Abstract
H2A.Z, a variant of H2A, is found at the promoters of inducible genes in both yeast and higher eukaryotes. However, its role in transcriptional regulation is complex since it has been reported to function both as a repressor and activator. We have previously found that mono-ubiquitylation of H2A.Z is linked to transcriptional silencing. Here, we provide new evidence linking H2A.Z deubiquitylation to transcription activation. We found that H2A.Z and ubiquitin-specific protease 10 (USP10) are each required for transcriptional activation of the androgen receptor (AR)-regulated PSA and KLK3 genes. USP10 directly deubiquitylates H2A.Z in vitro and in vivo, and reducing USP10 expression in prostate cancer cells results in elevated steady-state levels of mono-ubiquitylated H2A.Z (H2A.Zub1). Moreover, knockdown of USP10 ablates hormone-induced deubiquitylation of chromatin proteins at the AR-regulated genes. Finally, by sequential ChIP assays, we found that H2A.Zub1 is enriched at the PSA and KLK3 regulatory regions, and loss of H2A.Zub1 is associated with transcriptional activation of these genes. Together, these data provide novel insights into how H2A.Z ubiquitylation/deubiquitylation and USP10 function in AR-regulated gene expression.
INTRODUCTION
H2A.Z is a highly conserved variant of the canonical core histone H2A. Various functions have been attributed to H2A.Z, including prevention of heterochromatin spreading (1), suppression of antisense RNAs (2) and maintenance of chromosome stability and segregation (3–5). By far, the strongest evidence links H2A.Z function to regulation of gene transcription [reviewed in (6,7)]. For example, Chromatin immunoprecipitation (ChIP)-microarray and ChIP-sequencing (ChIP-seq) studies showed that most genes have H2A.Z deposited around their transcription start sites. Many studies have also highlighted a dual role for H2A.Z in transcriptional regulation, wherein it can act in either a negative or positive manner [discussed in (8)]. How a single histone variant can perform such contrasting functions is still unclear. However, its ability to either repress or promote transcription could be explained by differential post-translational modifications (PTMs) that occur on H2A.Z. For example, fractions of total H2A.Z are acetylated or mono-ubiquitylated, and we have previously hypothesized that these modifications distinguish H2A.Z’s functions in activation or repression (6). In this regard, the enzymes that modulate these respective modifications represent key regulatory components that dictate H2A.Z functions.
Mammalian H2A.Z is known to be multiply acetylated at several lysine residues (including K4, K7 and K11) at the N-terminus (9–11). In contrast, mono-ubiquitylation predominantly occurs at K120 at the C-terminus, although K121 and K125 can serve as alternative ubiquitylation sites (12). In yeast, H2A.Z is acetylated by the NuA4 and GCN5 acetyltransferases (13–15). Deficiency in H2A.Z acetylation leads to defects in chromosome transmission and cell proliferation. Furthermore, the acetylated form of H2A.Z is found at the 5′-end of active genes, suggesting a positive function for this modified form of H2A.Z in transcriptional regulation (15,16). Much less is known about H2A.Z ubiquitylation other than that ubiquitylated H2A.Z (H2A.Zub1) is enriched at facultative heterochromatin in mammalian cells (12). Ubiquitylation of H2A.Z is mediated by Ring1b, the E3 ligase member of the polycomb repressive complex 1, suggesting that this modified form of H2A.Z is part of the polycomb silencing pathway.
Like many PTMs, ubiquitylation is also a reversible reaction. Removal of ubiquitin is mediated by a specialized class of enzymes called isopeptidases. These deubiquitylating enzymes (DUBs) act by cleaving the isopeptide bond that links ubiquitin to its substrate. These enzymes are generally classified into five families based on their catalytic domains: ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumour proteases (OTUs), Josephins and Jab1/MPN/MOV34 metalloenzymes (JAMM/MPN+) [reviewed in (17)]. USPs, UCHs, OTUs and Josephins are all cysteine proteases, whereas the JAMM/MPN+ enzymes are zinc metalloproteases.
To date, several mammalian DUBs have been identified to target H2Aub1, and many have been linked to transcriptional activation [reviewed in (18)]. For example, USP16 was first described as a mitotic DUB targeting H2Aub1 (19), and a recent report suggested that this enzyme also regulates Hox gene expression (20). Similarly, USP21 was found to promote transcription of developmentally regulated genes in the liver presumably through its deubiquitylation of H2Aub1 (21). Finally, USP22 and the 2A-DUB (KIAA1915), both target H2Aub1 (although USP22 can also deubiquitylate H2Bub1), and both were described as co-activators of androgen receptor (AR)-mediated transcription (22,23).
Thus far, an H2A.Z deubiquitylase has not been reported. Given that H2A.Zub1 has also been linked to repression of gene expression, we predicted that the DUB for H2A.Z would have a transcriptional activation function. In this regard, ubiquitin-specific protease 10 (USP10) has been implicated as a transcription co-activator for AR-regulated genes (24) and, therefore, we tested whether it might target H2A.Z. Indeed, in this study, we found that USP10 deubiquitylates H2A.Zub1 and H2Aub1 in vitro and in vivo. Importantly, knockdown of USP10 in prostate cancer cells increases the global levels of H2A.Zub1 and H2Aub1, and prevents deubiquitylation of chromatin proteins associated with hormone-activated genes. USP10 and H2A.Z are each required for AR-regulated gene transcription, and ChIP assays showed that H2A.Z is directly associated with the promoter and enhancer of the prostate-specific antigen (PSA) and kallikrein-like 2 (KLK2) genes. Finally, knockdown of USP10 also impairs the loss of H2A.Z at the regulatory regions of PSA and KLK2 that normally occurs during their transcriptional activation. Collectively, our data support a novel role for H2A.Z in the regulation of AR-mediated transcription that also involves the deubiquitylating activity of USP10 towards histones.
MATERIALS AND METHODS
Cell culture, transfection, plasmids and antibodies
The 293T cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. LNCaP cells were obtained from ATCC and PC-3 cells stably expressing AR [PC-3(AR)] were courtesy of Dr Theodore J. Brown (Mount Sinai Hospital, Toronto, Canada). Both cell types were grown in RPMI 1640 media supplemented with 10% fetal bovine serum. For culturing in the absence of hormone, cells were grown in phenol-red free RPMI 1640 supplemented with 5% charcoal-stripped fetal bovine serum (Invitrogen) for 72 h prior to treatment with hormone. Dihydrotestosterone (DHT) was obtained from Sigma and re-suspended in absolute ethanol (EtOH); DHT was added to cells at a final concentration of 10 nM, or for control samples, an equivalent volume of EtOH was added. All transfections were carried out using Lipofectamine 2000 (Invitrogen). USP10 and histone expression constructs used were based on the pcDNA 3.1 (+) (Invitrogen) backbone with the Flag tag cloned in-frame. In co-transfection experiments, the ratio of the Flag-H2A/Flag-H2A.Z to Flag-USP10 plasmids were either 1:1 or 1:3. The HA (hemagglutinin)-tagged ubiquitin construct, cloned into the pMSCV vector, was courtesy of Dr Dwayne Barber (Princess Margaret Hospital, Toronto, ON, Canada). H2A.Z antibody directed against the L1 loop was described earlier (12), H2A acidic patch antibody was from Upstate, H3 (ab1791) and USP10 antibodies were from Abcam, H2Bub1 monoclonal antibody (NRO3) was from MédiMabs, Flag M2 and anti-His monoclonal antibodies were from Sigma, and AR antibody (PG-21) was from Millipore. Anti-HA Affinity Matrix (Roche), containing rat monoclonal antibody (clone 3F10) conjugated to agarose beads, was used for HA ChIP experiments.
In vitro deubiquitylation assay
Deubiquitylases were cloned into the pVL1393 vector (BD Biosciences) and His tag sequences were added in-frame to the DUB cDNA. His-tagged recombinant protein was generated using BaculoGold (BD) packaging vector in the baculovirus expression system using standard techniques, and purified via Ni-NTA agarose beads. Equal amounts of purified DUBs (1.5 µg each) were mixed with 2 µg of total acid-soluble protein harvested from cells expressing either Flag-tagged H2A.Z or Flag-tagged H2A in a buffer containing 60 mM HEPES (4-2 [hydroxyethyl]-1-piperazineethanesulfonic acid, pH 7.5), 5mM MgCl2, 4% glycerol, 1mM EDTA, 1mM DTT plus fresh protease inhibitors. Reactions were incubated at 37°C for 30 min. The reactions were stopped by the addition of 2× sample buffer and the samples were boiled for 5 min.
Immunofluorescence analysis
LNCaP cells were seeded on glass coverslips and grown as described above. Immunofluorescence (IF) analysis was performed as described in (12) using anti-USP10 and anti-AR antibodies.
Small-scale biochemical fractionation
Following treatment of cells with EtOH or 10 nM DHT for 2 h, subcellular fractions of LNCaP cells were harvested using the Subcellular Protein Fractionation Kit (Thermo Scientific) according to manufacturer’s instructions. Equal percentages (by volume) of each fraction were analysed by SDS–PAGE and Western blotting using standard techniques.
Stable shRNA and protein expression via retroviral transduction of LNCaP cells
H2A.Z (cagctgtccagtgttggtg) and USP10 (gaatatcagagaattgagt) shRNA (short hairpin RNA) target sequences were cloned into the pSUPER–retro–puro vector (Oligoengine) as per manufacturer’s instructions. To generate stable LNCaP cells, 7.0 × 105 cells were seeded per well in 6-well dishes in RPMI 1640, 10% fetal bovine serum. Twenty-four hours later, culture media was replaced with 4 ml of viral supernatant (containing 4 µg/ml of polybrene) and plates were centrifuged at 1500g for 4 h at 20°C. Following centrifugation, viral supernatant was removed and replaced with fresh culture media. Twenty-four hours later, cells were selected in 0.9 µg/ml puromycin for a minimum of 4 days, after which cells were maintained in media containing 0.6 µg/ml puromycin.
Luciferase assays
USP10 or P/CAF expression constructs, or their respective vector controls (pcDNA3.1 or pLNCX2), were transfected into PC-3(AR) cells along with the AR-dependent luciferase reporter, which contains three repeats of the androgen response element (ARE) in its promoter. Cells were lysed 48 h after transfection in cell culture lysis reagent (Promega). Luciferase activities were measured using the Luciferase assay system (Promega) according to the manufacturer’s instructions. Co-transfection of a β-gal-expressing plasmid, with subsequent measurement of β-gal activity, was used to normalize luciferase data.
RT-quantitative polymerase chain reactions analysis
LNCaP cells were grown in the absence of hormone and then treated with DHT or EtOH as described above. Twenty-four hours following treatment, RNA was harvested in TRIzol reagent (Invitrogen) according to manufacturer’s instructions. RNA was re-suspended in molecular biology grade ddH2O and 500 ng was used in a reverse transcription reaction to synthesize cDNA using Oligo (dT)12–18 and SuperScript II (Invitrogen). Quantitative polymerase chain reactions (qPCR) were assembled in triplicate using PerfeCta SYBR Green SuperMix (Quanta Biosciences) and transcript-specific primers. Reactions were run on an Applied Biosystems SDS7900HT thermal cycler in a 384-well format. Gene expression was normalized to expression of the housekeeping gene RPL27 as described (25). Data are presented as means ± SDs and are representative of at least three independent experiments using independently generated batches of stable cells. Primers for the PSA gene were described earlier (22). KLK2 primers were as follows: forward 5′ gctgggagtgtgagaagattc 3′, reverse 5′ gtttcaggctcaaacaggttgtg 3′. Primers for RPL27 were described in (25).
ChIP and sequential ChIP assays
LNCaP cells were grown in 15 cm plates, in the absence of hormone, and then treated with DHT or EtOH as described above. Cells were fixed by adding formaldehyde directly to the culture medium to a final concentration of 1% and incubated at room temperature for 10 min. Formaldehyde was quenched by adding glycine to a final concentration of 125 mM with incubation at room temperature for an additional 5 min. Cells were washed three times in cold 1 × PBS plus protease inhibitors. Cells were then re-suspended in a nuclei isolation buffer comprising 50 mM HEPES (pH 8.0), 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 10% glycerol, 0.5% Igepal CA-630, 0.25% Triton X-100, plus protease inhibitors. Following a 10 min incubation on ice, nuclei were pelleted via centrifugation and re-suspended in a buffer composed of 10 mM HEPES (pH 8.0), 1mM EDTA, 0.5mM EGTA, 200 mM NaCl and incubated on ice for 10 min. Nuclei were pelleted via centrifugation and re-suspended in sonication buffer containing 50 mM HEPES (pH 8.0), 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholic acid. Chromatin was sonicated using a Branson Sonifier 450 to produce chromatin fragments with an average size of 600 bp. Samples were then centrifuged at maximum speed for 10 min, 4°C. Supernatant was used for IPs after pre-clearing with Protein G Dynabeads (Invitrogen). 2 × 106 cells were used per IP in a total volume of 200 µl of sonication buffer at 4°C overnight, with rotation. The following amounts of antibodies were used per IP: AR (PG-21), 1.2 µg; H3 1.5 µg; H2A.Z L1 loop, 2 µl serum. Chromatin–antibody complexes were captured using Protein G Dynabeads for 1h at 4°C with rotation. Captured chromatin was washed sequentially, for 10 min each, in the following buffers containing fresh protease inhibitors: Radioimmunoprecipitation assay buffer (‘RIPA’) [150 mM NaCl, 50 mM Tris–HCl (pH 8.0), 0.1% SDS, 0.5% Na–deoxycholic acid, 1% Igepal CA-630], ‘High Salt’ [500 mM NaCl, 50 mM Tris–HCl (pH 8.0), 0.1% SDS, 1% Igepal CA-630], ‘LiCl buffer’ [250 mM LiCl, Tris–HCl (pH 8.0), 0.5% Na–deoxycholic acid, 1% Igepal CA-630], followed by two washes in ‘TE buffer’ [10 mM Tris–HCl (pH 8.0), 1mM EDTA]. For sequential ChIP assays, following the final TE wash, chromatin complexes were eluted in 1% SDS, 50 mM Tris–HCl pH 7.5, 10 mM EDTA for 30 min at room temperature, then diluted 10-fold in sonication buffer (minus SDS), and subjected to a second round of ChIP using anti-HA affinity matrix. Following overnight incubation, chromatin complexes were subjected to four 10-min washes in sonication buffer, followed by two washes in TE buffer.
For both ChIP and re-ChIP, chromatin was eluted in 2 × 200 µl of ‘elution buffer’ (2% SDS, 10 mM DTT, 0.1M sodium bicarbonate). Sixteen microliters of 5M NaCl were added to each sample followed by reverse cross-linking at 65°C for 4–6 h. One milliliter of 95% EtOH was added to each sample and placed at −20°C overnight. Chromatin was pelleted via centrifugation, washed once in 70% EtOH and re-suspended in TE buffer. Following treatment of samples with RNase A and Proteinase K, DNA was extracted using standard phenol–chloroform procedures and re-dissolved in 100 µl of molecular biology-grade water. Five microliters of each sample were used for qPCR analysis, in triplicate, as described above. The following primers were used for ChIP analyses: PSA Enhancer, forward 5′ agatccaggcttgcttactgtcct 3′, reverse 5′ acctgctcagcctttgtctctgat 3′; PSA (–)2 kb forward 5′ caaccaaaacctgacccaac 3′, reverse 5′ tttgcctggcccgtagt 3′; PSA Promoter, forward 5′ tgggtcttggagtgcaaaggatct 3′, reverse 5′ agacacgcccaggatgaaacagaa 3′; KLK2 Enhancer forward 5′ gttgaaagcagacctactctgga 3′, reverse 5′ ctggaccatcttttcaagcat 3′; KLK2 Promoter, forward 5′ gggaatgcctccagactgat 3′, reverse 5′ cttgccctgttggcacct 3′. Data are presented as means ± SDs and are representative of at least three independent experiments. H2A.Z ChIP data are presented as ‘H2A.Z enrichment’, to account for nucleosome density, which was calculated by dividing the % INPUT of the H2A.Z IPs by the % INPUT of the total H3 IPs.
RESULTS
USP10 specifically deubiquitylates H2A.Z and H2A
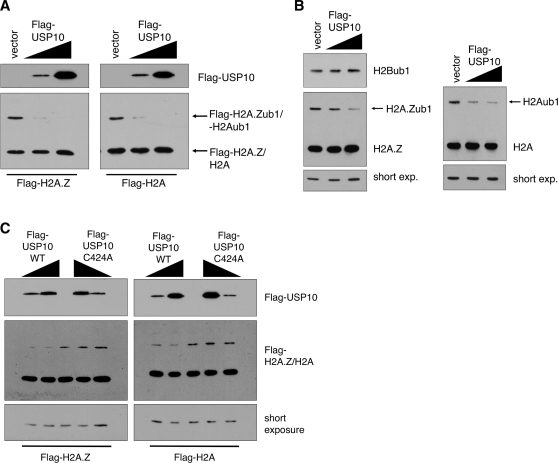
To test whether USP10 targets histones as substrates, we co-expressed Flag-tagged USP10 (Flag-USP10) with tagged H2A.Z or H2A in 293T cells and measured the levels of ubiquitylated histones by Western blot. We have previously determined that the mono-ubiquitylated form of H2A.Z can be detected as a slower migrating band using our H2A.Z-L1 antibody in Western blots (12). We have also observed the slower migrating form of H2A using antibodies against both the endogenous and tagged versions of H2A. For both H2A.Z and H2A Western blots, we further confirmed that the slower migrating species are the mono-ubiquitylated forms of the respective histones by conducting HA-tagged ubiquitin immunoprecipitations (Supplementary Figure S1). Increasing expression of Flag-USP10 resulted in a corresponding reduction of the ubiquitylated histone levels in a dose-dependent manner (Figure 1A). Moreover, expression of USP10 alone effectively reduced the endogenous H2A.Zub1 and H2Aub1 levels (Figure 1B). Importantly, increased USP10 expression did not affect the levels of endogenous H2Bub1 (Figure 1B), indicating that USP10 specifically targets the H2A family of histones. Finally, this effect was dependent on the enzymatic activity of USP10 since expression of the catalytically inactive mutant containing the cysteine 424-to-alanine substitution (C434A) mutation (26) did not alter the levels of ubiquitylated histones (Figure 1C). Together, these findings show that expression of USP10 leads to a specific reduction of H2A.Zub1 and H2Aub1 levels in vivo.
Figure 1.
Expression of USP10 causes a reduction in H2A.Zub1 and H2Aub1 levels. (A) A Flag-tagged version of USP10, or empty vector, was transiently co-transfected with either Flag-H2A.Z or Flag-H2A, in 293T cells. Total protein lysate was harvested and used for western blotting using the Flag M2 antibody. (B) Flag-USP10, or empty vector, was transiently transfected into 293T cells and levels of endogenous mono-ubiquitylated H2A.Z and H2A were assessed using antibodies that recognize both the unmodified forms (lower band) as well as the mono-ubiquitylated forms (upper band) of the endogenous histones (see Supplementary Figure S1). As in (A), increasing expression of Flag-USP10 causes a reduction in the levels of the mono-ubiquitylated forms of endogenous H2A.Z and H2A. However, Flag-USP10 does not affect the levels of mono-ubiquitylated H2B, as assessed using an antibody specifically recognizing the mono-ubiquitylated form of H2B (after probing the membrane with anti-H2A.Z antibody, the membrane was stripped and re-probed with anti-H2Bub1 antibody). (C) In contrast to wild-type Flag-USP10, co-transfection of the catalytically inactive C424A mutant of USP10 with either Flag-H2A.Z or Flag-H2A does not affect levels of the mono-ubiquitylated forms of the tagged histones.
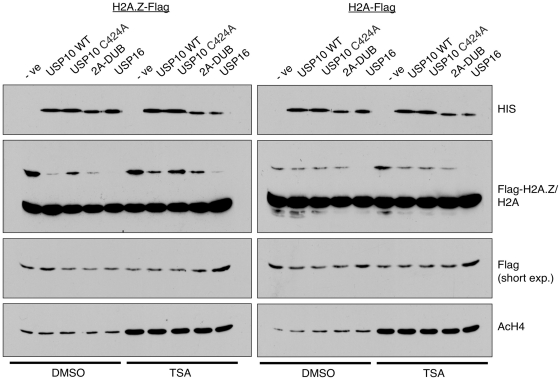
We next tested whether USP10 directly targets H2A.Zub1 and H2Aub1 by using an in vitro deubiquitylation assay. To do so, we incubated acid-extracted histones from 293T cells expressing either H2A-Flag or H2A.Z-Flag with a panel of recombinant deubiquitylases generated using the baculovirus expression system. Using an anti-Flag antibody in Western blots, we then measured the changes in H2A.Zub1 and H2Aub1 levels (upper band detected by the Flag antibody, Figure 2) brought about by the activities of the recombinant enzymes. As shown, His-tagged USP10 directly deubiquitylated H2A.Z in this assay (Figure 2, left panel). Consistent with the in vivo results (Figure 1C), the enzymatically inactive C424A mutant was impaired in its ability to deubiquitylate the incubated histones. In addition to USP10, we also tested the in vitro activities of two other DUBs, USP16 and 2A-DUB, previously identified to target H2Aub1 (20,22). Of all the enzymes tested, USP16 consistently displayed the highest efficiency in deubiquitylating both H2Aub1 and H2A.Zub1. USP10 and 2A-DUB have similar activities towards H2A.Zub1, but both are less efficient than USP16. Interestingly, while USP16 has similar activity towards both H2A.Zub1 and H2Aub1, neither USP10, nor 2A-DUB, showed significant activities towards H2Aub1 in this assay. The previous report on 2A-DUB suggested that, in vitro, H2A deubiquitylation was enhanced by hyper-acetylation of the histone substrates via trichostatin A (TSA) treatment (22). We therefore examined the deubiquitylation activities of the DUBs on histone substrates harvested from TSA-treated cells. Interestingly, we found that TSA treatment not only increased acetylation levels of H4, but it also led to a small but consistent increase in the overall H2A.Zub1 and H2Aub1 levels (Figure 2). However, in contrast to the previous report, hyper-acetylation of H2A or H2A.Z did not enhance the deubiquitylation of these substrates in our assay conditions.
Figure 2.
USP10 deubiquitylates H2A.Zub1 and H2Aub1 in vitro. In vitro deubiquitylation assays were assembled using a panel of His-tagged deubiquitylases that were generated using the baculovirus protein expression system. Purified deubiquitylases were incubated with acid-soluble protein extracts from nuclei of 293T cells expressing either Flag-tagged H2A.Z or H2A. Levels of the various deubiquitylases were compared by Western blotting using an anti-His antibody (top panel) and levels of the mono-ubiquitylated tagged histones were assessed using the Flag M2 antibody—short exposures of the Flag blots show the unmodified band of the tagged histones for loading purposes. In order to determine if hyper-acetylated histones are better substrates for in vitro deubiquitylation, acid-soluble protein was harvested from cells treated with TSA and hyper-acetylation of histones was confirmed using an anti-acetyl H4 antibody (bottom panel). See text for discussion.
In summary, our in vitro assay showed that H2A.Zub1 is a direct substrate for three histone deubiquitylases: USP16, USP10 and 2A-DUB. While USP16 deubiquitylates both H2A.Zub1 and H2Aub1, both USP10 and 2A-DUB prefer H2A.Zub1 in vitro. It is possible that additional co-factors not present in our assay system are needed for these enzymes to target H2A. Moreover, given that both USP10 and 2A-DUB were reported to function in activating AR-regulated genes, whereas USP16 was found to be important for deubiquitylating H2Aub1 during mitosis, their differences in activities and substrate preferences may reflect their different functions in vivo.
USP10 is localized to both the cytoplasm and nucleus
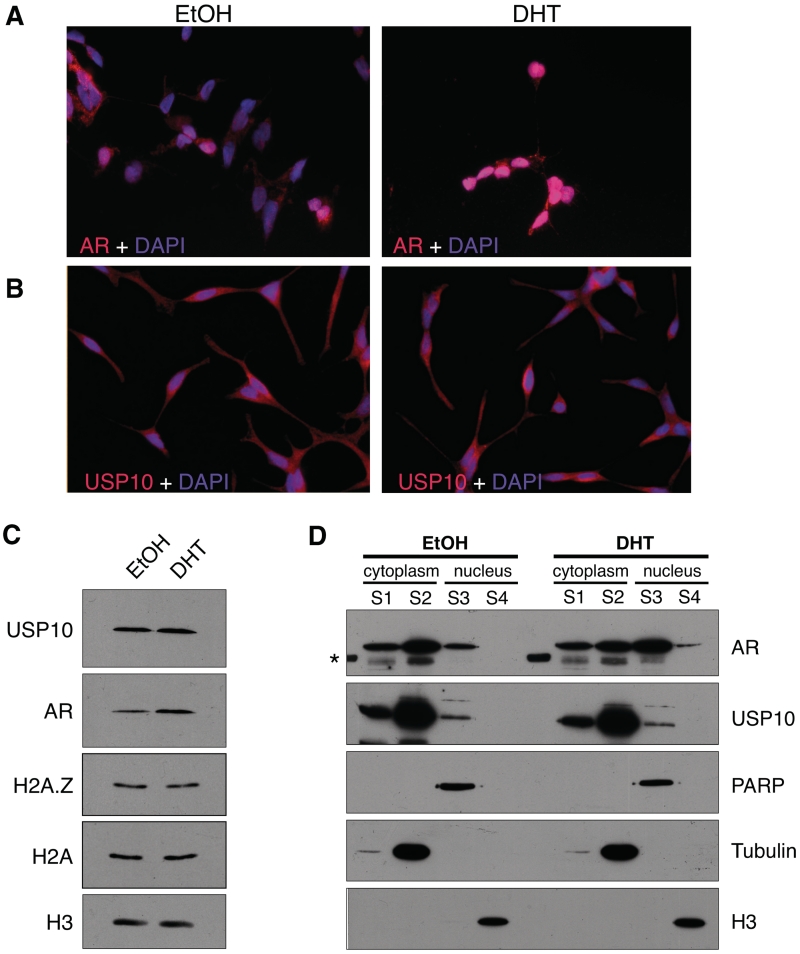
Insofar as USP10 was previously identified as a transcriptional co-activator for AR-regulated genes, we wanted to examine the subcellular distribution of this enzyme, particularly in the androgen-responsive LNCaP cell line. By IF, we found that USP10 is present in both the cytoplasm and the nuclei of LNCaP cells. USP10 was also previously suggested to physically interact with AR and, therefore, we examined the localization of this hormone receptor in our IF studies as well. While the majority of AR is in the cytoplasm of the hormone-deprived cells, treatment with 10 nM DHT dramatically re-localized the majority of AR to the nucleus. In contrast, DHT did not significantly alter USP10′s distribution in the cytoplasm and nuclei of the hormone-treated cells. Our data suggest that if USP10 and AR do physically interact, this interaction only occurs in a small nuclear fraction of USP10.
To examine the sub-cellular localization of USP10 with a more quantitative method, we used a biochemical fractionation assay to separate the cytoplasmic and nuclear protein fractions from LNCaP cells. We first compared total protein levels of AR and USP10 in EtOH- or DHT-treated LNCaP cells (Figure 3C). Additionally, total levels of H2A and H2A.Z, both substrates of USP10, were also assessed. Hormone treatment induced a slight but consistent increase in the total levels of AR, whereas it did not have any effect on the levels of USP10, H2A or H2A.Z in these cells. Consistent with the IF results, USP10 remained mostly cytoplasmic under both conditions, whereas AR was re-localized to the nucleus upon DHT treatment. However, a small but distinct fraction of USP10 does reside in the nucleus and its abundance is not affected by hormone treatment (Figure 3D). Insofar as significant amounts of USP10 are in the cytoplasm, it likely has cytoplasmic substrates and functions. Indeed, USP10 regulates the stability of transmembrane ion channels in epithelial cells (27,28), and it has recently been reported to target cytoplasmic p53 and regulate its protein stability (29). In contrast, the nuclear fraction of USP10 likely has a different function, such as transcription regulation, and since very little USP10 fractionates with chromatin, it suggests that the interaction between USP10 and chromatin is highly dynamic and transient.
Figure 3.
USP10 localizes to the cytoplasm and nucleus. LNCaP cells were grown on glass coverslips in charcoal-stripped, phenol red-free RPMI 1640 media for 72 h. Cells were then treated for 2 h with either DHT at a final concentration of 10 nM, or an equivalent volume of absolute EtOH. Cells were fixed and co-stained with DAPI (4′,6-diamidino-2-phenylindole) and antibodies recognizing either AR (A) or USP10 (B). (C) Total protein levels of AR and USP10 were compared, via Western blot, between LNCaP cells treated with DHT or EtOH for 2 h (H3 used as a loading control). (D) Sub-cellular localization of USP10 and AR was determined by Western blot using cytoplasmic and nuclear fractions (S1––membrane-associated fraction; S2––soluble cytoplasmic fraction; S3––soluble nuclear fraction; S4––chromatin-associated fraction) of LNCaP cells treated with EtOH or DHT for 2 h. Tubulin, PARP (poly ADP-ribose polymerase) and H3 were used as loading controls for the cytoplasmic, soluble nuclear and chromatin fractions, respectively. Sub-cellular fractions were harvested as described in ‘Materials and Methods’ section. Although AR shows a significant redistribution to the nuclear fraction upon treatment with hormone, USP10 does not redistribute in a similar manner. However, a distinct fraction of USP10 does reside in the nucleus. The asterisk marks a band in the ladder that cross-reacted with the AR antibody.
USP10 and H2A.Z regulate AR-mediated transcription
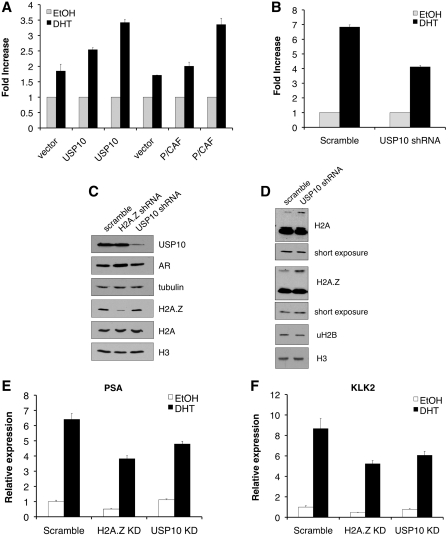
Since we did not observe a change in the distribution of USP10 following stimulation with DHT, we wanted to confirm that it does affect AR-regulated gene expression as reported. We first used a luciferase-based reporter assay in which the reporter gene is under the control of three AREs. This is a conserved DNA sequence to which AR binds, and is found in the regulatory regions of androgen-responsive genes (30). In prostate cancer, PC3 cells that stably express the AR [PC-3(AR)], expression of the AR-dependent luciferase reporter gene was indeed induced following treatment with DHT (Figure 4A). Co-transfection of USP10 along with the reporter construct showed that USP10 enhances the DHT-induced luciferase expression in a dose-dependent manner and its effect is comparable to that of another AR co-activator, the histone acetyltransferase P/CAF. More importantly, when the PC-3(AR) cells were pre-treated with USP10 shRNA to knockdown expression of endogenous USP10, a clear reduction in the DHT-induced luciferase level was observed (Figure 4B). These results replicate the previous findings by Faus et al. (24), confirming that USP10 is a bonafide co-activator of AR-regulated transcription.
Figure 4.
USP10 and H2A.Z are required for full expression of AR-regulated genes. (A) PC-3 cells, stably expressing AR, were grown in the absence of hormone, then transiently transfected with a firefly luciferase-expressing plasmid containing 3X AREs in the promoter, along with a plasmid expressing either USP10, the histone acetyltransferase P/CAF, or empty vector. In all cases, an additional plasmid expressing β-galactosidase (β-gal) was co-transfected for normalization. Cells were then stimulated with DHT (10 nM) or EtOH for 24 h, after which lysates were collected for measuring luciferase activity. Both USP10 and P/CAF stimulate the transcriptional activity of AR in a dose-dependent manner. Since USP10 and P/CAF are in different vector backbones, their respective empty vectors were used as negative controls. (B) Cells were grown as in (A) then co-transfected with the luciferase reporter plasmid, β-gal-expressing plasmid, and either a plasmid expressing an shRNA targeting USP10 mRNA, or a scrambled shRNA sequence control, followed by treatment with hormone or EtOH. Luciferase activity was measured 24 h after treatment. Knockdown of USP10 reduces the transcriptional activity of AR compared with the scramble control. For (C–F), LNCaP cells were transduced with retrovirus expressing shRNAs targeting USP10 or H2A.Z mRNA, or a non-targeting scrambled control sequence. (C) Whole-cell lysates were used to assess USP10 and H2A.Z knockdown efficiencies by Western blot. Additionally, AR and H2A protein levels were examined in the knockdown cells. Tubulin and H3 were used as loading controls. (D) Total protein lysates from LNCaP cells with stable knockdown of USP10 were used to examine the levels of H2A.Zub1, H2Aub1 and H2Bub1 by Western blot. Stable knockdown of USP10 increases the amount of both H2A.Zub1 and H2Aub1 but not H2Bub1. Shorter exposures of the H2A.Z and H2A blots, showing the unmodified band only, as well as H3, are used to demonstrate equal loading. (E) and (F) Transduced cells were grown in the absence of hormone for 72 h then treated with 10 nM DHT or an equivalent volume of EtOH for 24 h. RNA was harvested using TRIzol, and cDNA was synthesized and used as template for quantitative PCR (qPCR) using primers specific for PSA (E) or KLK2 (F) mRNA. Knockdown of either USP10 or H2A.Z results in decreased expression of both PSA and KLK2. Each PCR reaction was performed in triplicate with each experiment repeated at least three times independently, using different batches of stable cells, also generated independently. Values are presented as means ± SDs.
To further test the dependency of endogenous AR-regulated gene expression on USP10 we examined the expression of the well-characterized AR-activated PSA (KLK3) gene, as well as the KLK2 gene, in LNCaP cells. Additionally, since USP10 targets H2A.Z for deubiquitylation, we asked if H2A.Z is involved in regulating hormone-induced gene expression mediated through AR. We first generated stable LNCaP cell lines expressing shRNAs that target USP10 or H2A.Z, and confirmed that USP10 and H2A.Z expression were knocked down in these cells by Western blot (Figure 4C). Endogenous USP10 expression levels in the USP10 knockdown cells were <20% of the wild-type control cells, whereas the H2A.Z expression was ∼50% in the H2A.Z knockdown cells (quantification shown in Supplementary Figure S2). Our H2A.Z shRNA is designed to target the 3′-UTR of the H2A.Z-1 transcript. Consequently, the residual amount remaining could be contributed by the recently discovered additional isoform of H2A.Z, H2A.Z-2 (31,32), which may not be targeted by our shRNA construct. Supporting our finding that USP10 targets histones, cells with stable knockdown of USP10 consistently have higher levels of H2A.Zub1 and H2Aub1, whereas the levels of H2Bub1 remain unchanged (Figure 4D). As measured by reverse transcription–quantitative PCR (RT–qPCR, Figures 4E and F), knockdown of either H2A.Z or USP10 each resulted in reduction of the induced levels of PSA and KLK2 mRNA relative to the scrambled control. This effect was not due to a reduction in AR expression (Figure 4C), but suggests that H2A.Z and USP10 are each required in the transcriptional activation of these genes.
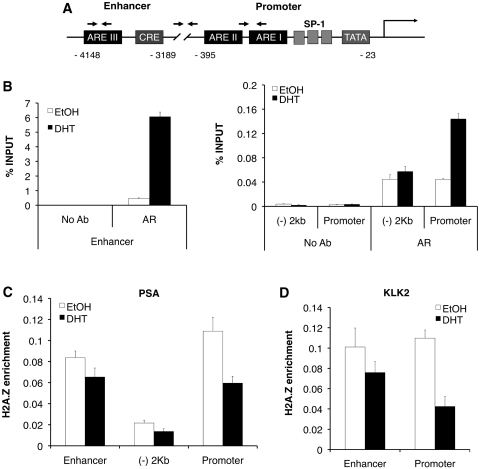
To test whether H2A.Z and USP10 directly regulate expression of the AR-responsive genes, we used ChIP assays to examine the localization of these proteins at the PSA and KLK2 genes. Consistent with current literature, we found that, upon DHT treatment, AR is recruited to the AREs present at the enhancers and promoters of the PSA and KLK2 genes (Figure 5B). Also, AR enrichment is much higher at the upstream enhancer than at the promoter of the PSA gene (note that the scale of the percentage input for the enhancer is much higher than that for the promoter), whereas AR levels are low and not increased upon DHT treatment at a control region located between these AREs. H2A.Z has been implicated in the regulation of other hormone-induced genes, such as glucocorticoid receptor (GR), and the estrogen receptor (ER), and is thought to localize at hormone receptor binding sites (33,34). We therefore examined the enrichment of H2A.Z, relative to total nucleosome density by also measuring the H3 signal, at the AREs before and after DHT treatment. We found that a significant amount of H2A.Z is present at the regulatory elements compared with the control region (Figure 5C). Interestingly, there is a consistent net loss of H2A.Z at the PSA and KLK2 enhancer and promoter regions upon DHT treatment. This transcription activation-induced loss of H2A.Z was confirmed using two additional commercially available antibodies against H2A.Z (data not shown), and is not due to a change in the total protein level in response to hormone (see Figure 3C). Such transcription-induced loss of H2A.Z was reported earlier for a number of other genes in mammalian cells (33,35), as well as in yeast cells (36–39), suggesting that this is a common feature of H2A.Z following gene activation. However, this observation is in contrast to the recent study by Gévry et al. (34) where hormone activation induced deposition of H2A.Z at the ER binding sites. At present, the exact role of H2A.Z in regulation of AR-regulated genes is not clear; however, our expression studies showed that H2A.Z is required for maximal induction of PSA and KLK2 expression upon DHT treatment.
Figure 5.
H2A.Z localizes to AREs at AR-regulated genes. LNCaP cells were grown in the absence of hormone for 72 h then treated with DHT (10 nM final concentration) or an equivalent volume of EtOH for 2 h. Cells were then fixed in 1% formaldehyde and chromatin was harvested for ChIP analysis. (A) Schematic of the PSA gene. Arrows represent approximate locations of primers used in qPCR. (B) ChIP analysis of AR recruitment to the PSA enhancer and promoter following stimulation of cells with DHT. The ‘(–) 2 kb’ label on the graphs represent the region in between the promoter and enhancer of the PSA gene and is ∼2 kb from the transcriptional start site. AR recruitment is specific to the AREs in the enhancer and promoter, the majority of which is recruited to the enhancer (compare scales of Y-axes). (C) and (D) H2A.Z is localized to the promoter and enhancer AREs in untreated cells. Following stimulation with DHT, a net loss of H2A.Z is observed at both regions. H2A.Z ChIP data are presented as ‘H2A.Z enrichment’, to account for nucleosome density, which was calculated by dividing the % INPUT of the H2A.Z IPs by the % INPUT of the total H3 IPs (data not shown). Each PCR reaction was performed in triplicate with each experiment repeated at least three times independently. Values are presented as means ± SDs.
In addition to AR and H2A.Z, we have made multiple attempts to ChIP USP10 to the PSA or KLK2 genes using two different commercially available antibodies. However, to date, we have not been able to consistently ChIP USP10 at the AREs of PSA and KLK2. One possibility is that the antibodies are not suitable for ChIP. On the other hand, given that our biochemical fractionation assay (Figure 3D) showed that only a small amount of USP10 resides in the nucleus, it is likely that the limited amount, as well as the transient nature of USP10 binding to chromatin preclude robust immunoprecipitation of this enzyme at the AR-regulated genes. Indeed a study of USP3, characterizing the enzyme as an H2Aub1 and H2Bub1 DUB, reached similar conclusions (40). Despite its prominent nuclear localization, wild-type USP3 could not immunoprecipitate its histone substrates, and by fluorescence recovery after photobleaching (FRAP) USP3 was shown to have a very transient and dynamic interaction with chromatin.
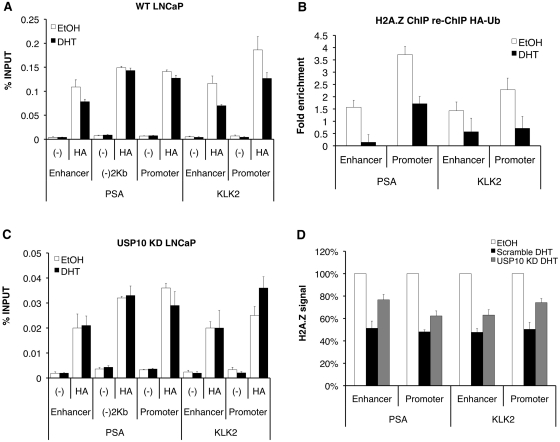
Since we do not have an antibody that specifically detects H2A.Zub1, we generated LNCaP cells stably expressing HA-tagged ubiquitin to assess changes in the levels of ubiquitylated proteins, including H2A.Zub1, at the PSA and KLK2 genes before and after hormone induction. ChIP analyses using anti-HA antibody showed that ubiquitylated proteins are present at all of the upstream regions of both PSA and KLK2 (Figure 6A). Interestingly, total levels of ubiquitylated proteins decreased at the PSA enhancer and both the promoter and enhancer regions of KLK2 following stimulation with hormone treatment. This suggests that hormone induction resulted in deubiquitylation of chromatin proteins, such as H2A and H2A.Z, and that this process is linked to transcriptional activation. Since we were able to readily immunoprecipitate H2A.Zub1 using HA-tagged ubiquitin (Supplementary Figure S1), we took advantage of this strategy to test whether H2A.Zub1 is present at the PSA and KLK2 genes using a sequential ChIP assay by first immunoprecipitating chromatin using an H2A.Z antibody, followed by a second ChIP using the HA antibody to immunoprecipitate ubiquitin-conjugated H2A.Z. Relative to the negative control region (between the enhancer and promoter of the PSA gene), we observed an enrichment of the HA–ubiquitin signal from H2A.Z-containing chromatin at the regulatory regions of the PSA and KLK2 genes, suggesting that H2A.Zub1 is present at these regions in the absence of hormone (Figure 6B). More importantly, the H2A.Zub1 levels are reduced following hormone induction, suggesting that deubiquitylation of H2A.Z is part of the transcriptional activation process.
Figure 6.
USP10 regulates levels of ubiquitylated proteins, including ubiquitylated H2A.Z, at AR-regulated genes. (A) LNCaP cells stably expressing HA-tagged ubiquitin were used for ChIP analysis following treatment of cells with EtOH or 10 nM DHT for 2 h, as described in ‘Materials and Methods’ section. Ubiquitylated proteins were detected at both regulatory regions of the PSA and KLK2 genes, as well as the control region, which lies between the PSA enhancer and promoter [‘(–)2 kb’]. Following treatment of cells with hormone, total levels of ubiquitylated proteins decrease at the PSA enhancer, and KLK2 enhancer and promoter. (B) Sequential ChIP was performed as described in ‘Materials and Methods’ section using LNCaP cells stably expressing HA-tagged ubiquitin with an anti-H2A.Z antibody followed by anti-HA antibody. The HA–ubiquitin signal co-localizes with regions of H2A.Z enrichment at the PSA and KLK2 genes and decreases following stimulation with hormone. Fold enrichment represents the % INPUT expressed relative to the control region, (–)2 kb. (C) ChIP was performed as in (A) using cells stably expressing both USP10 shRNA and HA–ubiquitin. Although a similar trend is observed in the levels of ubiquitylated proteins at the various regions of PSA and KLK2 under vehicle-treated conditions (EtOH), a similar drop in the HA–ubiquitin signal is not observed following stimulation with hormone. (D) H2A.Z ChIP was performed using Scramble- or USP10 shRNA-expressing cells as described. Compared with the control cells, knockdown of USP10 leads to an increase in the retention of H2A.Z at the PSA and KLK2 genes following stimulation of cells with hormone. Data were first calculated as % INPUT then expressed as a percentage relative to the EtOH-treated samples. ‘(–)’ represents the no antibody, negative control samples.
In order to test whether USP10 affects the levels of ubiquitylated proteins at the PSA and KLK2 genes, we generated LNCaP cells stably expressing both HA-tagged ubiquitin and USP10 shRNA for use in ChIP assays (Figure 6C). In the absence of hormone (white bars) the relative enrichment of HA–ubiquitin at the various regions was comparable to the trend observed in cells expressing HA–ubiquitin alone (Figure 6A). In contrast, knockdown of USP10 expression increased the retention of ubiquitylated proteins at both the PSA and KLK2 genes following stimulation with hormone, suggesting that USP10 directly affects the levels of ubiquitylated proteins at these genes in response to hormone stimulation. Finally, compared to cells expressing a scrambled control shRNA, we found a consistent defect in the eviction of H2A.Z (normally seen following hormone stimulation) from both the PSA and KLK2 regulatory regions in the USP10 knockdown cells (Figure 6D). Since expression of H2A.Z is unaffected by knockdown of USP10 (see Figure 4C) this finding strongly suggests that USP10 is required for the transcription-induced loss of H2A.Z at AR-regulated genes. Collectively, these data suggest that USP10 regulates the levels of ubiquitylated proteins at AR-regulated genes, such as H2A.Zub1 and H2Aub1, and that eviction of H2A.Z upon transcriptional activation is dependent on this deubiquitylase.
DISCUSSION
In this study, we have identified USP10 as a DUB that targets both H2A.Zub1 and H2Aub1. Co-transfection studies showed that USP10 specifically deubiquitylates H2A.Zub1 and H2Aub1, but not H2Bub1, in a dose dependent manner. More significantly, stable knockdown of USP10 expression in LNCaP cells led to a global increase in the total H2A.Zub1 and H2Aub1 levels in these cells, as well as an increase in the levels of ubiquitylated proteins at AR-regulated genes. In addition, knockdown of USP10 prevented full eviction of H2A.Z at the regulatory elements of AR-regulated genes during their transcriptional activation. Finally, in vitro assays confirmed that H2A.Z is a direct target of USP10, as well as other DUBs such as USP16 and 2A-DUB. Interestingly, while USP16 has robust activity towards both H2A.Zub1 and H2Aub1 in this assay, neither USP10, nor 2A-DUB deubiquitylated H2Aub1 in vitro. USP16, also known as Ubp-M, was first described as the enzyme responsible for deubiquitylating H2A during mitosis (19). Early studies showed that the bulk of H2A is deubiquitylated during mitosis and it was hypothesized that removal of the ubiquitin group is required to allow maximal condensation of chromatin into chromosomes during this stage of the cell cycle. In contrast, USP10 and 2A-DUB have been functionally linked to transcriptional regulation, a process that requires a large number of activators and co-activators. Since USP10 and 2A-DUB affect H2A ubiquitylation levels in vivo, but not significantly in our in vitro assay, it is likely that additional cofactors are required to increase the efficiencies or expand the substrate specificities of these enzymes in the cellular context.
H2A.Z is a highly conserved variant of H2A specifically deposited at the promoters of a large proportion of genes in the human and yeast genomes. This variant has been directly linked to both transcriptional activation and repression; however, its exact functions in these processes have not yet been clearly elucidated. We have previously found that a fraction of H2A.Z in mammalian cells is mono-ubiquitylated and such modified form of the variant is associated with facultative heterochromatin (12). We also found that the transcription-repressing PRC1 component Ring1b mediates mono-ubiquitylation of H2A.Z, and hypothesized that the enzyme that removes this modification likely has an opposite function, such as that of a transcriptional co-activator (6). In this regard, USP10 fulfills this prediction since we showed that USP10 is needed for maximal induction of AR-regulated genes. For example, knockdown of USP10 not only reduced induction of the AR-dependent luciferase reporter gene, but it also reduced expression of the endogenous AR-regulated PSA and KLK2 genes in DHT-treated LNCaP cells. Recently, Zhu et al. (22) showed that the 2A-DUB enzyme also functions in the activation of AR-regulated genes by deubiquitylating H2Aub1. Our findings support this model and further suggest that H2A.Z deubiquitylation is also part of this process. Moreover, three separate DUBs, USP10, USP22 and 2A-DUB are involved in this pathway. At present, we do not know whether they have redundant or separate functions in AR-regulated gene expression. In addition, it is possible that their specific functions may differ temporally or in the recruitment of unique biding partners or co-activators. This concept of histone modifying enzymes with overlapping functions mediating AR-induced transcription is not without precedence. The histone demethylases LSD1 and JHDM2A (JMJD1A) can both catalyse the removal of mono- and di-methyl groups from H3 lysine 9, yet both function prominently in the transcriptional activation of AR-regulated genes (42,43). The large number of co-activators identified in this pathway attests to the fact that AR-mediated transcriptional regulation is highly complex and that multiple histone-modifying activities are functionally required for this process [see (44) for review].
To further elucidate the role of USP10 in the transcription regulation of the DHT-induced PSA and KLK2 genes, we have made multiple attempts to ChIP this enzyme to the enhancers and promoters of these genes. We have used two different commercially available antibodies against USP10 for these experiments, but neither gave consistent results. Our IF and biochemical fractionation analyses showed that although a fraction of USP10 is in the nucleus of LNCaP cells, very little is stably associated with the chromatin fraction, which suggested its interaction with chromatin is highly dynamic and transient. These factors likely contributed to the difficulties of ChIP’ing this enzyme to the PSA and KLK2 genes. A large fraction of USP10 is localized to the cytoplasm and indeed a recent publication showed that USP10 deubiquitylates p53 and regulates its stability (29). Interestingly, many histone-modifying enzymes, such as the histone acetyltransferases P/CAF and p300 (45–47), the deacetylase SirT1 (48) and the de-methylase LSD1 (49), have all been shown to target and modify p53 as well. The utilization of the same enzyme on both histone, and non-histone substrates, is a common theme in biology and illustrates the multi-functionality of PTMs in many cellular pathways.
By ChIP analysis, we found that H2A.Z is enriched at the enhancers and promoters of the PSA and KLK2 genes under non-inducing conditions. Our sequential ChIP analysis of HA-tagged ubiquitin from H2A.Z-enriched chromatin further confirms that the mono-ubiquitylated form of H2A.Z is present at these regions in the absence of hormone, as is the case with H2Aub1. This is the first direct evidence showing that ubiquitylated H2A.Z is associated with repressed genes. Moreover, induction of these genes upon DHT treatment led to a significant loss of H2A.Z at these regulatory regions, a process that is at least partially dependent on USP10. Consistent with our conclusion that H2A.Z is directly involved in AR-regulated gene expression, its requirement has also been reported for other hormone-regulation pathways such as GR- and ER-induced genes (33,34). In addition, the chromatin remodeling complex, Snf2-Related CBP Activator Protein (SRCAP), which catalyses incorporation of H2A.Z into chromatin (50), has also been described as a co-activator of AR-mediated transcription (51,52). Interestingly, while our findings are consistent with the GR-regulated genes in that a substantial amount of H2A.Z is enriched at the regulatory regions of these genes prior to hormone induction, they are opposite to findings by Gévry et al. (34) that showed that H2A.Z is deposited to the ER-binding sites only after hormone induction. In that case, they found that p400, another H2A.Z deposition complex, is required for the hormone-induced recruitment of H2A.Z to the ER-binding sites. This differential usage of the chromatin remodeling complexes (SRCAP versus p400) in the AR- versus ER-regulated gene activation may explain the disparate timing of H2A.Z deposition in these hormone receptor-signalling pathways. However, the functional roles of H2A.Z in these two different hormone regulation pathways are currently unclear. Furthermore, given that there are two isoforms of H2A.Z that differ by only three amino acids (31,32), it would be interesting to determine the contribution of each isoform in the regulation of these pathways.
Our observed loss of H2A.Z upon DHT treatment and AR recruitment is consistent with multiple studies that showed a loss of H2A.Z upon gene activation (33,35–39). Genome-wide studies showed that, in yeast, H2A.Z preferentially regulates inducible genes and this variant is often deposited at regions that flank nucleosome-free regions at the promoters (15,39,53–55). In mammalian cells, H2A.Z is also frequently found at regulatory elements such as promoters, enhancers and insulators, which often display DNase I hypersensitivity (33,56,57). Moreover, for GR-regulated genes, enrichment of H2A.Z at the DNase I hypersensitive sites is lost upon GR recruitment. This frequent loss of H2A.Z upon transcriptional activation may be related to biochemical studies that showed that H2A.Z-containing nucleosomes are less stable (58–61). Collectively, these and other studies suggest that H2A.Z functions to maintain a chromatin structure that is amenable to rapid remodelling and poised for transcriptional activation (15,16). It is of further interest to note that acetylation of H2A.Z has been suggested to correlate with transcriptional activation and eviction of H2A.Z from promoter elements. Thus far, it is not known how H2A.Z ubiquitylation might affect this process. One hypothesis is that ubiquitylation of H2A.Z may alter nucleosome stability and impede H2A.Z eviction and thus deubiquitylation of H2A.Zub1 is a pre-requisite step for transcription activation. Further studies testing this hypothesis will be important for determining how H2A.Z deubiquitylation might function in transcriptional regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Cancer Society Research Institute, Operating (grant # 19228 to P.C.); Ontario Graduate Scholarship (to R.D.). Funding for open access charge: Canadian Cancer Society Research Institute operating grant (# 19228).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 2.Zofall M, Fischer T, Zhang K, Zhou M, Cui B, Veenstra TD, Grewal SI. Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature. 2009;461:419–422. doi: 10.1038/nature08321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rangasamy D, Greaves I, Tremethick DJ. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat. Struct. Mol. Biol. 2004;11:650–655. doi: 10.1038/nsmb786. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed S, Dul B, Qiu X, Walworth NC. Msc1 acts through histone H2A.Z to promote chromosome stability in Schizosaccharomyces pombe. Genetics. 2007;177:1487–1497. doi: 10.1534/genetics.107.078691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hou H, Wang Y, Kallgren SP, Thompson J, Yates JR, III, Jia S. Histone variant H2A.Z regulates centromere silencing and chromosome segregation in fission yeast. J. Biol. Chem. 2010;285:1909–1918. doi: 10.1074/jbc.M109.058487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Draker R, Cheung P. Transcriptional and epigenetic functions of histone variant H2A.Z. Biochem. Cell. Biol. 2009;87:19–25. doi: 10.1139/O08-117. [DOI] [PubMed] [Google Scholar]
- 7.Svotelis A, Gevry N, Gaudreau L. Regulation of gene expression and cellular proliferation by histone H2A.Z. Biochem. Cell. Biol. 2009;87:179–188. doi: 10.1139/O08-138. [DOI] [PubMed] [Google Scholar]
- 8.Guillemette B, Gaudreau L. Reuniting the contrasting functions of H2A.Z. Biochem. Cell. Biol. 2006;84:528–535. doi: 10.1139/o06-077. [DOI] [PubMed] [Google Scholar]
- 9.Beck HC, Nielsen EC, Matthiesen R, Jensen LH, Sehested M, Finn P, Grauslund M, Hansen AM, Jensen ON. Quantitative proteomic analysis of post-translational modifications of human histones. Mol. Cell. Proteomics. 2006;5:1314–1325. doi: 10.1074/mcp.M600007-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Bonenfant D, Coulot M, Towbin H, Schindler P, van Oostrum J. Characterization of histone H2A and H2B variants and their post-translational modifications by mass spectrometry. Mol. Cell. Proteomics. 2006;5:541–552. doi: 10.1074/mcp.M500288-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Ishibashi T, Dryhurst D, Rose KL, Shabanowitz J, Hunt DF, Ausio J. Acetylation of vertebrate H2A.Z and its effect on the structure of the nucleosome. Biochemistry. 2009;48:5007–5017. doi: 10.1021/bi900196c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarcinella E, Zuzarte PC, Lau PN, Draker R, Cheung P. Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Mol. Cell. Biol. 2007;27:6457–6468. doi: 10.1128/MCB.00241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babiarz JE, Halley JE, Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 2006;20:700–710. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruce K, Myers FA, Mantouvalou E, Lefevre P, Greaves I, Bonifer C, Tremethick DJ, Thorne AW, Crane-Robinson C. The replacement histone H2A.Z in a hyperacetylated form is a feature of active genes in the chicken. Nucleic Acids Res. 2005;33:5633–5639. doi: 10.1093/nar/gki874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell. Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 18.Vissers JH, Nicassio F, van Lohuizen M, Di Fiore PP, Citterio E. The many faces of ubiquitinated histone H2A: insights from the DUBs. Cell Div. 2008;3:8. doi: 10.1186/1747-1028-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai SY, Babbitt RW, Marchesi VT. A mutant deubiquitinating enzyme (Ubp-M) associates with mitotic chromosomes and blocks cell division. Proc. Natl Acad. Sci. USA. 1999;96:2828–2833. doi: 10.1073/pnas.96.6.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, Chang C, Wang H. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa T, Kajitani T, Togo S, Masuko N, Ohdan H, Hishikawa Y, Koji T, Matsuyama T, Ikura T, Muramatsu M, et al. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008;22:37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol. Cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 24.Faus H, Meyer HA, Huber M, Bahr I, Haendler B. The ubiquitin-specific protease USP10 modulates androgen receptor function. Mol. Cell. Endocrinol. 2005;245:138–146. doi: 10.1016/j.mce.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 25.de Jonge HJ, Fehrmann RS, de Bont ES, Hofstra RM, Gerbens F, Kamps WA, de Vries EG, van der Zee AG, te Meerman GJ, ter Elst A. Evidence based selection of housekeeping genes. PLoS One. 2007;2:e898. doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soncini C, Berdo I, Draetta G. Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene. 2001;20:3869–3879. doi: 10.1038/sj.onc.1204553. [DOI] [PubMed] [Google Scholar]
- 27.Boulkroun S, Ruffieux-Daidie D, Vitagliano JJ, Poirot O, Charles RP, Lagnaz D, Firsov D, Kellenberger S, Staub O. Vasopressin-inducible ubiquitin-specific protease 10 increases ENaC cell surface expression by deubiquitylating and stabilizing sorting nexin 3. Am. J. Physiol. Renal. Physiol. 2008;295:F889–F900. doi: 10.1152/ajprenal.00001.2008. [DOI] [PubMed] [Google Scholar]
- 28.Bomberger JM, Barnaby RL, Stanton BA. The deubiquitinating enzyme USP10 regulates the post-endocytic sorting of cystic fibrosis transmembrane conductance regulator in airway epithelial cells. J. Biol. Chem. 2009;284:18778–18789. doi: 10.1074/jbc.M109.001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140:384–396. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 31.Dryhurst D, Ishibashi T, Rose KL, Eirin-Lopez JM, McDonald D, Silva-Moreno B, Veldhoen N, Helbing CC, Hendzel MJ, Shabanowitz J, et al. Characterization of the histone H2A.Z-1 and H2A.Z-2 isoforms in vertebrates. BMC Biol. 2009;7:86. doi: 10.1186/1741-7007-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eirin-Lopez JM, Gonzalez-Romero R, Dryhurst D, Ishibashi T, Ausio J. The evolutionary differentiation of two histone H2A.Z variants in chordates (H2A.Z-1 and H2A.Z-2) is mediated by a stepwise mutation process that affects three amino acid residues. BMC Evol. Biol. 2009;9:31. doi: 10.1186/1471-2148-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, et al. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol. Cell. 2008;29:611–624. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 34.Gevry N, Hardy S, Jacques PE, Laflamme L, Svotelis A, Robert F, Gaudreau L. Histone H2A.Z is essential for estrogen receptor signaling. Genes Dev. 2009;23:1522–1533. doi: 10.1101/gad.1787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farris SD, Rubio ED, Moon JJ, Gombert WM, Nelson BH, Krumm A. Transcription-induced chromatin remodeling at the c-myc gene involves the local exchange of histone H2A.Z. J. Biol. Chem. 2005;280:25298–25303. doi: 10.1074/jbc.M501784200. [DOI] [PubMed] [Google Scholar]
- 36.Santisteban MS, Kalashnikova T, Smith MM. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- 37.Adam M, Robert F, Larochelle M, Gaudreau L. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 2001;21:6270–6279. doi: 10.1128/MCB.21.18.6270-6279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larochelle M, Gaudreau L. H2A.Z has a function reminiscent of an activator required for preferential binding to intergenic DNA. EMBO J. 2003;22:4512–4522. doi: 10.1093/emboj/cdg427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicassio F, Corrado N, Vissers JH, Areces LB, Bergink S, Marteijn JA, Geverts B, Houtsmuller AB, Vermeulen W, Di Fiore PP, et al. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr. Biol. 2007;17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 41.Jaworski T. Degradation and beyond: control of androgen receptor activity by the proteasome system. Cell. Mol. Biol. Lett. 2006;11:109–131. doi: 10.2478/s11658-006-0011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 43.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 44.Heemers HV, Tindall DJ. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr. Rev. 2007;28:778–808. doi: 10.1210/er.2007-0019. [DOI] [PubMed] [Google Scholar]
- 45.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 46.Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 49.Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 50.Ruhl DD, Jin J, Cai Y, Swanson S, Florens L, Washburn MP, Conaway RC, Conaway JW, Chrivia JC. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry. 2006;45:5671–5677. doi: 10.1021/bi060043d. [DOI] [PubMed] [Google Scholar]
- 51.Monroy MA, Schott NM, Cox L, Chen JD, Ruh M, Chrivia JC. SNF2-related CBP activator protein (SRCAP) functions as a coactivator of steroid receptor-mediated transcription through synergistic interactions with CARM-1 and GRIP-1. Mol. Endocrinol. 2003;17:2519–2528. doi: 10.1210/me.2003-0208. [DOI] [PubMed] [Google Scholar]
- 52.Slupianek A, Yerrum S, Safadi FF, Monroy MA. The chromatin remodeling factor SRCAP modulates expression of prostate specific antigen and cellular proliferation in prostate cancer cells. J. Cell. Physiol. 2010;224:369–375. doi: 10.1002/jcp.22132. [DOI] [PubMed] [Google Scholar]
- 53.Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl Acad. Sci. USA. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 57.Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 58.Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausio J. Characterization of the stability and folding of H2A.Z chromatin particles: implications for transcriptional activation. J. Biol. Chem. 2001;276:41945–41949. doi: 10.1074/jbc.M108217200. [DOI] [PubMed] [Google Scholar]
- 59.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Placek BJ, Harrison LN, Villers BM, Gloss LM. The H2A.Z/H2B dimer is unstable compared to the dimer containing the major H2A isoform. Protein Sci. 2005;14:514–522. doi: 10.1110/ps.041026405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Henikoff S. Labile H3.3 + H2A.Z nucleosomes mark ‘nucleosome-free regions’. Nat. Genet. 2009;41:865–866. doi: 10.1038/ng0809-865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.