known coding genes within 100 kb in the genome region and thus were functionally predicted. The FANTOM3 data set is well annotated (21,48,49) and the functionality of 34 030 ncRNAs listed in FANTOM3 is also supported by computational evidence, for example, they are more conserved, more likely to be expressed, and have lower free-energy scores than random sequences (50). However, whether or not these lncRNAs are independent transcriptional units is still a controversial issue. For example, van Bakel et al. (51) recently proposed that many of these transcripts might be experimental artifacts or the result of background transcription. Especially the intervening non-coding transcripts that are located nearby the protein-coding genes may be fragments of mRNAs or associated with alternative cleavage or polyadenylation site usage or unannotated UTR extensions of neighboring protein-coding genes (51). However, a number of studies have suggested that independently transcribed lncRNAs may still be co-expressed and functionally related to neighboring coding genes (11,21,40,41). Besides, Jia et al. (52) has recently shown that most lncRNAs located within 10 kb of protein-coding genes are independent transcriptional units. Based on the above facts, it is reasonable to assume most non-coding transcripts are transcribed independently of the nearby coding genes, and we have therefore consider all non-coding transcripts after CSF score filtering as independent transcriptional units.
Taken together, our study is the first large-scale bioinformatics prediction of lncRNA functions, and the results are an important resource for further biological research. The study demonstrates that re-annotation of expression profiles from multiple experimental environments is a powerful method for functional analysis of lncRNAs that should warrant wider usage, and similar re-annotation pipelines as that used in this study can probably be applied to other microarray platforms to further mine lncRNA functions in other organisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National High Technology Research and Development of China (No. 2008AA02Z306); Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-EW-R-01); 2010 Innovation Program of Beijing Institutes of Life Sience, the Chinese Academy of Sciences; National Program on Key Basic Research Project (No. 2010CB530004); National Natural Science Foundation of China (No. 31071137, No. 30771888 and No.30972574). Funding for open access charge: National High Technology Research and Development of China (No.2008AA02Z306).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to express thanks to the anonymous reviewers’ comments. Supplementary material, including raw data and R script, can be found at http://ebiomed.org/pub/ncfan/.
REFERENCES
- 1.Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 2.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 3.Tupy JL, Bailey AM, Dailey G, Evans-Holm M, Siebel CW, Misra S, Celniker SE, Rubin GM. Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2005;102:5495–5500. doi: 10.1073/pnas.0501422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J. Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 5.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 8.Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Nesterova TB, Barton SC, Surani MA, Brockdorff N. Loss of Xist imprinting in diploid parthenogenetic preimplantation embryos. Dev. Biol. 2001;235:343–350. doi: 10.1006/dbio.2001.0295. [DOI] [PubMed] [Google Scholar]
- 10.Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen W, Mattick JS. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J. Immunol. 2009;182:7738–7748. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- 11.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl Acad. Sci. USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu J, Lee JC, Salit ML, Cam MC. Transcript-based redefinition of grouped oligonucleotide probe sets using AceView: high-resolution annotation for microarrays. BMC Bioinformatics. 2007;8:108. doi: 10.1186/1471-2105-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Finney RP, Clifford RJ, Derr LK, Buetow KH. Detecting false expression signals in high-density oligonucleotide arrays by an in silico approach. Genomics. 2005;85:297–308. doi: 10.1016/j.ygeno.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Harbig J, Sprinkle R, Enkemann SA. A sequence-based identification of the genes detected by probesets on the Affymetrix U133 plus 2.0 array. Nucleic Acids Res. 2005;33:e31. doi: 10.1093/nar/gni027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo F, Yang Y, Zhong J, Gao H, Khan L, Thompson DK, Zhou J. Constructing gene co-expression networks and predicting functions of unknown genes by random matrix theory. BMC Bioinformatics. 2007;8:299. doi: 10.1186/1471-2105-8-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharan R, Ulitsky I, Shamir R. Network-based prediction of protein function. Mol. Syst. Biol. 2007;3:88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wren JD. A global meta-analysis of microarray expression data to predict unknown gene functions and estimate the literature-data divide. Bioinformatics. 2009;25:1694–1701. doi: 10.1093/bioinformatics/btp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 22.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin MF, Carlson JW, Crosby MA, Matthews BB, Yu C, Park S, Wan KH, Schroeder AJ, Gramates LS, St Pierre SE, et al. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res. 2007;17:1823–1836. doi: 10.1101/gr.6679507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 27.Liu WM, Mei R, Di X, Ryder TB, Hubbell E, Dee S, Webster TA, Harrington CA, Ho MH, Baid J, et al. Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics. 2002;18:1593–1599. doi: 10.1093/bioinformatics/18.12.1593. [DOI] [PubMed] [Google Scholar]
- 28.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 29.Bono H, Yagi K, Kasukawa T, Nikaido I, Tominaga N, Miki R, Mizuno Y, Tomaru Y, Goto H, Nitanda H, et al. Systematic expression profiling of the mouse transcriptome using RIKEN cDNA microarrays. Genome Res. 2003;13:1318–1323. doi: 10.1101/gr.1075103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pang KC, Stephen S, Engstrom PG, Tajul-Arifin K, Chen W, Wahlestedt C, Lenhard B, Hayashizaki Y, Mattick JS. RNAdb–a comprehensive mammalian noncoding RNA database. Nucleic Acids Res. 2005;33:D125–D130. doi: 10.1093/nar/gki089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mondal T, Rasmussen M, Pandey GK, Isaksson A, Kanduri C. Characterization of the RNA content of chromatin. Genome Res. 2010;20:899–907. doi: 10.1101/gr.103473.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Louro R, Smirnova AS, Verjovski-Almeida S. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics. 2009;93:291–298. doi: 10.1016/j.ygeno.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 35.Arima T, Wake N. Establishment of the primary imprint of the HYMAI/PLAGL1 imprint control region during oogenesis. Cytogenet Genome Res. 2006;113:247–252. doi: 10.1159/000090839. [DOI] [PubMed] [Google Scholar]
- 36.Yazgan O, Krebs JE. Noncoding but nonexpendable: transcriptional regulation by large noncoding RNA in eukaryotes. Biochem. Cell Biol. 2007;85:484–496. doi: 10.1139/O07-061. [DOI] [PubMed] [Google Scholar]
- 37.Woo J, Kwon SK, Choi S, Kim S, Lee JR, Dunah AW, Sheng M, Kim E. Trans-synaptic adhesion between NGL-3 and LAR regulates the formation of excitatory synapses. Nat. Neurosci. 2009;12:428–437. doi: 10.1038/nn.2279. [DOI] [PubMed] [Google Scholar]
- 38.Lin JC, Ho WH, Gurney A, Rosenthal A. The netrin-G1 ligand NGL-1 promotes the outgrowth of thalamocortical axons. Nat. Neurosci. 2003;6:1270–1276. doi: 10.1038/nn1148. [DOI] [PubMed] [Google Scholar]
- 39.Kim S, Burette A, Chung HS, Kwon SK, Woo J, Lee HW, Kim K, Kim H, Weinberg RJ, Kim E. NGL family PSD-95-interacting adhesion molecules regulate excitatory synapse formation. Nat. Neurosci. 2006;9:1294–1301. doi: 10.1038/nn1763. [DOI] [PubMed] [Google Scholar]
- 40.Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pu S, Vlasblom J, Emili A, Greenblatt J, Wodak SJ. Identifying functional modules in the physical interactome of Saccharomyces cerevisiae. Proteomics. 2007;7:944–960. doi: 10.1002/pmic.200600636. [DOI] [PubMed] [Google Scholar]
- 43.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gory-Faure S, Windscheid V, Bosc C, Peris L, Proietto D, Franck R, Denarier E, Job D, Andrieux A. STOP-like protein 21 is a novel member of the STOP family, revealing a Golgi localization of STOP proteins. J. Biol. Chem. 2006;281:28387–28396. doi: 10.1074/jbc.M603380200. [DOI] [PubMed] [Google Scholar]
- 46.Pellissier F, Gerber A, Bauer C, Ballivet M, Ossipow V. The adhesion molecule Necl-3/SynCAM-2 localizes to myelinated axons, binds to oligodendrocytes and promotes cell adhesion. BMC Neurosci. 2007;8:90. doi: 10.1186/1471-2202-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun YM, Da Costa N, Chang KC. Cluster characterisation and temporal expression of porcine sarcomeric myosin heavy chain genes. J. Muscle Res. Cell Motil. 2003;24:561–570. doi: 10.1023/b:jure.0000009895.03111.b3. [DOI] [PubMed] [Google Scholar]
- 48.Furuno M, Pang KC, Ninomiya N, Fukuda S, Frith MC, Bult C, Kai C, Kawai J, Carninci P, Hayashizaki Y, et al. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006;2:e37. doi: 10.1371/journal.pgen.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maeda N, Kasukawa T, Oyama R, Gough J, Frith M, Engstrom PG, Lenhard B, Aturaliya RN, Batalov S, Beisel KW, et al. Transcript annotation in FANTOM3: mouse gene catalog based on physical cDNAs. PLoS Genet. 2006;2:e62. doi: 10.1371/journal.pgen.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lebenthal I, Unger R. Computational evidence for functionality of noncoding mouse transcripts. Genomics. 2010;96:10–16. doi: 10.1016/j.ygeno.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 51.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA. 16:1478–1487. doi: 10.1261/rna.1951310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.