Abstract
The stem cell protein Lin28 functions to inhibit the biogenesis of a group of miRNAs but also stimulates the expression of a subset of mRNAs at the post-transcriptional level, the underlying mechanism of which is not yet understood. Here we report the characterization of the molecular interplay between Lin28 and RNA helicase A (RHA) known to play an important role in remodeling ribonucleoprotein particles during translation. We show that reducing Lin28 expression results in decreased RHA association with polysomes while increasing Lin28 expression leads to elevated RHA association. Further, the carboxyl terminus of Lin28 is necessary for interaction with both the amino and carboxyl termini of RHA. Importantly, a carboxyl terminal deletion mutant of Lin28 that retains RNA-binding activity fails to interact with RHA and exhibits dominant-negative effects on Lin28-dependent stimulation of translation. Taken together, these results lead us to suggest that Lin28 may stimulate translation by actively recruiting RHA to polysomes.
INTRODUCTION
Lin28 is an evolutionarily conserved RNA-binding protein that plays important roles in the timing of development, pluripotency and oncogenesis (1). As a multi-functional protein Lin28 acts as a post-transcriptional regulator of the biogenesis of a group of miRNAs. These include the let-7 family miRNAs shown to participate in the regulation of expression of genes involved in cell growth and differentiation (2). Lin28 binds to the loop regions of miRNA precursors, leading to inhibition of their processing into mature miRNAs (3–5), and/or induction of uridylation of the precursors that are subsequently degraded (6–8).
However, Lin28 also exerts biological effects that are independent of let-7 miRNAs. Indeed, Lin28 was able to alter cell fates during neurogliogenesis by mechanisms distinct from those that are mediated by let-7 and to cause significant changes in gene expression before any effect on let-7 could be detected (9). Importantly, a mutant Lin28 that allowed let-7 production could still completely inhibit gliogenesis (9). Moreover, Zhu et al. (10) have recently demonstrated that transgenic mice that overexpress Lin28 exhibit overgrowth and delayed onset of puberty. However, no decrease in the level of let-7 was observed despite Lin28 overexpression in the hypothalamic-pituitary-gonadal axis that plays a critical role in controlling development and reproduction. Therefore, the biological changes occurred in these tissues most likely could not be attributed to let-7 effects.
The first evidence that Lin28 may act as a translational modulator came from two studies. Balzer and Moss (11) reported that in undifferentiated embryonal carcinoma cells Lin28 was present in polysomes and associated with actively translating mRNAs. Further, Polesskaya et al. (12) demonstrated that during muscle cell differentiation Lin28 drives IGF-2 mRNA to polysomes, thereby stimulating its translation. A role for Lin28 in translation was further suggested by our studies showing that it selectively binds to mRNAs of the key pluripotency factor Oct4 and a subset of cell cycle-related factors (including histone H2a, cyclins A and B, and cdk4) and promotes their expression (13–15). Lin28-binding sites have been mapped to the 5′-UTR of IGF-2 (12), ORFs of Oct4 and histone H2a (13,15), and 3′-UTRs of cyclins A, B, and cdk4 mRNAs (14). In human embryonic stem (ES) cells Lin28 associates with Oct4 mRNA in polysomes and inhibition of Lin28 leads to decreased Oct4 expression at the protein level (15). Importantly, a sequence element (369 nt in length) derived from the coding region of Oct4 mRNA and recognized specifically by Lin28 was able to stimulate translation of a reporter gene in a Lin28-dependent fashion (15).
Recently, using co-immunoprecipitation and proteomic analysis we have identified RNA helicase A (RHA) as a putative Lin28 functional partner (15). RHA is a member of the highly conserved DEAD-box RNA helicase family proteins. It is ubiquitously expressed, and participates in a wide variety of cellular processes ranging from transcription, mRNA splicing, nuclear export and translation, to RNA interference (16,17). It is believed that like most other RNA helicases RHA elicits its functions by catalyzing RNA–RNA and RNA–protein rearrangements in RNP complexes (17). Indeed, RHA has been shown to be required for the efficient translation of a number of mRNAs bearing highly structured RNA elements in their 5′-UTRs. These elements, called post-transcriptional control elements (PCEs), are found in some viral and cellular transcripts (18–21). It has been proposed that PCEs act as roadblocks for efficient ribosomal scanning and that RHA binds directly to the PCEs by recognition of their specific structural features, thus relieving the blockade and facilitating RNA–RNA and RNA–protein rearrangements necessary for efficient translation to occur.
In this report we further characterize the interaction between Lin28 and RHA. We provide evidence that RHA is an important co-factor of Lin28 and is likely recruited to the translational machinery by Lin28 to enhance the translation of Lin28 target mRNAs.
MATERIALS AND METHODS
Antibodies, siRNAs and plasmids
The polyclonal anti-Lin28 (Abcam, ab46020), anti-RPL22 (Novus, NBP1-06069), anti-β-tubulin (Abcam, ab6046), anti-Flag (Santa Cruz, sc-807), anti-Oct4 (Chemicon, AB3209), monoclonal anti-RHA (Abcam, ab54593), anti-NXF1 (Sigma, T1076), anti-PABP (Santa Cruz, sc-32318) and anti-Flag M2 (Stratagene, 200472) antibodies were purchased. The siLin28 (Dharmacon, ON-TARGETplus SMARTpool, L-018411-01) and siCon (Dharmacon, D-001810-10-05) were described earlier (15). Flag-Lin28 was built by cloning human Lin28 coding region (accession number NM_024674) into pFLAG-CMV-2 (Sigma, E7398) at the NotI and BamHI sites (15). The firefly luciferase reporter construct Oct4-R2 was previously described (15). Flag-N300 of RHA was constructed by cloning the RHA residues 1-300 from pcDNA-FL RHA (C.G Lee, University of Dentistry and Medicine of New Jersey) into the EcoRV and NotI sites of the pcDNA3.1 vector (Invitrogen). GST-N300 was created by inserting the RHA residues 1-300 into the EcoRI site of the pGEX-2T vector (GE Healthcare). GST-DEIH (411-767), GST-RGG (1161-1269) and GST-DEIH extension (953-1160) were made by cloning the RHA residues 411-767, 1161-1269, 953-1160, respectively, into the BamHI and EcoRI sites of the pGEX-2T vector.
Cell culture and siRNA transfection
Human HEK293 and PA-1 cells were cultured using standard protocols provided by the ATCC. Cell transfections were carried out as described (22).
Protein extraction and western blot analysis
These were carried out as described previously (14).
Sucrose gradient polysome fractionation
These were carried out in the absence of cycloheximide as described previously (15). Briefly, HEK293 (3 × 107) or PA-1 (2.5 × 107) cells were collected, washed with PBS, and resuspended in 0.5 ml of freshly prepared MCB buffer [100 mM KCl, 0.1% Triton X-100, 50 mM HEPES, pH 7.4, 2 mM MgCl2, 10% glycerol, 1 mM DTT, 20 U/ml Protector RNase Inhibitor (Roche), 1× complete mini EDTA-free protease inhibitor cocktail (Roche)]. After incubation on ice for 10 min, the lysate was centrifuged at 1300g at 4°C for 10 min to remove insoluble materials. The supernatant was applied onto the top of a 15–55% (W/W) linear sucrose gradient made by Density Gradient Fractionation System (Teledyne ISCO Inc.), and centrifuged at 150 000 g for 3 h in a Beckman ultracentrifuge (Beckman, CA, USA). Fractions (0.2 ml) were collected and used for RNA extraction or protein analysis.
Luciferase assays
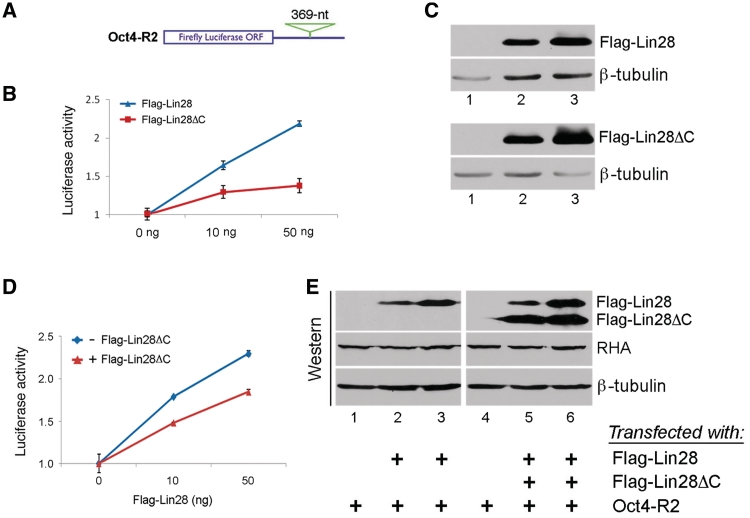
These were done essentially as previously described (14). In Figure 6B, Oct4-R2 was transfected into HEK293 cells, together with 0, 10 or 50 ng of Flag-Lin28 or Flag-Lin28ΔC. In Figure 6D, Oct4-R2 was transfected into HEK293 cells, together with 0, 10 or 50 ng of Flag-Lin28, with (+) or without (−) co-transfection of 50 ng of Flag-Lin28ΔC. In all transfections a Renilla reporter was also included for normalization purposes. The amount of total plasmid DNA per well of a 48-well plate was 400 ng that included an appropriate amount of empty vector (pFLAG-CMV-2), 100 ng of Oct4-R2, 2 ng of Renilla, and the indicated amount of Flag-Lin28 and/or Flag-28ΔC. Luciferase activities and protein and mRNA levels were measured 24 h post-transfection. Relative firefly luciferase activities were plotted after normalization against firefly luciferase mRNA levels.
Figure 6.
Effects of C-terminal deletion on translation of a reporter gene. (A) Schematic of the reporter construct (Oct4-R2) showing a 369-nt sequence from Oct4 ORF inserted at its 3′-UTR. (B) C-terminal deletion mutant has a reduced ability to stimulate translation. Oct4-R2, together with increasing amounts of Flag-Lin28 or Flag-Lin28ΔC, was transfected into HEK293 cells. Luciferase activities and mRNA levels were measured 24 h later. Relative luciferase activities were plotted after normalization against luciferase mRNA levels. Luciferase activities in the absence of wild-type or mutant Lin28 expression were set as 1. Numbers are mean ± SD (n = 3). (C) Western blot analysis showing expression levels of the indicated proteins from transfected cells. β-tubulin was used as a loading control. (D) C-terminal deletion has a dominant-negative effect on the translation of the reporter RNA. Oct4-R2, together with 0, 10 or 50 ng of Flag-Lin28, were transfected into HEK293 cells, with (red line) or without (blue line) co-transfection of 50 ng of Flag-Lin28ΔC. Luciferase assays were performed 24 h later. Relative luciferase activities are presented after normalization against luciferase mRNA levels. Luciferase activities in the absence of Flag-Lin28 were set as 1. Numbers are mean ± SD (n = 3). (E) Western blot results of aliquot samples from D. A single western blot membrane was cut into three pieces with each containing Flag-Lin28/Flag-Lin28ΔC, RHA and β-tubulin, respectively, and probed using anti-Flag (for Flag-Lin28 and Flag-Lin28ΔC), anti-RHA, and anti-β-tubulin, respectively.
Co-immunoprecipitation
To examine the interactions between Lin28 and RHA, 8 × 106 HEK293 (or PA-1) cells were transfected with 2 µg of Flag-Lin28 (Flag-28ΔC, Flag-28ΔN or empty vector), with or without co-transfection of 6 µg of Flag-N300 in a 6 cm plate scale (total DNA per plate was 8 µg). Thirty-eight hours later, cells were collected by manual scraping using a rubber policeman and pelleted by centrifugation. Cell pellet was resuspended in 400 µl of gentle lysis buffer [10 mM Tris–HCl at pH 7.5, 10 mM NaCl, 10 mM EDTA, 0.5% Triton X-100, 1 mM PMSF, 1× protease inhibitor cocktail (Calbiochem), 1 mM DTT and 10 µg/ml of RNase A (Roche)] and incubated on ice for 15 min. Insoluble materials were removed by centrifugation at 13 400 g in a microcentrifuge at 4°C for 15 min. NaCl was added to the cleared lysate to a final concentration of 200 mM, and 350 µl of the lysate incubated with 20 µl of protein-A sepharose beads pre-bound with 10 µl of anti-Lin28 antibody, pre-immune IgG (Figure 4C), or 10 µg of anti-Flag M2 antibody (Figure 5B) at 4°C overnight. The next day, beads were washed and bound fractions eluted by 3× SDS-sample buffer by heating at 95°C for 5 min. Proteins were resolved by SDS–PAGE, followed by western blot analysis.
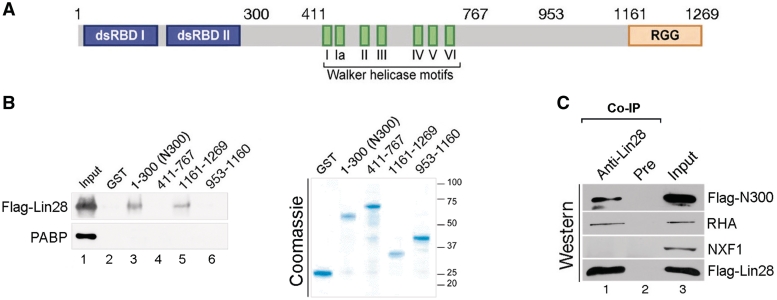
Figure 4.
The N- and C-terminal regions of RHA interact with Lin28. (A) Domain organization of human RHA protein. Double-stranded RNA binding domain I and II (dsRBD I and II), C-terminal domain rich in arginine-glycine-glycine (RGG) repeats and the Walker helicase motifs of the conserved DEAD-box RNA helicases are depicted. Numbers indicate corresponding amino acid residue. (B) GST pulldown results. HEK293 cell lysate containing Flag-Lin28 was incubated with bacterial lysate containing the indicated recombinant RHA domains or GST alone in the presence of RNase A, followed by GST pulldown assays. Left panel, anti-Flag and anti-PABP antibodies were used in the upper and lower blots, respectively. Input was 0.5% of the total amount of proteins used for each GST pulldown. Right panel, Coomassie staining determined comparable amounts of the recombinant proteins used in the GST pulldown assays. 1% of the input was loaded in each lane. Molecular size markers are on the right. (C) Flag-Lin28 and Flag-N300 were co-transfected into HEK293 cells. Co-IP was carried out in the presence of RNase A 24 h later using anti-Lin28 antibody to bring down Flag-Lin28 together with its associated proteins, followed by western blot analysis. Antibodies used in the western blot were anti-RHA (top two blots, note, this antibody recognizes both full-length RHA and Flag-N300), anti-NXF1 (third blot from top), and anti-Flag M2 (bottom blot). Total proteins (2%) used for each immunoprecipitation was loaded as input.
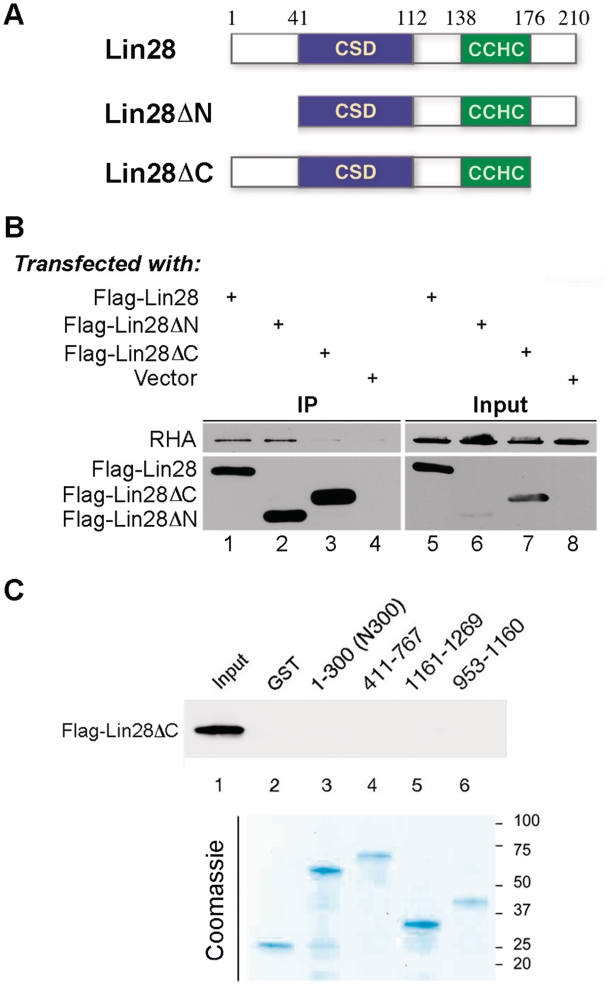
Figure 5.
C-terminus deletion reduces Lin28′s ability to interact with RHA. (A) Schematic of wild-type and mutant Lin28 protein. Numbers are in amino acids. (B) Flag-Lin28, Flag-Lin28ΔN, Flag-Lin28ΔC or empty vector were each transfected into HEK293 cells. Co-IP was carried out in the presence of RNase A using anti-Flag antibody. Resulting protein complexes were resolved by SDS–PAGE, followed by western blot analysis. Anti-RHA and polyclonal anti-Flag antibody were used in the top and bottom blots, respectively. Three percent of input was loaded. (C) Flag-Lin28ΔC was transfected into HEK293 cells. Cell lysate containing Flag-Lin28ΔC was incubated with bacterial lysate containing the indicated recombinant RHA domains or GST, followed by GST pulldown assays. Top panel, anti-Flag M2 antibody was used to detect Flag-Lin28ΔC. About 0.5% of the input was loaded in lane 1. Bottom panel, Coomassie staining of the recombinant proteins used in the GST pulldown assays. About 1% of the input was loaded in each lane. Molecular size markers are on the right.
Purification of GST fusion proteins and in vitro GST pulldown assays
Escherichia coli BL21 cells (Stratagene, 230240) transformed with plasmids of GST-RHA domains were grown overnight in LB medium supplemented with carbenicilin. The next morning, cells were diluted 1:1000 into 500 ml LB with 75 µg/ml carbenicilin, grown to an optical density at 600 nm of 0.4 and induced with 0.5 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG). After 3 h of induction, cells were harvested, pelleted at 800 g for 10 min, resuspended in 10 ml phosphate-buffered saline (PBS) containing 100 µl of protease inhibitor cocktail (Sigma P8340). Cells were broken by two passages through a French pressure cell. Cell lysates were centrifuged at 13 400 g for 10 min to remove insoluble materials.
To produce Flag-Lin28-containing lysates, HEK293 cells in a 10 cm plate were transfected with 4 µg of Flag-Lin28 (or Flag-28ΔC) plasmid. Cells were harvested 48 h post transfection and lysed in 1 ml of cell lytic buffer (Sigma, C2978) supplemented with 2 mM PMSF, 10 µl of protease inhibitor cocktail (Sigma P8340), and 100 µg/ml of RNase A (Sigma R6513). Lysates containing GST fusion proteins were incubated with 40 µl of glutathione-agarose beads slurry (Thermo Scientific, 15160) with continuous rocking. Glutathione-agarose beads incubated with GST were used as a negative control. After 1 h of incubation at 4°C, beads were pelleted at 800 g for 1 min, washed once with NETN low salt buffer (0.5% NP40, 0.1 mM EDTA, 20 mM Tris–HCl, pH 7.4, 150 mM NaCl), once with NETN high salt buffer (0.5% NP40, 0.1 mM EDTA, 20 mM Tris–HCl, pH 7.4, 1 M NaCl), and once with PBS for 4 min each. Mammalian lysates containing Flag-Lin28 (or Flag-Lin28ΔC) were added to the washed beads and the volume was brought up to 500 µl using PBS in the presence of 2 mM PMSF, 10 µl protease inhibitor cocktail, and 100 µg/ml of RNase A. After 3 h of incubation at 4°C with continuous rocking, beads were pelleted at 800 g for 1 min and washed twice with NETN low salt buffer and once with NETN medium salt buffer (0.5% NP40, 0.1 mM EDTA, 20 mM Tris–HCl, pH 7.4, 200 mM NaCl) for 4 min each. The beads were then pelleted, boiled in 20 µl of 2× SDS loading dye. The samples were subjected to western blot analysis.
RESULTS
Lin28 affects the polysome profile of Oct4 mRNA
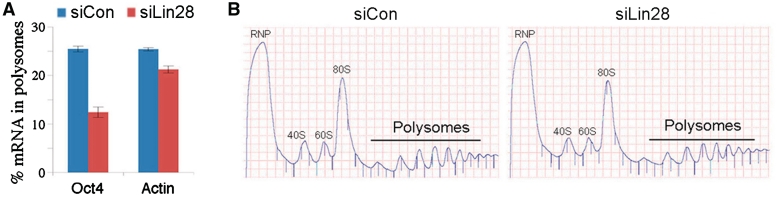
We have previously shown that reducing Lin28 levels by siRNA in undifferentiated embryonal carcinoma PA-1 cells led to decreased Oct4 expression at the protein level while its mRNA level was not affected (15). These results are consistent with a role of Lin28 in promoting Oct4 mRNA translation. It is generally believed that mRNAs actively being translated are associated with polysomes and that an increased polysome association indicates an increase in translation efficiency. To provide further evidence supporting the role of Lin28 in Oct4 mRNA translation, we performed polysome profile analysis. PA-1 cells were transfected with a siRNA specific for Lin28 (siLin28; ref. 15) or a control siRNA (siCon), followed by sucrose gradient fractionation of cytoplasmic extracts collected 48 h after transfection. Total RNAs were isolated from polysome or non-polysome fractions (that included RNP, 40S, 60S and 80S), and polysome distributions of Oct4 and β-actin mRNAs measured by quantitative PCR. As shown in Figure 1A, while 25% of Oct4 mRNA was present in polysomes from siCon-transfected cells, this percentage decreased to 12.5% in cells transfected with siLin28. On the other hand, polysome association of β-actin mRNA decreased only slightly in cells transfected with siLin28 (21.5% polysome association) compared to siCon (25% polysome association). We speculate that the decreased polysome association of Oct4 mRNA observed was likely due to reduced translation.
Figure 1.
Lin28 influences polysome association of Oct4 mRNA. PA-1 cells were transfected with siCon or siLin28. Cytoplasmic extracts were collected 48 h after transfection and subjected to sucrose gradient fractionation. (A) Quantifications of Oct4 and β-actin mRNAs in the polysomes. Following sucrose gradient sedimentation, RNAs were extracted from each fraction and subjected to RT–qPCR using primers specific for Oct4, β-actin (negative control) or β-tubulin (loading control) mRNA. The efficiency of translation was then reported as percentage mRNA in polysomes, which was calculated, after normalization to β-tubulin mRNA, by comparing the RNA level in polysomes with total fractions (combining the polysome and non-polysome fractions). Numbers are mean ± SD (n = 3), P < 0.01. (B) Representative polysome profiles of PA-1 48 h after transfection of siCon (left) or siLin28 (right).
Lin28 influences RHA association with polysomes
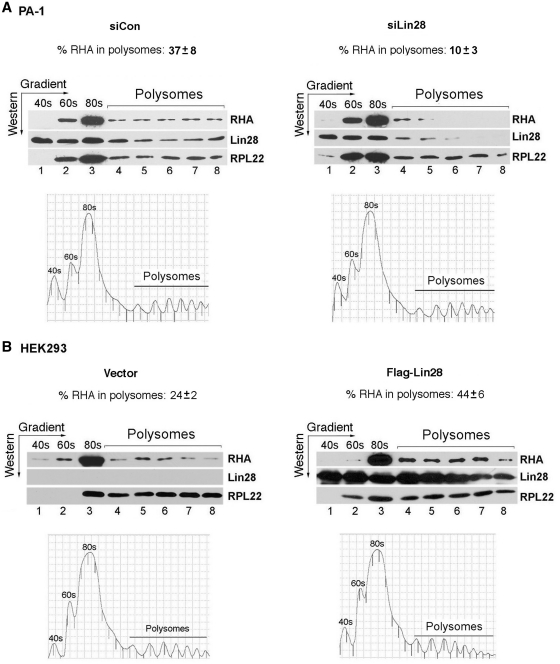
Lin28 and RHA interact with each other in both human ES cells (15) and PA-1 cells (Figure 2) assessed by co-immunoprecipitation (co-IP). In addition, reducing RHA levels impedes Lin28-dependent stimulation of translation of a reporter gene (15). To gain further mechanistic insights into the molecular interplay between Lin28 and RHA, we analyzed the distribution patterns of Lin28 and RHA proteins on polysomes. We hypothesized that Lin28 selectively binds to its target mRNAs and subsequently recruits RHA to the translational machinery to facilitate RNP remodeling during translation, thereby enhancing translation efficiency. To test this hypothesis, we performed sucrose gradient fractionation of cytoplasmic extracts from PA-1 cells 48 h after transfection with siLin28 or siCon. We asked whether changes in the Lin28 level affect the pattern of RHA association with polysomes. Indeed, while there was a significant decrease in the association of Lin28 with polysomes in cells transfected with siLin28 compared to siCon, a concomitant decrease in polysome association of RHA was also observed. Results shown in Figure 3A are representative of three independent experiments. In cells transfected with siCon, 37% of RHA was present in the polysomes, while the amount of RHA in polysomes decreased to 10% in cells transfected with siLin28 (Figure 3A, compare the left with the right panel). The polysome shift in response to Lin28 reduction was specific to RHA, as the polysome profile of ribosomal protein RPL22 was not altered (45% polysome association in siCon versus 44% in siLin28-transfected cells).
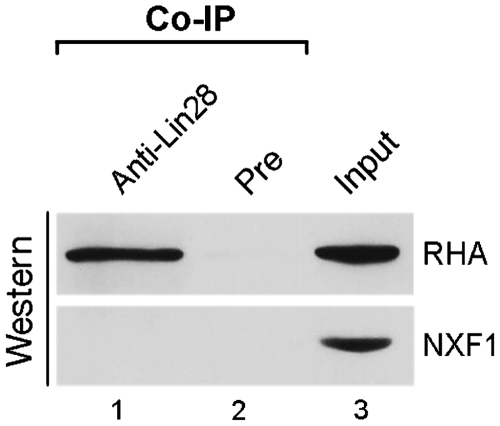
Figure 2.
Lin28 interacts with RHA in PA-1 cells. Lin28-containing protein complexes were immunoprecipitated in the presence of excess amounts of RNase A from PA-1 cells using anti-Lin28 or pre-immune IgG (as a negative control for non-specific binding). Co-IP complexes (lanes 1 and 2) and 3% input (lane 3) were resolved by SDS–PAGE, followed by western blot analysis using anti-RHA (top blot) and anti-NXF1 (bottom blot) antiboddies, respectively.
Figure 3.
Lin28 influences RHA association with polysomes. (A) PA-1 cells were transfected with siCon or siLin28. Cytoplasmic extracts were collected 48 h later. (B) HEK293 cells were transfected with empty vector or Flag-Lin28. Cytoplasmic extracts were collected 24 h later. In both A and B, cytoplasmic extracts were separated on sucrose gradients and fractionated to the indicated ribosomal species (polysome profiles are shown beneath the western blot gels). Aliquots of the indicated fractions were resolved by SDS–PAGE, followed by western blot analysis. Antibodies used in the western blot analysis are labeled on the right. Protein bands on western gels were quantitated using the Bio-Rad Quantity One software. Ribosomal protein L22 (RPL22) was used as a loading control. The percentages of RHA in polysomes were calculated, after normalization against RPL22, by comparing fractions 4 through 8 with the total fractions (fractions 1 through 8). In A, numbers are mean ± SD (n = 3), P < 0.01. In B, numbers are averages of two experiments.
Next, we performed reciprocal experiments using HEK293 cells that do not express endogenous Lin28. In the absence of Lin28, 24% of RHA was associated with polysomes (Figure 3B, left panel). The presence of RHA in polysomes in cells that do not express Lin28 has been documented and is consistent with its role in translational stimulation of a class of endogenous mRNAs in these cells (20). RHA has been shown to stimulate the translation of a number of messages, including the cellular JunD mRNA, that contain a structured PCE within the 5′-UTR (20). It has been proposed that the PCE poses a barrier to efficient ribosome scanning and that direct binding of RHA to the PCE induces RNA–protein and RNA–RNA rearrangements, resulting in enhanced polysome incorporation (20). Importantly, when Lin28 was expressed by Flag-Lin28 transfection, the amount of RHA associating with polysomes increased to 44% (Figure 3B, right panel), while such a shift was not observed with RPL22 (73% polysome association in vector-transfected versus 69% in Flag-Lin28-transfected cells). Collectively, these results reveal the association of Lin28 with actively translating mRNAs and are consistent with the view that Lin28 may actively recruit RHA to polysomes during translation of Lin28 target mRNAs.
The carboxyl terminus of Lin28 is required for interaction with RHA at both its amino and carboxyl terminal regions
We were previously able to capture the interaction between Lin28 and RHA using co-IP in human ES cells (15). Figure 2 recapitulates this interaction in PA-1 cells. To learn more about how Lin28 interacts with RHA, we set out first to map the interaction domains on RHA. Four recombinant fragments of RHA were tested for their interaction with Lin28 by in vitro GST pulldown assays. The full-length RHA protein contains two double-stranded RNA binding domains (dsRBD I and II) at its amino terminus, a DEAD-box helicase domain (Walker helicase motifs) in the middle, and a domain rich in arginine-glycine-glycine repeats (RGG) at the carboxy terminus (Figure 4A). Four regions (amino acids 1–300, 411–767, 953–1160 and 1161–1269) were each fused to GST at their N-termini and the recombinant proteins expressed in bacteria. To assess the interactions between Lin28 and the RHA domains, Flag-Lin28 was transfected into HEK293 cells. Bacterial lysates containing GST alone or the individual recombinant RHA fragments were purified with glutathione agarose beads and then incubated with cell lysates containing Flag-Lin28. As both Lin28 and RHA are RNA-binding proteins, an excess amount of RNase A was included during the incubation to ensure disruption of any RNA-bridged interactions. Following GST pulldown, eluants were subjected to western blot analysis. Figure 4B shows a representative of three independent pulldown experiments. The interactions of Lin28 with the N-terminal domain (N300) and the C-terminal domain (amino acids 1161–1269) of RHA were readily detected (Figure 4B, left panel, lanes 3 and 5), but those with the central domains (amino acids 411–767 and 953–1160), as well as GST alone were not (Figure 4, left panel, lanes 2, 4 and 6). Importantly, the absence of PABP [poly (A)-binding protein which binds to poly (A) tails of most mRNAs in the cell, ref. 15] in the GST pulldown samples (Figure 4B, left panel, bottom blot) suggests that the RNase A treatment was effective. GST alone and all of the recombinant proteins were expressed at comparable levels (Figure 4B, right panel).
To test whether these interactions also occur in vivo, we transfected Flag-Lin28 together with Flag-N300 into HEK293 cells and performed co-IP using anti-Lin28 antibody in the presence of RNase A, followed by western blot analysis. Flag-N300 could be easily detected in the Lin28-containing complexes brought down by the anti-Lin28 antibody (Figure 4C, top blot, lane 1), as was the case of endogenous full-length RHA (second blot from the top, lane 1). The absence of the general mRNA nuclear export factor NXF1 (an RNA-binding protein) in the same complexes (second blot from the bottom, lane 1) confirmed that RNase A treatment was efficient and that the interaction between Lin28 and N300/full-length RHA was not mediated by RNA. These results, however, do not allow us to exclude the possibility that an additional protein may bridge these interactions. Although our inability to stably express the C-terminal domain of RHA in HEK293 cells has precluded an examination of its interaction with Lin28 in vivo, the above results nevertheless are in line with the notion that both the N- and C-terminal domains of RHA interact with Lin28.
We next carried out domain mapping on Lin28. Lin28 harbors two types of RNA-binding motifs—a cold shock domain (CSD) and a pair of retroviral-type CCHC zinc fingers (Figure 5A) (23,24). While point mutations in either domain abolished the ability of Lin28 to interact with RNA (11), the same mutations did not affect its interaction with RHA (data not shown). Thus, two Lin28 deletion mutants were made. Lin28ΔN and Lin28ΔC contain 41- and 35-amino acid deletions, respectively, at the amino and carboxyl terminus of Lin28 (Figure 5A). Attempting to express mutant proteins with larger deletions has been unsuccessful due to poor expression in transfected cells (data not shown). To examine the effects of the N- and C-terminal deletions on RHA interaction, Flag-Lin28, Flag-Lin28ΔN, Flag-Lin28ΔC or empty vector were each transfected into HEK293 cells. Co-IP was carried out (in the presence of RNase A) using a monoclonal anti-Flag M2 antibody, followed by western blot analysis. The amino terminal deletion (Lin28ΔN) did not affect the ability of Lin28 to interact with RHA (Figure 5B, top blot, compare lane 2 to lane 1), but the carboxyl terminal deletion (Lin28ΔC) did (top blot, compare lane 3 to lane 1). Our in vitro GST pulldown analysis also indicated lack of interactions between Flag-Lin28ΔC and the RHA domains (Figure 5C). Together these results suggest that the C-terminus of Lin28 plays a critical role in interaction with RHA. However, we cannot exclude the possibility that the C-terminal domain of Lin28 affects protein folding in such a way as to promote interaction between another region and RHA.
A C-terminal deletion mutant of Lin28 exerts dominant-negative effects on Lin28-dependent stimulation of translation
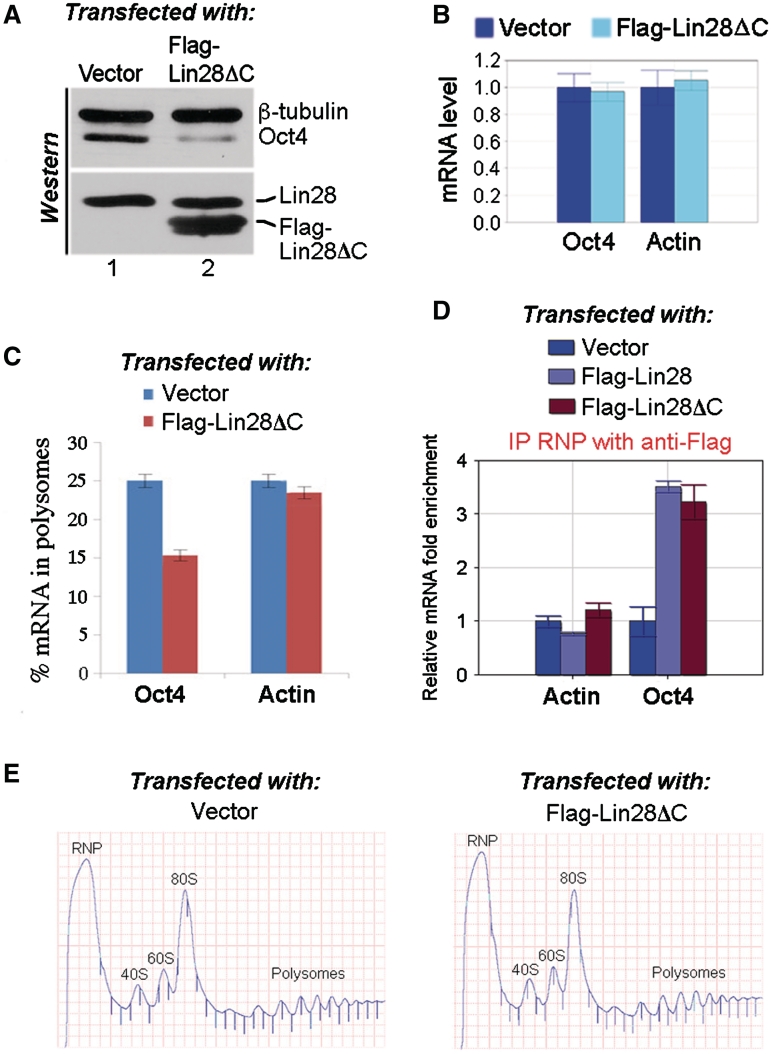
As Lin28ΔC exhibited a weakened interaction with RHA compared to wild-type Lin28 (Figure 5B), we speculated that its ability to stimulate translation might also be compromised. To test this hypothesis, we performed reporter assays using a luciferase construct (Oct4-R2) that contains a 369-nt Lin28-binding sequence from Oct4 mRNA at its 3′-UTR (Figure 6A, and ref. 15). To ask whether Lin28ΔC has an impaired ability to stimulate translation of the reporter gene, Oct4-R2 was transfected into HEK293 cells, together with increasing amounts of Flag-Lin28ΔC. Flag-Lin28 instead of Flag-Lin28ΔC was used as a positive control in parallel experiments. Luciferase activities were measured 24 h after transfection. Relative luciferase activities were presented after normalization against luciferase mRNA levels. Consistent with our previous report (13,15), the luciferase activity increased in response to Flag-Lin28 (Figure 6B, blue line), indicating stimulation of translation by Lin28. Western blot analysis confirmed increased Flag-Lin28 levels in response to transfection of increasing amounts of Flag-Lin28 plasmid (Figure 6C, top blots). In contrast, the ability of Flag-Lin28ΔC to stimulate luciferase expression was significantly reduced (Figure 6B, red line) despite the fact that its expression level was comparable to that of Flag-Lin28 (Figure 6C, bottom blots). We next asked whether the presence of Flag-Lin28ΔC might interfere with wild-type Lin28 function. Indeed, when Flag-Lin28ΔC was co-transfected into HEK293 cells, the stimulation of luciferase activity by Flag-Lin28 was significantly inhibited (Figure 6D, compare red line with blue line). Western blot analysis revealed that the reduced stimulation was not due to decreased expression levels of Flag-Lin28 and/or RHA as a result of Flag-Lin28ΔC expression (Figure 6E). These results thus suggested that Flag-Lin28ΔC (which has an impaired ability to interact with RHA) may compete with Flag-Lin28 for binding to the reporter RNA, thus interfering with translation. If this were the case, Flag-Lin28ΔC expression should interfere with the translation of endogenous Oct4 mRNA. To test this possibility, we transfected Flag-Lin28ΔC into PA-1 cells and examined its effect on Oct4 expression. Oct4 protein level in Flag-Lin28ΔC-transfected cells was reduced to 42 ± 8% (P < 0.01) of that of vector-transfected cells (Figure 7A, top blot, compare lane 2 to lane 1), while Oct4 mRNA level was not affected (Figure 7B). The conclusion that translation was inhibited was further supported by our polysome profile analysis, showing that in Flag-Lin28ΔC-transfected cells, the association of Oct4 mRNA with polysomes decreased to 15% compared to that in vector-transfected cells (25%) (Figure 7C, compare the blue bar with the red bar on the left), while the percentages in polysome association of β-actin mRNA were similar in both cases (Figure 7C, compare the blue bar with the red bar on the right). Surprisingly, while a significant amount of Flag-Lin28ΔC associated with polysomes in these assays, we did not observe a corresponding decrease in polysomal RHA association in the presence of this mutant protein (data not shown). There are several possible reasons for this observation. First, the level of expression of Flag-Lin28ΔC in our assays may have been insufficient to result in an RHA change great enough to detect using western analysis. Indeed, we could only express Flag-Lin28ΔC ∼2-fold above the level seen in cells transfected with Flag-Lin28 (Figure 6E) or above the level of expression of endogenous Lin28 (Figure 7A, bottom blot). In addition, a significant fraction of RHA may associate with polysomes through interactions independent of Lin28. Some RHA is associated with polysomes even in cells not expressing Lin28 (Figure 3B, and ref. 20). Second, it is possible that some Lin28 target mRNAs are more sensitive to Lin28 levels than others are. The fact that we were able to detect an Oct4 mRNA polysome shift (Figure 7C) as well as a change in protein level (Figure 7A) in response to Flag-Lin28ΔC expression is consistent with the possibility that Oct4 expression is particularly responsive to the level of Lin28.
Figure 7.
The C-terminal deletion of Lin28 has a dominant-negative effect on the translation of endogenous Oct4 mRNA. Flag-Lin28ΔC or empty vector was transfected into PA-1 cells. Forty-eight hours later, protein and RNA were extracted and levels determined by western blot (A) and RT–qRCR (B), respectively. In (A), top blot, polyclonal antibodies against β-tubulin and Oct4 were simultaneously used to probe a single membrane piece that contained both Oct4 and β-tubulin. In the bottom blot, polyclonal anti-Lin28 antibody was used to detect a single piece of membrane containing both endogenous Lin28 and the transfected Flag-Lin28ΔC. Protein bands on western gels were quantitated using the Bio-Rad Quantity One software. (C) Distribution of Oct4 and β-actin mRNAs in the polysomes. Flag-Lin28ΔC or empty vector was transfected into PA-1 cells, and polysome fractionation carried out 48 h later. RNAs were extracted from each fraction and subjected to RT–qPCR using primers specific for Oct4, β-actin or β-tubulin (loading control) mRNA. The efficiency of translation was calculated as described in Figure 1 legend. Numbers are mean ± SD (n = 3), P < 0.01. (D) The C-terminus deletion of Lin28 does not affect its RNA-binding. Flag-Lin28, Flag-Lin28ΔC, or empty vector was transfected into PA-1 cells, IP RNP experiments using monoclonal anti-Flag M2 antibody were carried out 30 h after transfection. RNAs were extracted from isolated RNPs and subjected to RT–qPCR analysis. Levels of β-actin and Oct4 mRNAs in IP samples derived from vector-transfected cells were arbitrarily set as 1. Numbers are mean ± SD (n = 3). (E) Representative polysome profiles of PA-1 48 h after transfection of empty vector or Flag-Lin28ΔC.
To exclude the possibility that the C-terminal deletion may somehow weaken the binding of Lin28 to target mRNAs, we performed transfection and IP RNP experiments. Thus, Flag-Lin28, Flag-Lin28ΔC or empty vector were individually transfected into PA-1 cells, and RNPs isolated using monoclonal anti-Flag M2 antibody. RNAs were extracted from the RNPs and subjected to RT–qPCR analysis. We observed ∼3-fold enrichment of Oct4 mRNA in both Flag-Lin28- and Flag-Lin28ΔC-containing RNPs versus control RNPs derived from vector-transfected cells, while no enrichment of β-actin mRNA was observed (Figure 7D). Therefore we suggest that the C-terminal deletion of Lin28 affects its interaction with RHA but not with target mRNAs.
DISCUSSION
Lin28 influences cellular growth and gene expression in multiple ways. While one important function of Lin28 is to downregulate the expression of a subset of miRNAs including let-7, we have focused on let-7-independent functions and have studied the mechanism by which Lin28 modulates the translation of target mRNAs, using Oct4 mRNA as a model system. Here we report a number of new findings. In response to decreased levels of Lin28, Oct4 protein levels decreased and a fraction of Oct4 mRNA was shifted from polysomes to non-polysome fractions. These results are consistent with the notion that Lin28 acts as a translational enhancer for a specific subset of mRNAs. There further exists a positive relationship between Lin28 and the polysome association of RHA. We have also mapped Lin28-interacting domains to both the N- and C-termini of RHA, and showed that the C-terminus of Lin28 is required for these interactions. Importantly, a mutant Lin28 missing the C-terminus not only has an impaired function in stimulation of translation but also acts as a dominant-negative inhibitor of Lin28-dependent stimulation of translation of both reporter and endogenous target mRNAs. Collectively, these studies suggest that Lin28 functions to stimulate the translation of its target mRNAs through interaction with RHA which is capable of facilitating RNP remodeling during the process of translation.
Based on our observation that both the N- and C-termini of RHA interact with Lin28, it is tempting to speculate that simultaneous interaction of both domains with Lin28 might be necessary for translational stimulation. This would be reminiscent of the mode of action of eukaryotic translation initiation factor eIF4A, a prototypic member of the DEAD box family of helicases. It is believed that the main function of eIF4A is to promote pre-initiation complex scanning during translation initiation by unwinding RNA secondary structures in the 5′-UTR of an mRNA (25). However, free eIF4A is a poor helicase and requires accessory proteins to stimulate its activity. One such protein is eIF4G, a large protein that provides docking sites for several initiation factors including eIF4A. It has been demonstrated that simultaneous interaction of both the N- and C-termini of eIF4A with eIF4G is required to ensure an active conformation of eIF4A, hence its helicase activity (25). It is possible that RHA functions in an analogous way to eIF4A in that two domains need to be simultaneously anchored onto Lin28 in order to promote function (Figure 8). Herein RHA is an additional accessory protein to supplement the activity of eIF4A in the context of selected complex mRNAs (20).
Figure 8.
A model for Lin28-RHA interaction in translation. Lin28 binds to target mRNA and recruits RHA through its C-terminal domain. The N- and C-termini of RHA interact simultaneously with Lin28 to ensure exposure of its helicase domain that is capable of interaction with the translational machinery.
RHA has been shown to facilitate translation of a group of viral and cellular mRNAs that are otherwise inefficiently translated because they harbor highly structured motifs or RNP complexes in their 5′-UTRs that block ribosomal scanning (17,20). Direct binding of RHA to the structured motifs relieves the blockade, likely owing to its ability to remodel the RNPs and allow the ribosomes to scan through. Similarly, we hypothesize that Lin28 target mRNAs may share yet unknown common structural or sequence features that reduce their efficiency of translation. This could be due to presence of highly structured RNA elements, or elements that are specifically recognized by inhibitory proteins. Binding of Lin28 and subsequent recruitment of RHA to these mRNAs would overcome the inhibition by removing the inhibitory structures/proteins, allowing efficient translation to occur. Genome-wide identification of Lin28 mRNA targets and detailed characterization of their interactions will be required to test this hypothesis.
Accumulating evidence suggests that Lin28 plays important roles in ES cell function and oncogenesis (1,6,10,13–15,26–28). Our recent finding that Lin28 modulates Oct4 mRNA expression at the post-transcriptional level further highlights its importance in ES cell biology (ref. 15, and this report), given that Oct4 is a transcription factor essential for maintaining ES cell self-renewal and pluripotency (29). While overexpression of Oct4 triggers human ES cell differentiation into primitive endoderm and mesoderm, a <2-fold decrease in expression leads to differentiation towards trophectoderm (30), suggesting that a precisely optimal level of Oct4 is required for maintaining the pluripotency. Likewise, we have recently reported that a sub-population of ovarian cancer cells co-expresses Lin28 and Oct4 and that their combined expression in tumor samples correlates with advanced tumor grade. Importantly, when the expression of these two proteins is repressed, there is significant reduction in cancer cell growth and survival (28). We propose that one important function of Lin28 via RHA is to facilitate the expression of genes whose translation is otherwise inefficient and that appropriate levels of expression of these genes are required for maintaining the health of ES cells as well as for tumor progression.
FUNDING
Funding for open access charge: Connecticut Stem Cell Research Grants (09SCAYALE14 to Y.H.); National Institutes of Health (RO1CA108882, P01CA100730 to K.B.-L.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Eric Moss for point mutation Lin28 constructs. The contents of this material are solely the responsibility of the authors and do not necessarily represent the official views of the State of Connecticut, the Department of Public Health of the State of Connecticut or Connecticut Innovations, Incorporated.
REFERENCES
- 1.Viswanathan SR, Daley GQ. Lin28: a microRNA regulator with a Macro Role. Cell. 2010;140:445–449. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz S, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 5.Trabucchi M, Briata P, Carcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heo I, Joo C, Kim Y-K, Ha M, Yoon M-J, Cho J, Yeom K-H, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA Uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balzer E, Heine C, Jiang Q, Lee VM, Moss EG. Lin28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development. 2010;137:891–900. doi: 10.1242/dev.042895. [DOI] [PubMed] [Google Scholar]
- 10.Zhu H, Shah S, Shyh-chang N, Shinoda G, Einhorn WS, Viswanathan SR, Takeuchi A, Grasemann C, Rinn JL, Lopez MF, et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat. Genet. 2010;42:626–630. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balzer E, Moss EG. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007;4:16–25. doi: 10.4161/rna.4.1.4364. [DOI] [PubMed] [Google Scholar]
- 12.Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu B, Huang Y. Histone H2a mRNA interacts with Lin28 and contains a Lin28-dependent posttranscriptional regulatory element. Nucleic Acids Res. 2009;37:4256–4263. doi: 10.1093/nar/gkp372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu B, Zhang K, Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA. 2009;15:357–361. doi: 10.1261/rna.1368009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu C, Ma Y, Wang J, Peng S, Huang Y. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010;38:1240–1248. doi: 10.1093/nar/gkp1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol. Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 17.Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol. Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 18.Hull S, Boris-Lawrie K. RU5 of Mason-Pfizer monkey virus 5' long terminal repeat enhances cytoplasmic expression of human immunodeficiency virus type 1 gag-pol and nonviral reporter RNA. J. Virol. 2002;76:10211–10218. doi: 10.1128/JVI.76.20.10211-10218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts TM, Boris-Lawrie K. Primary sequence and secondary structure motifs in spleen necrosis virus RU5 confer translational utilization of unspliced human immunodeficiency virus type 1 reporter RNA. J. Virol. 2003;77:11973–11984. doi: 10.1128/JVI.77.22.11973-11984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartman TR, Qian S, Bolinger C, Fernandez S, Schoenberg DR, Boris-Lawrie K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- 21.Bolinger C, Sharma A, Singh D, Yu L, Boris-Lawrie K. RNA helicase A modulates translation of HIV-1 and infectivity of progeny virions. Nucleic Acids Res. 2010;38:1686–1696. doi: 10.1093/nar/gkp1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma Y, Jin J, Dong C, Cheng E-C, Lin H, Huang Y, Qiu C. High-efficiency siRNA-based gene knockdown in human embryonic stem cells. RNA. 2010;16:2564–2569. doi: 10.1261/rna.2350710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 24.Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev. Biol. 2003;258:432–442. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- 25.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 27.Darr H, Benvenisty N. Genetic analysis of the role of the reprogramming gene LIN-28 in human embryonic stem cells. Stem Cells. 2009;27:352–362. doi: 10.1634/stemcells.2008-0720. [DOI] [PubMed] [Google Scholar]
- 28.Peng S, Maihle NJ, Huang Y. Pluripotency factors Lin28 and Oct4 identify a subpopulation of stem cell-like cells in ovarian cancer. Oncogene. 2010;29:2153–2159. doi: 10.1038/onc.2009.500. [DOI] [PubMed] [Google Scholar]
- 29.Pei D. Regulation of pluripotency and reprogramming by transcription factors. J. Biol. Chem. 2009;284:3365–3369. doi: 10.1074/jbc.R800063200. [DOI] [PubMed] [Google Scholar]
- 30.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]