Abstract
Forum domains are stretches of chromosomal DNA that are excised from eukaryotic chromosomes during their spontaneous non-random fragmentation. Most forum domains are 50–200 kb in length. We mapped forum domain termini using FISH on polytene chromosomes and we performed genome-wide mapping using a Drosophila melanogaster genomic tiling microarray consisting of overlapping 3 kb fragments. We found that forum termini very often correspond to regions of intercalary heterochromatin and regions of late replication in polytene chromosomes. We found that forum domains contain clusters of several or many genes. The largest forum domains correspond to the main clusters of homeotic genes inside BX-C and ANTP-C, cluster of histone genes and clusters of piRNAs. PRE/TRE and transcription factor binding sites often reside inside domains and do not overlap with forum domain termini. We also found that about 20% of forum domain termini correspond to small chromosomal regions where Ago1, Ago2, small RNAs and repressive chromatin structures are detected. Our results indicate that forum domains correspond to big multi-gene chromosomal units, some of which could be coordinately expressed. The data on the global mapping of forum domains revealed a strong correlation between fragmentation sites in chromosomes, particular sets of mobile elements and regions of intercalary heterochromatin.
INTRODUCTION
Transcription in eukaryotes may be regulated at the gene level by mechanisms using adjacent DNA sequences (local regulation). Another type of regulation may involve mechanisms operating at the chromosomal domain level using distant DNA sequences (distant regulation). In Drosophila, about 200 groups of genes have been found to show similar expression profiles (1). Each group spans regions between 20 and 200 kb and shows no correlation with polytene banding patterns. The mechanisms underlying this type of coordinated expression are not yet known.
A number of years ago, we attempted to identify higher order chromosomal structures that could be excised from chromosomes during spontaneous fragmentation of chromosomes upon incubation of cells in low-melt agarose (2,3). Non-random fragmentation of eukaryotic chromosomes was detected; the corresponding domains, possessing mainly 50–250 kb DNA segments, were denoted as forum domains. Following detection of the SuUR protein, a modifier of polytene chromosomes in Drosophila and a ubiquitous marker of heterochromatin in various cell types (4,5), binding of the protein at the terminal regions of forum domains was studied. Two forum domain termini were found to bind to SuUR, and it was suggested that forum domains may correspond to units of chromosomal silencing shaped by the regular distribution of heterochromatin islands along eukaryotic chromosomes (6).
Heterochromatin regions in Drosophila are localized mainly around centromeres (pericentromeric heterochromatin, P-HC) and telomeres and scattered along chromosomes (intercalary heterochromatin, I-HC) (7). The most important components required for silencing in P-HC are SU(VAR)3–9 histone methyltransferase, which controls methylation specific for H3K9, and its interaction partner HP1 protein, which bind to methylated nucleosomes via its chromo domain (8–11). To date, the silencing mechanisms in islands of I-HC are not characterized. However, there are indications that some targets of SuUR are associated with Polycomb-group (PcG) proteins (5).
PcG protein complexes bind to chromatin and are required to establish and maintain chromatin states leading to epigenetic silencing (12–15). Polycomb repressive complex 2 (PRC2) possesses a SET domain histone methyltransferase that is specific for H3K27 and that creates the heritable repressive histone methylation marks. PRC1 recognizes these marks via the chromodomain of the Polycomb (Pc) protein and also produces epigenetic repressive H2A-ubiquitin marks via the RING1B catalytic subunit (16). Previous genome-wide profiling of binding regions of different Pc proteins and the H3K27me3 marks in Drosophila revealed regions (mainly 10–30 kb in length) associated with the Pc protein (Pc domains) and more than 200 PcG target genes (17–19). The binding sites of PcG proteins were found to colocalize to presumptive polycomb response elements (PREs), whereas H3K27 marks were found to form broad domains that include the entire transcription unit and regulatory regions (18).
Trithorax-group (trxG) proteins are important expression activators of numerous developmental genes (20). Some of them bind to specific DNA sequences called trithorax response elements (TREs), whereas others (SET domain proteins) produce histone methylation marks associated with transcriptional activation. A number of trx proteins are involved in the formation of chromatin remodeling complexes that use the energy of ATP to remodel nucleosome structure and/or position and form open chromatin structures (SWI/SNF complex, NURF complex) by reading the methylation marks produced by SET domain trxG proteins (21). The SuUR protein, which is associated with repressed I-HC regions, has homology with the bromodomain and ATPase/helicase domain of the Brahma protein, which is a component of the SWI/SNF complex (6). Similarly, some PcG and trxG proteins share a SET domain. Recent data showed that the presence of PcG proteins at a target gene does not necessarily result in gene repression and that the binding of PcG complexes is more dynamic than previously thought (22). These facts taken together with the data on close neighborhood of PREs and TREs (23) suggest that epigenetic mechanisms of repression and activation are dynamic and tightly linked and that even the functional division into PcG repressors and trxG activators may be oversimplified (24,25).
To better understand the nature of forum domains, we conducted genome-wide mapping of forum domain terminal regions. A method for rapid amplification of forum termini (RAFT) was developed, and the RAFT probes were used to map domains on Drosophila polytene chromosomes and on a genomic tiling array. Our data indicate that forum domains do not correspond to the known types of chromosomal domains (i.e. bands/interbands, Pc domains, H3K27 methylated domains or looped domains delimited by scaffold attachment regions (SARs)/matrix attachment regions (MARs) and are domains confined by small I-HC islands that separate coordinately expressed complex genetic loci or groups of genes.
MATERIALS AND METHODS
Preparation of forum domains
DNA-agarose plugs were prepared as described before (3). Schneider 2 cells were pelleted by centrifugation at 2000 rpm, washed with a phosphate-buffered saline (PBS) solution (125 mM NaCl/25 mM sodium phosphate buffer, pH 7–7.2), resuspended to a concentration of 5 × 108 cells per ml, gently mixed at 43°C with an equal volume of a 1% agarose L (LKB) in PBS solution, and distributed on a mold containing 100 µl wells. The mold was placed on ice for 2–5 min. The agarose plugs then were placed in Petri dishes containing 0.5 M EDTA (pH 9.5), 1% sodium laurylsarcosine and 1–2 mg of proteinase K solution per ml for 40–48 h at 50°C. The samples were stored at 4°C in the same solution. Each DNA-agarose plug contained about 20 µg of DNA corresponding to about 107 cells.
To test the quality of the isolated DNA, fractionation in the pulsed-field gels was conducted as described previously (3). Portions of the original DNA-agarose plugs (5–50 µl) containing 1–10 µg of DNA were used for electrophoresis without any restriction enzyme digestion. The DNA samples were run in 0.8% agarose gels on an LKB Pulsaphor system using a hexagonal electrode and switching times of 25 or 450 s.
To elute the DNA preparations, fractionation in 1% agarose conventional mini-gels was performed. One-half of a DNA-agarose plug was washed in 1 × TE three times (for 15 min each) followed by three washings in the same solution containing 17.4 µg/ml PMSF. After fractionation in the mini-gel, the ethidium bromide-stained DNA band was excised and electoeluted inside a dialysis cellulose membrane bag. After overnight dialysis without stirring against 1 l of 0.01 × TE at 4°C, the DNA was concentrated with PEG (4°C) and redialyzed.
RAFT procedure
Figure 1B illustrates the RAFT procedure. About 1.5 µg of isolated DNA (see above) were ligated with 70 ng of double-stranded oligonucleotide (25 bp long 5′-phosphorylated 5′ pCCCCTGCAGTATAAGGAGAATTCGGG 3′ oligonucleotide annealed with 26 bp long 5′ biotinylated 5′ bio-CCGAATTCTCCTTATACTGCAGGGG 3′ oligonucleotide) in 150 µl of solution containing 0.1 M NaCl, 50 mM Tris–HCl (pH 7.4), 8 mM MgCl2, 9 mM 2-mercaptoethanol, 7 µM ATP, 7.5% PEG and 40 U of T4 DNA ligase at 20 C for 16 h. After heating at 65°C for 10 min, the DNA preparation was digested with the Sau3A enzyme to shorten the forum domain to the termini attached to the ligated oligonucleotide. The selection of such termini was performed in 0.5 ml Eppendorf tubes using 300 µl of suspension containing streptavidin magnesphere paramagnetic particles (SA-PMP; Promega) according to the manufacturer’s recommendations. After extensive washing with 0.5 × SSC to remove DNA fragments corresponding to internal parts of forum domains, the forum termini (FT) DNA preparation was eluted from the SA-PMP using digestion with the EcoRI enzyme in a final volume of 50 µl (double-stranded FT). The FT were then ligated with 100 × molar excess of the double-stranded Sau3A adaptor (5′-phosphorylated 5′ pGATCGTTTGCGGCCGCTTAAGCTTGGG 3′ oligonucleotide annealed with 5′ CCCAAGCTTAAGCGGCCGCAAAC 3′ oligonucleotide). In some experiments, the FT DNA preparation was eluted from the SA-PMP via heating at 100°C for 3 min in 50 µl of 0.01 × TE (single-stranded FT). Before heating the FT, preparation was ligated with a 100 × molar excess of the double-stranded Sau3A adaptor in suspension with SA-PMP (see above). Both final DNA samples (double-stranded FT or single-stranded FT) were used for PCR amplifications. Forty-cycle PCR amplification in 30 µl of a solution containing 67 mM Tris–HCl (pH 8.4); 6 mM MgCl2; 10 mM 2-mercaptoethanol; 16.6 mM ammonium sulfate; 6.7 µM EDTA; 5 µg/ml BSA; 1 mM dNTPs; 1 µg of primer corresponding to the Sau3A adaptor (5′ CCCAAGCTTAAGCGGCCGCAAAC 3′); 1 µg of primer corresponding to the biotinilated oligonucleotide (5′ CCGAATTCTCCTTATACTGCAGGGG 3′) and 1 U of Taq polymerase was performed using Eppendorf Mastercycler Personal. Amplification conditions were 90°C for melting, 65°C for annealing and 72°C for extension for 1 min each.
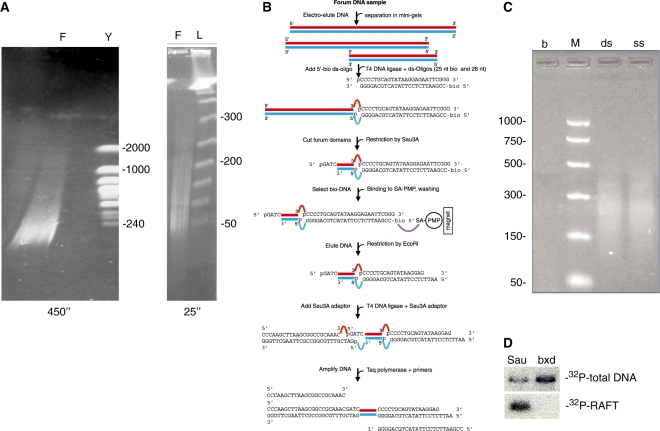
Figure 1.
RAFT procedure. (A) Elecrophoretic separation of DNA from DNA-agarose plugs containing DNA from fragmentized chromosomes. F, separation of forum domains; Y, separation of S. cerevisiae chromosomes; L, separation of lambda ladder. Numbers indicate the length of the DNA fragments in kb. The pulsed-field 1% agarose gels were run under switching times of 25 or 450 s. (B) The steps of the RAFT procedure (see ‘Materials and Methods’ section). (C) Separation of the RAFT preparations using the isolated single-stranded (ss) or double-stranded (ds) DNAs. b, amplification on distilled water; M, DNA marker, length in bp. (D) Southern blot hybridization using either a [32P]-labeled RAFT probe or a [32P]-labeled total DNA probe. Sau, the fragment from LCR of the cut locus containing PRE/TRE motifs and the fragmentation site; bxd, the fragment containing the characterized PRE from BX-C.
Labeling and hybridization of microarrays
After PCR amplification, about 4 µg of the RAFT product generated either on single-stranded FT or double-stranded FT was used for labeling with Alexa Fluor 3 using the BioPrime total genomic labeling system (Invitrogen) according to the manufacturer’s recommendations. The total DNA isolated from Schneider 2 cells was digested with the Sau3A enzyme and then labeled using the same system with Alexa Fluor 5. The specific activities (% of labeled nucleotides) reached 0.6–1.2. In four independent experiments, 2 µg of RAFT probe were mixed with 2 µg of total DNA probe and hybridized on a Drosophila genome tiling array (http://furlonglab.embl.de/methods/tools/tiling_array) using the Agilent hybridization protocol and Agilent hybridization chambers, as described previously (29). The microarray slides were scanned using a GenePix 4100A scanner. Each microarray image was analyzed with GenePix Pro 4.0 image analysis software to derive the Alexa 3 and Alexa 5 fluorescent intensity and background noise for all spots on the array. The intensities of the Alexa 3 and Alexa 5 channels were adjusted by subtracting the background intensity of each spot, and then the Alexa3/Alexa 5 signal intensity ratio was measured for each spot. Initially, spots with P < 0.01 and an Alexa 3/Alexa 5 ratio > 1.6 (log2 > 0.7) were considered to be genomic fragments containing putative FT. A total of 1330 forum domains were detected in 50% of the Drosophila genome under these cut-off criteria, and the average size of a forum domain was estimated to be 46 kb. However, the average size of fractionated domains is higher (Supplementary Figure S1). Therefore, for the remaining analysis we used a log2 > 0.85 cut-off; this generated 652 domains with an average size of 94 kb, which corresponds to the average size of fractionated domains. These 652 domains were from the 50% of the Drosophila melanogaster genome that included the entire 2L, 2R, 3R arms, Chromosome 4 and the proximal part of the X chromosome from section 11. The data from four independent experiments were processed, combined and evaluated using the CoCo program package (http://furlonglab.embl.de/coco/, EMBL) and GenePix Pro analysis software. We deposited the microarray data in the ArrayExpress database under accession number E-MEXP-2468.
FISH
A mixture containing 2 µg of the Alexa 3-labeled RAFT probe was mixed with 2 µg of the Alexa 5-labeled total DNA probe (see above) and hybridized on Drosophila polytene chromosomes of the Oregon R line. Squashes of polytene chromosome were incubated in 2 × SSC for 30 min at 70°C and then 2 × SSC for 30 min at 65°C, denatured in 2 × SSC, 0.07 N NaOH for 1.5 min, dehydrated in cold 70% ethanol for 15 min twice, rinsed in cold 96% ethanol for 10 s and then air dried. Mixed labeled probes (0.25 µg per slide) and salmon sperm DNA (10 µg per slide) were heat denatured (99°C, 3 min) and cooled in ice. Denatured DNA was added to the hybridization mix (75% formamide, 15% dextran sulfate, 3 × SSC) at 65°C to attain a final volume of 21 µl per slide. Hybridization was performed overnight (16 h) at 37°C. Unbound probe was removed with two 10 min washes in 4 × SSC at 65°C, two 15 min washes in 2 × SSC at 65°C and two 15 min washes in 0.2 × SSC at 65°C. Chromosomes on slides were stained with Hoechst 322 (0.5 μg/ml in 0.1 × SSC) at room temperature for 5 min and then dried for 10 s by air blast. Finally, 8 µl of antifade reagent VECTASHIELD (Vector Laboratories, Inc.) were added before examination using an Axioplan 2 Imaging E-mot fluorescence microscope (ZEISS). Images were created by the ISIS4 (MetaSystems GmbH) program for fluorescence analysis. Interference filters were used: N43 (ZEISS) for Alexa 3 (pseudocolor red), NSP103v1 (CHROMA) for Alexa 5 (pseudocolor green) and N49 (ZEISS) for Hoechst 322 (pseudocolor blue).
Statistical treatments
The FISH data were treated using 2 × 2 contingency table as a model for contingency testing, Cramer’s φ coefficient and Pearson chi-square test in calculating the correlation and confidence level, correspondingly. The data were obtained on analysis of distribution of the sites shown in Supplementary Table S2 in 188 bands of 3R chromosome arm (61A–100F).
RESULTS
RAFT and characterization of the amplified product
In this study, we used forum domains isolated from D. melanogaster cultured Schneider 2 cells. Figure 1A shows the pattern of separation of forum domains in pulsed-field gels under different switching times. DNA was electroeluted and ligated with annealed 26 nt long phosphorylated and 25 nt long biotinilated oligonucleotides (Figure 1B). The long domains were shortened by digestion with Sau3A endonuclease. The terminal stretches of forum domains then were purified on streptavidin paramagnetic particles, followed by strong washing, elution and ligation with the Sau3A adapter. The final product was eluted from the paramagnetic particles either in double-stranded or in single-stranded form (see ‘Materials and Methods’ section) and used for PCR. Separation of RAFT samples in agarose gel revealed that amplified DNA was mainly between 50 and 300 bp in length (Figure 1C). These fragments should correspond to the regions confined between a fragmentation site in the chromosomal DNA and the nearest Sau3A site inside a domain. Initially, analysis of the RAFT product was performed by cloning and sequencing about 45 random RAFT fragments. Supplementary Table S1 shows that about one-third of the RAFT fragments correspond to mobile elements.
We expected that random fragmentation of chromosomal DNA is inevitable during manipulation in solution with very long DNA molecules (up to 1 Mb in length). Nevertheless, we believed that the specific fragmentation sites would be concentrated at forum domain termini, whereas sites corresponding to random degradation during the procedure would be scattered along the molecules. If random degradation was not high, the RAFT probes should contain much higher specific activity of a label at the forum domain termini and could allow mapping of domains on Drosophila polytene chromosomes, on genomic microarrays or on regular Southern blots.
A [32P]-labeled RAFT probe was used for Southern blot analysis of two short DNA fragments that possessed PRE/TRE motifs from two homeotic loci: the Sau10 fragment from the distal LCR of the cut locus (26) and bxd from the bithorax complex (27). The [32P]-labeled probes corresponding to RAFT or total Drosophila DNA digested with Sau3A endonuclease were hybridized (Figure 1D). The forum domain terminus previously had been mapped inside the Sau10 fragment (our unpublished data). The data strongly indicated that the RAFT probe was enriched in forum domain termini: under the conditions used, the 268 bp Sau10 fragment efficiently hybridized only with the RAFT probe, whereas no hybridization signal was detected from the 588 bp bxd fragment with this probe. The data also indicated that the RAFT probe corresponds to the non-randomly excised chromosomal fragments.
Forum domains often are delimited by intercalary heterochromatin regions
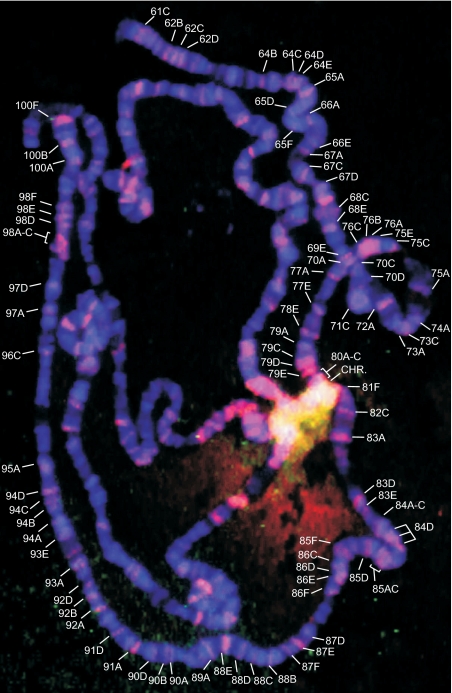
We used RAFT DNA samples or total Drosophila DNA digested with the Sau3A endonuclease that had been labeled with Alexa 3 or Alexa 5 dyes, respectively, for FISH analysis of salivary gland polytene chromosomes from Oregon-R stock. Figure 2 shows the merged image obtained after hybridization with mixture of the RAFT probe and total DNA probe. Signals from total DNA labeled by Alexa 5 (pseudocolor green) are not visible on chromosome arms but are present in chromocenter (CHR) and euheterochromatic junction regions. In these regions, Alexa 5 signals are colocalized with signals from the RAFT probe labeled by Alexa 3 (pseudocolor red). Colocalization regions are shown in yellow. Hybridization signals of the RAFT probe labeled by Alexa 3 are clearly visible in CHR and euheterochromatic junction regions, and these signals also are discretely visible on the chromosome arms. Supplementary Table S2 shows the mapping of the RAFT probe and the data on colocalization with I-HC regions. RAFT probe hybridization sites are scattered on chromosome arms and are extremely bright in the CHR and in the regions close to it. About 38% of the RAFT hybridization sites correspond to the sites where binding of the SuUR protein was described previously (28), and about 79% of RAFT hybridization sites correspond to sites where the SuUR protein binds in the line possessing four copies of SuUR. In all, 74% of regions of the late replication in polytene chromosomes hybridize with the RAFT probe in the 3L and 3R chromosome arms. As appeared, correlation was the most significant between RAFT hybridization sites and SuUR binding sites (P < 2,2E-5 for a normal line possessing two copies of SuUR and P < 2E-6 for the line possessing two extra copies of SuUR). The correlation between RAFT hybridization sites and late replication sites is rather high (P < 0.00133) (see ‘Materials and methods’ section). These data indicate that forum domain termini very often, but not always, are located in I-HC regions.
Figure 2.
FISH on Drosophila polytene chromosomes. Hybridization was performed using the Alexa 3-labeled RAFT probe mixed with the Alexa 5-labeled total DNA probe (see ‘Materials and Methods’ section). CHR, chromocenter. Hybridization sites of the RAFT probe in the 3L and 3R arms are indicated. The sites are listed in Supplementary Table S2.
Hybridization of the RAFT probes on the Drosophila melanogaster genomic tiling array
To conduct global mapping of forum domains, we used a genomic microarray created in EMBL in Furlong’s lab, EMBL (29). The microarray consists of overlapping about 35 000 spots of about 50% overlapping 3 kb DNA fragments that tile across about 40% of the Drosophila genome. The 1–10 sections of the X chromosome and the entire 3L chromosome are not covered by this array. In this analysis, we used a mixture of probes containing an Alexa 3-labeled RAFT preparation and an Alexa 5-labeled total DNA digested with the Sau3A enzyme. Four experiments were performed with independently isolated and amplified forum domain termini from Schneider 2 cells.
To test whether random fragmentation of the same chromosomes could yield a similar pattern of DNA fragments, we performed Monte Carlo simulations. Chromosomes present on the tiling array were ‘cleaved’ in random positions according to the numbers of FT actually mapped in each chromosome. Thus, we generated the random data sets in the same chromosomal regions where the real RAFT data set was observed using the same chromosome coordinates and the same number of fragments. The newly created random data sets then were compared with the real RAFT data set using the mean Pearson’s correlation coefficient and the Kolmogorov–Smirnov test. The mean Pearson’s correlation coefficient (for 10 000 experiments) was 0.010 928 (usually correlation begins to be significant at levels of 0.2–0.3), which indicated that the two data sets were not correlated. The Kolmogorov–Smirnov test for two independent groups (one was the group of randomly created data sets and the other was the real RAFT group) states that there are two different valid groups (P < 0.001). Hence, these results show that the pattern present in the RAFT data set cannot be obtained randomly. The data strongly suggest the non-randomness character of fragmentation of chromosomes giving rise to forum domains. The conclusion is in agreement with the data on separation of forum domains in the pulsed-field gels (3), Southern hybridization results and the FISH data (Figures 1D and 2, respectively).
Hox genes are located in large forum domains
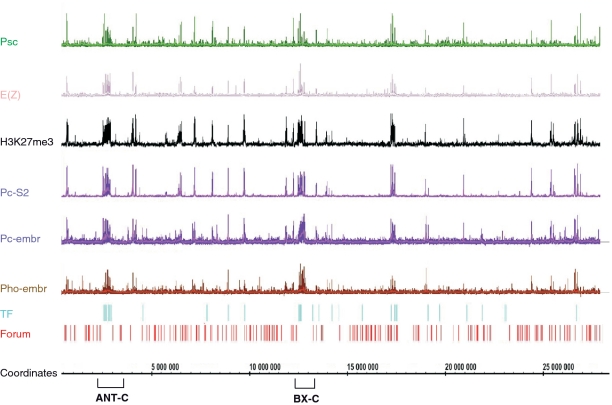
Figure 3 provides an overview of the forum domain termini mapping of chromosome 3R (overviews of other chromosomes are presented in Supplementary Figure S2). Also included in the figure is the mapping of transcription factor (TF) binding sites (30), binding profiles of Pc (a canonical member of the PRC1 complex) and binding profiles of pleiohomeotic (Pho; a DNA binding protein proposed to recruit the PRC2 complex) (31) in embryos (32). Additionally, the distributions of two other PcG proteins [E(Z) and Psc] and of the trimethylation of histone H3 Lys27 (H3K27me3) in Schneider 2 cells (18) are indicated. E(Z) is the SET domain histone methyltransferase that is specific for H3K27 in PRC2 (33). Psc is a DNA binding component of the PRC1 complex (34). As expected, FT are scattered along the chromosome. The 3R chromosome contains about 15 big forum domains (length 500–900 kb), and some of them have TF binding sites that coincide with the binding sites of different PcG proteins and H3K27me3 marks (e.g. the large forum domains possessing ANT-C and BX-C). The ability to detect very large forum domains by mapping of the RAFT probes strongly indicates that the random degradation of chromosomal DNA in our experiments did not interfere with the results.
Figure 3.
Overview of the 3R chromosome. Positions of FT along the chromosome are shown in red (Forum). The binding profiles of Psc, E(Z), Pc (in embryos and the Schneider 2 line), and Pho proteins and the distribution of H3K27me3 marks are shown (18,22). TF, transcription factor binding sites are shown in blue (30). The regions corresponding to Hox genes are indicated by the brackets.
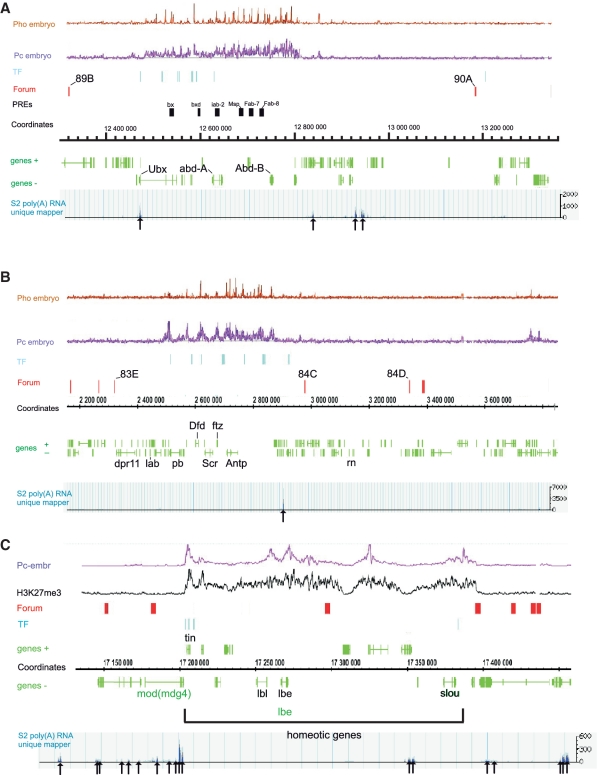
Figure 4A and B show in more detail the forum domains that possess two major Hox gene complexes in D. melanogaster: BX-C and ANT-C. BX-C is about 300 kb long, includes the Uxb, abd-A and abd-B genes, and is located inside an 860-kb forum domain. The region of the genes is covered by broad Pc domains and sharp Pho binding sites (17,32). A number of different TFs and characterized PREs are located inside this forum domain, which spans 89B–90A bands. Only some genes that are located outside of Pc domains are transcribed in S2 cells. The same organization is present inside a 540-kb domain that possesses the 300-kb homeotic ANT-C.
Figure 4.
Organization of forum domains at the Hox complexes in the 3R chromosome. Indications are the same as in Figure 3. (A) The forum domain possessing BX-C. The characterized PREs are shown by black bars. The arrows indicate positions of transcribed regions as visualized by the D. melanogaster Genome Browser using the S2 poly(A) RNA unique mapper (http://modencode.oicr.on.ca/fgb2/gbrowse/fly/). (B) The forum domains in the region of ANT-C. (C) Homeotic genes in the 90E region reside in two neighboring forum domains containing Pc domains.
Figure 4C shows two more examples of forum domains that possess homeotic genes. Two adjacent 115 and 100 kb forum domains from the 90E region contain the tin, lbl, lbe and slou genes, and a number of TFs are bound nearby the tin and slou genes. Both domains also are covered by Pc domains and by K27-methylated H3. In most chromosomes, the overlapping of FT with Pc domains was not observed. On the contrary, Pc domains were observed inside forum domains. Whole-genome analysis of Pc domains and forum domains revealed that 87% of Pc domains are located inside forum domains (SE = 3.3). These data indicate that forum domains possessing homeotic genes are repressed by PcG proteins.
Domains with a different type of organization also exist. For example, a ∼300-kb long forum domain spanning the 84C–84D region and located adjacent to one containing ANT-C includes a number of genes but does not exhibit prominent binding with Pc and Pho proteins in embryos (Figure 4B). The same is true and for the domains located around the homeotic forum domains shown in Figure 4C.
Forum domains contain coordinately expressed genes
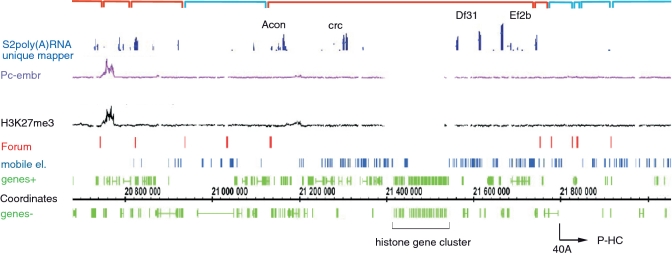
To better understand the nature of forum domains, we studied the organization of genes that are known to be coordinately regulated during development. Histone genes represent one of first discovered examples. They form a gene cluster in the 39D4 region of the 2L chromosome. Figure 5 shows that a histone gene cluster resides in a 690-kb forum domain. The data on PcG proteins binding are absent for the region corresponding to cluster itself presented by repeats. A slight peak of binding of Pc and a H3K27me3 mark exist in the distal portion of this domain. This domain in the sequenced genome line is enriched in insertions of mobile elements around the histone cluster. Four known genes (Acon, crc, Df31 and Ef2b) and six unique computed genes are actively transcribed along with the histone gene cluster in this forum domain. The co-expressed genes have different molecular functions and are involved in different biological processes. It is of interest, that close downstream of the cluster the P-HC is located, coinciding with poorly transcribed forum domains in S2 cells.
Figure 5.
Organization of forum domains at the histone gene cluster in the 2L chromosome. Indications are the same as in Figure 3. Positions of mobile elements are shown in blue. The data for Pc binding and distribution of H3K27me3 marks do not cover the site corresponding to the histone genes repeat (18,22). The red brackets indicate the forum domains possessing the clusters of actively transcribed genes in S2 cells, as visualized by the IGB using the S2 poly(A) RNA unique mapper (http://modencode.oicr.on.ca/fgb2/gbrowse/fly/). The blue brackets indicate the forum domains containing the silent or weakly expressed clusters of genes. The region corresponding to P-HC is indicated by the arrow.
In Supplementary Figure S3, the 56E region of the 2R chromosome is shown; it contains a 5S rRNA gene cluster that is transcribed by RNA polymerase I. The region has a 715-kb forum domain, in which a ∼50-kb long cluster of 5S rRNA genes are located. A ∼20-kb long Pc domain containing H3K27me3 marks in S2 cells is present about 400-kb upstream from the cluster.
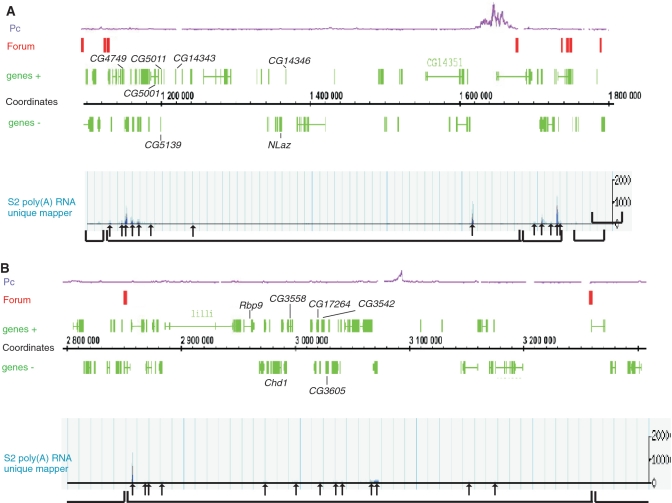
The same chromosome contains two regions for which coordinately expressed genes were described previously (1). We used the data presented in the ratiogram in Figure 2 of Spellman and Rubin (1), where two clusters containing different similarly expressed genes from the 2L chromosome are listed. We found that a coordinately expressed gene cluster in the 22A region, which contains NLaz and other genes, is located in a 500-kb forum domain (Figure 6A). Another described gene cluster from the 23C region, which contains Rbp9 and other genes, resides in a separate 400 kb forum domain (Figure 6B).
Figure 6.
Organization of forum domains at the 22A and 22C regions containing clusters of coordinately expressed genes in the 2L chromosome. Genes forming the coordinately expressed clusters are indicated by the brackets (1). The arrows indicate positions of transcribed regions as visualised by the D. melanogaster Genome Browser using S2 the poly(A) RNA unique mapper (http://modencode.oicr.on.ca/fgb2/gbrowse/fly/). (A) Cluster containing the NLaz gene. (B) Cluster containing the Rbp9 gene.
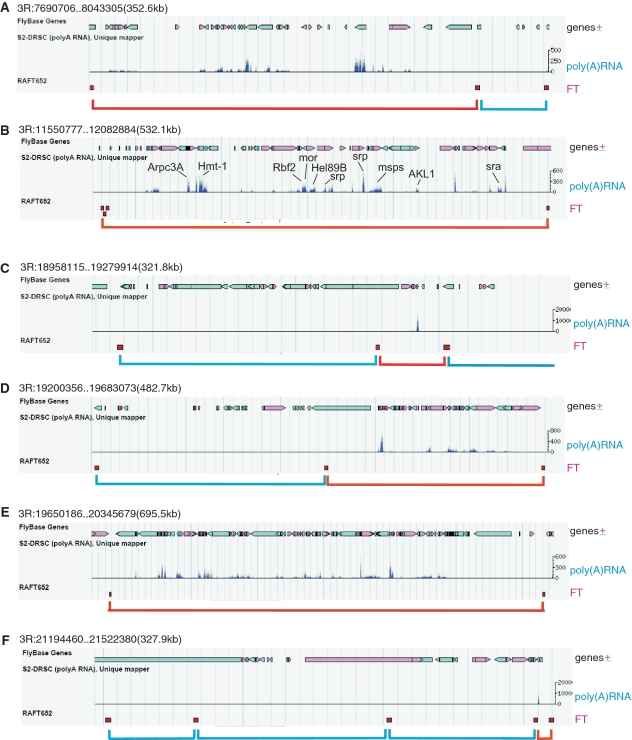
Figure 7 shows that six genomic regions in the 3R chromosome arm possess coordinately expressed genes. The forum domains shown in Figure 7B and E possess clusters of genes actively transcribed in S2 cells. In some regions, the silent or less active forum domains alternate with actively transcribed clusters of genes in the neighboring forum domains (Figure 7A, C and F). Supplementary Figure S4 presents six more regions from chromosomes 2L, 2R, X and 4. These regions also contain forum domains that have clusters of silent or actively transcribed genes with different molecular functions.
Figure 7.
Forum domains can possess the clusters of coordinately expressed genes. Six regions from the 3R chromosome are shown (A–F). The red brackets indicate the forum domains possessing the clusters of actively transcribed genes in S2 cells, as visualized by the D. melanogaster Genome Browser using the S2 poly(A) RNA unique mapper (http://modencode.oicr.on.ca/fgb2/gbrowse/fly/). The blue brackets indicate the forum domains containing the silent or weakly expressed clusters of genes. The red squares along the RAFT652 line show the positions of the mapped FT that delimit the forum domains.
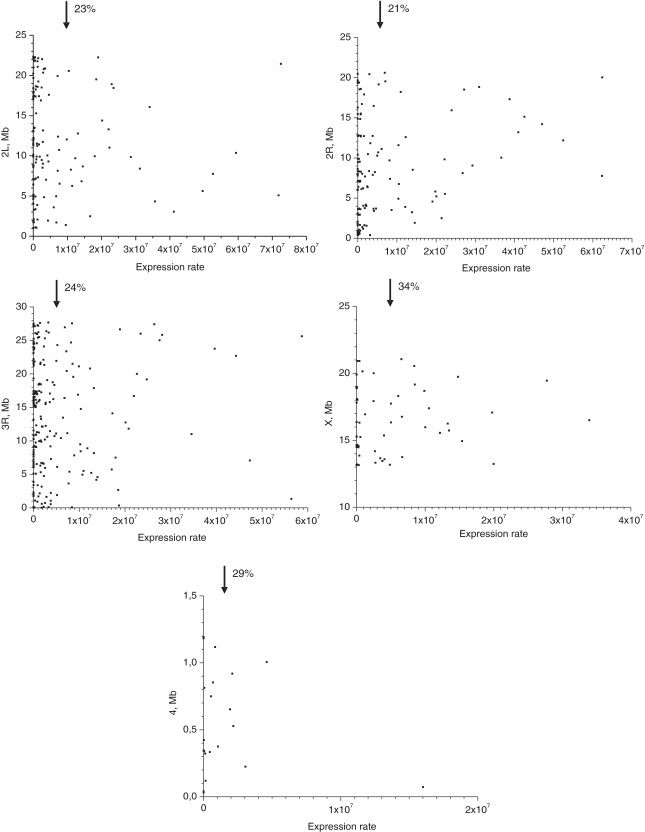
From 21% to 29% of forum domains in different chromosomes possess the clusters of coordinately expressed genes with an expression level that is higher than the average in S2 cells (Figure 8). Clearly, the major part of forum domains in all chromosomes contains the clusters of silent genes or genes with lower relative expression. Taken together, these data demonstrate that forum domains can possess coordinately regulated genes or complex genetic loci.
Figure 8.
Expression levels inside forum domains. The data for expression of all genes located inside of an individual forum domain (using the S2 poly(A) RNA unique mapper data, http://modencode.oicr.on.ca/fgb2/gbrowse/fly/) were summarized and plotted according to their coordinates along chromosomes 2L, 2R, 3R, X and 4. The arrows indicate the position of the average expression level of forum domains in a particular chromosome. The value to the right of arrow indicates the portion of forum domains that is more highly expressed. The dots on the Y axis indicate the forum domains that possess coordinately silent genes.
Forum domains and mobile elements
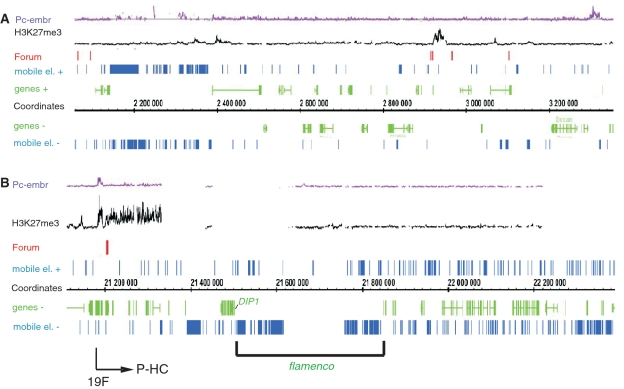
Our in situ hybridization data suggest that our RAFT probes often hybridized with the I-HC regions. Ananiev et al. (35) reported that mobile elements often are localized in these regions. About 4–9% of the D. melanogaster genome is composed of mobile elements (36,37). Of the 45 RAFT fragments we cloned, more than one-third of clones corresponded to mobile elements. Thus, it follows that chromosomal fragmentation sites are enriched with mobile elements. Analysis of the relationship between FT and mobile elements along chromosomes in the sequenced genome is difficult because the positions of many elements in the line used for genomic sequencing and in Schneider 2 cells used for RAFT preparation could differ. Nevertheless, clusters of different mobile elements in particular genomic regions exist, mainly in heterochromatin (38), that are observed in different Drosophila lines. We examined the relationship between forum domains and mobile elements in such regions. In the region 42A14, where one-third of D. melanogaster piRNAs reside (39), no FT correspond to the region occupied by fragments of different mobile elements (Figure 9A). On the contrary, the piRNA cluster is located in a large ∼800-kb forum domain. Similarly, the flamenco locus, which contains another piRNA gene cluster, has a region highly enriched by mobile elements, and this gene cluster corresponds to a huge forum >1000 kb in length (Figure 9B).
Figure 9.
Relationship between FT and mobile elements. (A and B) Large forum domains possessing the main piRNA clusters in the 42A14 (2R chromosome) and 20A4-5 (X chromosome) regions. The region corresponding to P-HC following 19F is indicated by the arrow.
Our sequencing data for 45 randomly cloned RAFT fragments show that the fragmentation sites often correspond to mobile elements. Mobile element sequences in the Alexa 3-labeled RAFT probe should have hybridized to all copies of the mobile elements represented on the array, but the corresponding signals should have been reduced because an Alexa 5-labeled total DNA was also present in the hybridization mixture. Cut-off criteria favoring selection of spots with a higher Alexa 3/Alexa 5 ratio probably removed the FT corresponding to mobile elements. This is why we did not observe FT in big regions containing clusters of different mobile elements (Figure 9A and B). On the other hand, some RAFT fragments could possess a part of a unique sequence and a part corresponding to the 5′- or 3′-end of a mobile element. This scenario offers the potential to map FT corresponding to insertion sites of mobile elements. Therefore, genomic arrays have a limitation for analysis of FT possessing scattered repetitive sequences. Thus, to better clarify the relations between fragmentation sites and mobile elements, we performed extensive analysis of the RAFT fragments using deep sequencing (these data will be published separately).
The data on separation of forum domains in the pulsed-field gels strongly indicate that no domains are present in the region below 50 kb (3). This result, along with previously collected data showing that the domains are located very close to each other in clusters of different mobile elements (38), suggest that only particular sets of mobile elements correspond to FT. Although mobile elements are preferred targets during fragmentation of chromosomes and more than one-third of cloned FT reside inside different mobile elements, long clusters of mobile elements located in large forum domains also exist. Consequently, the chromosomal context is important for the destination of a particular mobile element as a fragmentation target. Our recent data on deep sequencing of the RAFT fragments support the presence of a strong link between FT and mobile elements (these data will be published separately).
A link between FT and small RNAs
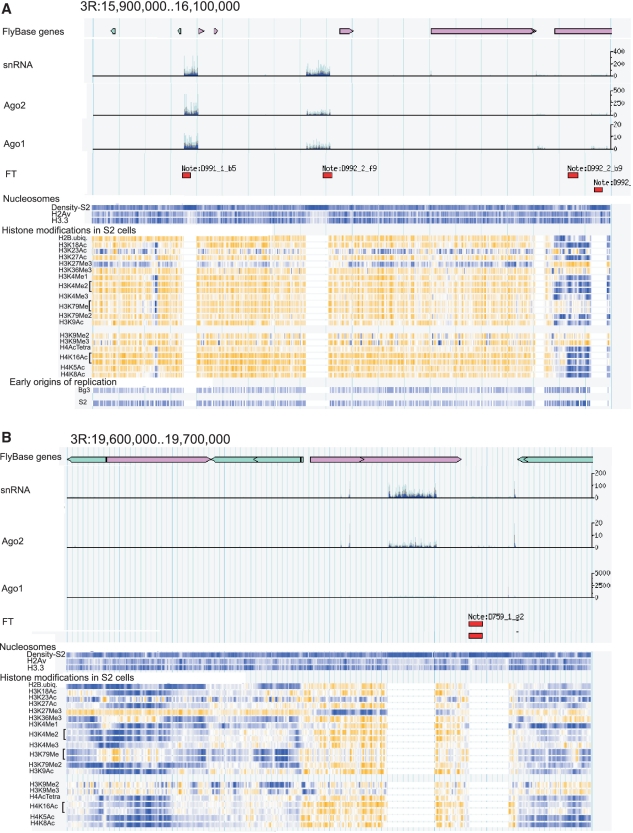
Our data suggesting that FT often correspond to I-HC regions encouraged us to make a comparison between some recently available epigenetic features in D. melanogaster chromosomes and the pattern of FT. We used the data obtained from genome-wide profiling of small non-coding 19–24 nt RNAs (snRNAs), Ago1 and Ago2 proteins, nucleosomes and chromatin marks, including some histone modifications, variants and early replication sites. Whole-genome analysis revealed that about 20.1% of FT are located exactly in or close to the regions where snRNAs, Ago1 and Ago2 are present inside rather short 3–10 kb chromosomal regions (Kendall’s τ = 0.9837). Figure 10A presents an example inside a 200-kb region in chromosome 3R, where a central 50 kb forum domain is delimited by both FT and small RNA-guided sites corresponding to non-coding regions. These 5–10 kb sites in S2 cells also possess high-density nucleosomal arrays, have a low content or even absence of H2Av and H3.3 histone variants and modified histones H3 and H4, and lack early replicated sites. We denoted such ‘holes’ in chromatin landscapes as heterochromatin islands or HIs. Supplementary Figure S5 shows an interesting example of HIs: four FT corresponding to the region of αγ-element and sharp signals coming from the ncRNA probe. In some cases, FT correspond to such HIs without detectable signals coming from RNAi-related protein complexes or the snRNA probe (the rightmost FT in Figure 10A, FT in Figure 10B and FT in Supplementary Figure S6). The data shown in Supplementary Figure S6 demonstrate that HIs are also depleted in 19 different chromatin proteins, including insulator proteins, HP1, Pc, GAF and sites of early replication. There are also many examples of HIs that are marked only by Ago1, Ago2 and snRNAs (Figure 10B).
These 3–10 kb HIs mostly correspond to sites where different mobile elements were found in different stocks inside genes or in the intergenic regions, and the mapped FT overlap with their sequences and/or genomic sequences in the integration sites (data not shown). As far as we mapped FT inside 3 kb regions, we cannot be more accurate in estimation of this overlapping. For more detailed analysis of relationships among FT, mobile elements and their integration sites, we will use the deep sequencing data (this work is in progress). Nevertheless, our currently available results suggest that a specific subset of FT and siRNA- or piRNA-guided DNA targets form HIs that are scattered along chromosomes.
Figure 10.
Relationships between FT, regions containing sequences of small non-coding 19–24 nt RNAs (snRNAs), Ago1 and Ago2 binding sites, nucleosomes and histone modifications in S2 cells (A–B). The Modencode genome browser was used (http://modencode.oicr.on.ca/fgb2/gbrowse/fly/) with the loaded tracks for snRNAs [‘S2-DRSC(Rubin)’, S2 smallRNA RNA-Seq (Lai project, Lai subgroup), GEO accession number GSM361908] and Ago1 and Ago2 [S2 cells Ago1-HA Immunoprecipitation (Lai project, Hannon subgroup), GEO accession numbers GSM280088 and GSM280087, respectively]. The data for nucleosomes and histone modifications correspond to the tracks referred to as ‘HenikoffNUCL:70001’ and ‘Karpen_HISMODS_S2:70001’, respectively.
DISCUSSION
Forum domains are excised form chromosomes as result of non-random fragmentation
Our data on Southern hybridization experiment with [32P]-labeled RAFT probe and FISH (Figure 1D and Figure 2, respectively), as well as the previously published data on separation of forum domains in the pulsed-field gels (3), strongly agues in favor of non-random character of fragmentation of DNA during excision of forum domains from chromosomes. Microarray data confirm this conclusion.
As far as the procedure used for isolation of domains starts with only few minutes incubation in PBS solution and immediately follows by the strongest possible treatment (0.5 M EDTA, 1% SDS and 1–2 mg/ml proteinase K at 50°C, see ‘Materials and Methods’ section), we believe that the fragmentation takes place in cells just before lysis, because it is difficult to imagine that some enzyme could digest DNA in the presence of concentrated EDTA, SDS and proteinase K reagents, quickly penetrating into thin 0.5% agarose plugs at 50°C. Thereafter, fragmentation sites should reflect the pattern of specific sites in chromosomes that are attacked first upon degradation of chromosomes and thus could reflect some feature in chromosomal organization. From this point of view, both a specific periodic distribution of some putative large chromosomal structures protecting DNA from degradation and rather small chromatin regions containing DNA sequences vulnerable for degradation are important as a novel information concerning organization of chromosomes. Really, presence of both protected and unprotected regions might reflect specific features in organization of chromosomes. Forum domains and total DNA possess that same fingerprinting patterns (3), suggesting that they are covering entire genome. Our microarray data clearly confirm this conclusion.
Forum domains do not correspond to Pc domains or looped domains formed by the nuclear scaffold
Our data indicate that forum domains do not correspond to the several known types of chromosomal domains [e.g. Pc domains formed by broad binding regions of Pc PcG protein (17–19)]; polytene bands and interbands; and looped domains formed by attachment of DNA regions to the nuclear SARs/MARs (40)]. Both Pc domains and forum domains generally are 50–150 kb in length, which is why originally we thought that forum domains should correspond to this type of domain. However, our data clearly demonstrate that broad Pc-bound trimethylated at histone H3 Lys27 regions reside inside clusters of genes and inside large forum domains. Pc domains often overlap with the binding sites of different TFs (Figures 3 and 4, Supplementary Figure S2), whereas these sites are inside forum domains.
SARs very often are much smaller than forum domains (41). Our new mapping data for the 690-kb forum domain containing the histone gene cluster strongly argue in favor of this conclusion. SARs in the histone cluster form rather small 4.8 kb looped domains (42). Moreover, 86 SARs were mapped inside an 800-kb segment in the 14B–15B region of the Drosophila X chromosome (43), and this segment corresponds to four forum domains (Supplementary Figure S2, around the 16.4 Mb coordinate).
Recently, Bushey et al. (44) described genome profiling of the Su(Hw) and dCTCF insulators in Drosophila. The pattern formed by this ‘insulator code’ in Drosophila is completely different from the pattern of forum domains, as the density of insulators along chromosomes is much higher and a great number of insulators correspond to each forum domain (Supplementary Figure S6). We also found no correlation between polytene bands and forum domains (data not shown).
Forum domains and coordinately expressed chromosomal regions
About 200 groups of Drosophila adjacent genes have similar expression profiles (1). Each group contains a cluster of 10–30 functionally unrelated genes and covers from 20 to 200 kb genomic regions. The mean size of such clusters is 100 kb, which is close to the mean size of forum domains (Supplementary Figure S1). We found that two previously described groups of genes reside in two distinct forum domains. The data suggest correspondence between clusters of coordinately expressed genes and forum domains. However, only 20% of the genes in the Drosophila genome appear to fall into such clusters. Thus, it may follow that only some parts of forum domains correspond to transcriptional territories that contain clusters of similarly expressed but functionally unrelated genes (1). Independent evidence in favor of coordinated regulation inside forum domains comes from analysis of clusters containing the same genes (e.g. similarly regulated histone genes and 5S rRNA genes that are located in separate forum domains) (Figure 5 and Supplementary Figure S3).
There are 37 large forum domains (spanning more than 500 kb) in the 2L, 2R, 3R and X chromosomes. We tried to address the question of preference for particular functional categories of genes located in the large forum domains. We found that Hox genes mainly reside in large forum domains. The chromosome overviews (Figure 3 and Supplementary Figure S2) show that the essential part of forum domains contains Pc domains and TF binding sites and that these correspond to homeotic genes. This finding supports the supposition that some part of a forum domain corresponds to large developmentally regulated chromosomal units controlled by PcG/trxG complexes. We speculate that the terminal regions of forum domains provide an additional mechanism of regulation. Our preliminary data for cloned FT using reporter genetic constructs support this view (the data will be published separately).
Most of the genes located in large forum domains correspond to computed genes with unknown functions. Nevertheless, we observed that the characterized genes in the large forum domains often correspond to housekeeping genes and genes with different molecular functions that are involved in development. For example, the domain containing the histone gene cluster (Figure 5) also contains four actively transcribed genes (Acon, crc, Df31 and Ef2b) that specify proteins that are involved in the tricarboxylic acid cycle, regulation of gene-specific transcription, nucleosome assembly and mitotic spindle organization, respectively. The same is true for the forum domain containing 5S rRNA genes and ribosomal proteins (Supplementary Figures S3 and S4). Similarly, the 532-kb long forum domain shown in the Figure 7B contains 10 characterized actively expressed genes (Arpc3A, Hmt-1, Rbf2, mor, Hel89B, srp, pnr, msps, Akt1, sra), most of which specify proteins involved in protein binding and in different biological processes of development.
Chromosomal fragmentation sites correspond to I-HC regions dispersed along eukaryotic chromosomes
It has been demonstrated that two cloned FT bind with SuUR protein in vitro (6). Our data for in situ hybridization of the RAFT probes on the polytene chromosomes strongly support this premise. The correspondence with the binding sites of SuUR antibodies in the line possessing two extra copies of SuUR gene is high—about 80%. About 74% of the hybridization sites of the RAFT probes also corresponded to regions of late replication, and sites of late replication and the binding sites of SuUR protein on polytene chromosomes are known to correspond to I-HC regions (28,45,46).
I-HC regions display chromosomal weak points or breaks, a high frequency of ectopic contacts and late replication sites and are enriched with mobile elements (7,35,47,48). These regions even can provoke the position effect in genes transposed into these regions by chromosomal rearrangements (49), and this phenomenon is known as the chromosomal position effect (CPE) (50). The CPE is connected with alterations of transgene expression that are associated with a distinct insertion into the epigenome milieu (51). The more extensively studied position effect variegation (PEV) is defined as the variegated pattern of expression from cell to cell when a gene is translocated into the proximity of dominant heterochromatin (52). Upon CPE and PEV, a gene’s expression is affected by distant changes in chromatin conformation, including histone deacetylation, H3K9me3 and others. In studies focused on understanding CPE phenomena, chromatin insulators have been used to protect transgenes from CPE (53,54).
In this study, we used several cloned FT in the reporter genetic constructs. Our data suggest that DNA sequences at FT could repress or activate the expression of a reporter gene and that the Su(Hw) insulator could protect a reporter gene from these FT effects. Using the 3C approach, we also found that FT can interact with distant promoters (these results will be published separately). These data are in agreement with our speculation that FT can provide additional distant regulation of gene expression in large chromosomal regions by an as yet unknown mechanism. The properties of I-HC (weak points, ectopic contacts, late replication, presence of mobile elements) clearly correlate with the properties of FT described here (fragmentation sites, 3C looping properties, SuUR colocalization and enrichment with mobile elements). In any case, our in situ hybridization data suggest that the essential parts of chromosomal fragmentation sites do correspond to I-HC regions and, thereafter, this type of domains is formed by regular distribution of I-HC islands along chromosomes. Recently, the Hi-C approach for mapping the dynamic conformations of a whole genome was described (55). The method detects the folding of each chromosome by long-range intrachromosomal looping interactions that form a distinct chromosome territory. SuUR mutation was found to dramatically reduce the frequency of ectopic contacts between I-HC regions in polytene chromosomes (56,57). Therefore, we speculate that I-HC regions and FT, where the SuUR protein binds, are good candidates for elements involved in chromosome folding.
Relations between fragmentations sites and human fragile sites
Many years ago, it was suggested that the I-HC of Drosophila is a good model to study fragile sites in human chromosomes (58). A fragile X syndrome is associated with delayed replication of the FMR1 gene (59). Others demonstrated that the ATR gene is required for maintenance of the stability of fragile sites, that mutations in the gene lead to chromosomal fragmentation, and that caspase-independent chromosome breaks occur prior to widespread apoptosis, suggesting that apoptosis is caused by a loss of genomic integrity (60,61). The possibility that spontaneous non-random fragmentation leading to excision of forum domains has the same nature as the fragmentation observed in ATR(–/–) cells cannot be excluded. These facts may indicate that fragile sites and FT have at least two properties in common: the association with fragmentation sites and the late replication. In any case, the possibility of identifying and isolating FT biochemically using the RAFT approach provides a new tool for studying the properties of fragile sites and I-HC at the molecular level.
The nature of the enzyme that cleaves chromosomal DNA at FT regions remains of interest. We analyzed 29 sites within the random RAFT clones (Supplementary Table S1) and found that no site corresponds to the consensus sequence of Drosophila topoisomerase II—GTNA/TAC/T ATTNATNNA/G (62). Thus, at present we cannot explain the nature of this enzyme. Deep sequencing data from RAFT preparations could address this and other questions about forum domains. The possibility that some RNA-mediated mechanisms could recognize sequences in I-HC regions cannot be excluded.
Forum domains and mobile elements
Mobile elements are believed to serve as nucleation sites in silent chromatin formation (63,64). Our data indicate that FT often correspond to mobile elements. Among 45 random RAFT clones, 17 corresponded to different mobile elements, mainly to LTR elements (Supplementary Table S1). This association is not simply due to the high content of mobile elements in the Drosophila genome. In our FISH and microarray experiments, we hybridized simultaneously the RAFT and total DNA probes, and the final results revealed higher hybridization ratios of the RAFT probes. Moreover, long forum domains possess clusters of mobile elements (Figures 5 and 9A, B). Thus, the following question arises: why were no FT detected inside the 690-kb histone gene domain, in the 800–1000-kb flamenco domain or in the 42A14 piRNA domain, all of which are enriched with mobile elements? How this could be possible? One explanation is that the sequenced Drosophila line, used for the Integrated Genome Browser (IGB) presentations, and the Schneider 2 line, used for preparation of the RAFT probes, have the same mobile elements in particular genomic regions. In other words, there may be genomic regions in which mobile elements are always present in different Drosophila lines (e.g. in piRNA clusters). Some of these regions belong to I-HC and others do not. We surmise that regularly dispersed I-HC regions form a specific chromosomal regulatory framework. Copies of LTR elements and long interspersed nuclear elements (LINEs) remain unmobilized in the integration sites. Thereafter, the most ancient integrated copies should occupy the same sites in different Drosophila lines. We speculate that cellular mechanisms that originally evolved for repression of mobile elements by formation of silent chromatin structures were used for dynamic regulation of the host genes. In this way, some of the repressed areas containing mobile elements perpetuated the heterochromatin marks triggering epigenetic modifications and became I-HC. However, it is also possible that mobile elements were guided by some unknown cellular mechanisms predominantly to the already existing sites of HC and I-HC, which resulted in their repression in chromosomes.
The presence of 3–10 kb HIs that are marked by siRNAs, piRNAs and FT suggests that RNAi-related mechanisms play an important role in shaping the I-HC in Drosophila. siRNA-dependent or miRNA-dependent HC formation was described previously in fungi and plants (65–67). From their study of mutation of AGO2 or piwi in D. melanogaster, Moshkovich and Lei (68) concluded that HC forms independently of the endo-siRNA and piRNA pathways. The data shown in Figure 10 and Supplementary Figure S5 strongly suggest that siRNAs and/or piRNAs are involved in the formation of HIs in Drosophila. However, careful analysis of individual HI sites must be conducted.
It is of interest to study the evolutionary plasticity and conservation of forum domains. Internal regions of forum domains are conserved, as illustrated by the observation that the genes located in D. melanogaster forum domains have orthologous genes across Drosophilids and non-Drosophilidae organisms, including pea aphids, mosquitos and silk worm (Supplementary Figure S7). Our preliminary analysis of sequences of several FT sequences collected from the deep sequencing reads of D. melanogaster RAFT samples shows that FT are conserved in the D. melanogaster group (one example is shown in Supplementary Figure S8). To address this question in more detail, we plan to study deep sequencing results (this work is in progress).
Recently, five principal chromatin types were described in the D. menalogaster Kc167 cell line (69). These data suggest that rather large regions covered by specific combinations of proteins exist in Drosophila chromosomes. In total, 8428 domains were identified. The domain types, defined as repressed BLUE, GREEN and BLACK chromatin and active RED and YELLOW chromatin, have a median length of 6.5 kb. It follows that much larger forum domains should contain different types of color domains. Direct comparison of forum domain and color domain patterns inside a 1-Mb region from the 2L chromosome confirmed this supposition (Supplementary Figure S9A). However, about 600 color domains (mainly BLACK, GREEN and YELLOW) span more than 50 kb each, and many of these large individual color domains include multiple neighboring genes [Filion et al. (69)]. Thus, it seems that the large color domains correspond to the forum domains in which they reside. However, we observed no correlation between the borders of large color domains and FT (Supplementary Figure S9B) and therefore conclude that forum domains contain different types of color domains. We currently are studying the binding of nuclear proteins with individual FT and total RAFT preparations from Drosophila and human cells. Our data suggest that forum domains are shaped by the distribution of particular genomic sequences binding with specific proteins that are not present among 53 broadly selected chromatin components that shape color domains (work in progress).
At present, we can only speculate about the nature of the forum domains that are characteristic of all tested eukaryotes in somatic cells and in germ lines (3). Taken together with the available data on genome-wide profiling of forum domains, the correspondence of FT to I-HC regions and their enrichment with mobile elements suggest that they are involved in the regulation of expression in the large chromosomal regions. The central point in understanding of the nature of the forum domains is the nature of this type of regulation, which controls mainly 50–150 kb segments. We believe that our current analysis of the looping properties of FT could address some questions raised by this study.
ACCESSION NUMBER
E-MEXP-2468.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Foundation for Basic Research (grants No. 08-04-00058-a, 09-04-12059-ofi_m, 11-04-00091). Funding for open access charge: Russian Foundation for Basic Research (grant No. 09-04-12059-ofi_m).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Eileen Furlong (EMBL) for providing the tiling arrays and valuable advice, and members of your lab, Martina Braun and Charles Girardot, for array hybridizations and initial processing of the raw data, respectively. We thank Robert White for sharing the data on Pc targets in embryos; Vincenzo Pirrotta for sharing the data obtained in S2 cells; Bas van Steensel, Bas Tolhius and Maarten van Lohuizen for sharing the data on Pc domains; Alexey Pindyurin for sharing data on SuUR and HP1 targets; and Dmitri Petrov and Anna-Sophie Fiston-Lavier for sharing data on mobile elements. The authors are very grateful to Dr V.N. Babenko for help with the statistical treatments of the in situ hybridization results and to E.D. Moiseeva for help with the cloning experiments. Conceived and designed the experiments: N.A.T. Performed experiments: N.A.T., O.V.K., D.V.S., I.A.Z., I.F.Z. Analyzed the data: N.A.T., Y.V.K. Wrote the paper: N.A.T.
REFERENCES
- 1.Spellman PT, Rubin GM. Evidence for large domains of similarly expressed genes in the Drosophila genome. J. Biol. 2002;1:5. doi: 10.1186/1475-4924-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tchurikov NA, Ponomarenko NA, Airich LG. Isolation and characterization of specific fraction of human chromosomal DNA – forum DNA. Dokl. Akad. Nauk USSR. 1988;303:491–493. [PubMed] [Google Scholar]
- 3.Tchurikov NA, Ponomarenko NA. Detection of DNA domains in Drosophila, human and plant chromosomes possessing mainly 50- to 150-kilobase stretches of DNA. Proc. Natl Acad. Sci. USA. 1992;89:6751–6755. doi: 10.1073/pnas.89.15.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belyaeva ES, Boldyreva LV, Volkova EI, Nanaev RA, Alekseenko AA, Zhimulev IF. Effect of the Suppressor of Underreplication (SuUR) gene on position-effect variegation silencing in Drosophila melanogaster. Genetics. 2003;165:1209–1220. doi: 10.1093/genetics/165.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pindyurin AV, Moorman C, Wit E, Belyakin SN, Belyaeva ES, Christophides GK, Kafatos FC, Steensel B, Zhimulev IF. SuUR joints separate sunsets of PcG, HP1 and B-type lamin targets in Drosophila. J. Cell Sci. 2007;120:2344–2351. doi: 10.1242/jcs.006007. [DOI] [PubMed] [Google Scholar]
- 6.Tchurikov NA, Kretova OV, Chernov BK, Golova YB, Zhimulev IF, Zykov IA. SuUR protein binds to the boundary regions separating forum domains in Drosophila melanogaster. J. Biol. Chem. 2004;279:11705–11710. doi: 10.1074/jbc.M306191200. [DOI] [PubMed] [Google Scholar]
- 7.Kaufmann BP. Distribution of induced breaks along the X chromosome of Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 1939;25:571–577. doi: 10.1073/pnas.25.11.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Re S, Jenuwein T, Reuter G. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craig JM. Heterochromatin – many flavours, common themes. BioAssays. 2005;27:17–28. doi: 10.1002/bies.20145. [DOI] [PubMed] [Google Scholar]
- 10.Ebert A, Schotta G, Lein S, Kubicek S, Krauss V, Jenuwein T, Reuter G. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev. 2004;18:2973–2983. doi: 10.1101/gad.323004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greil F, van der Kraan I, Delrow J, de Wit E, Bussemaker H, van Dreil R, Henikoff S, van Steensel B. Distinct HP1 and Su(var)3-9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Genes Dev. 2003;17:2825–2838. doi: 10.1101/gad.281503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 13.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 14.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 16.Fang J, Chen T, Chadwick B, Li E, Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J. Biol. Chem. 2004;279:52812–52815. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- 17.Tolhuis B, Muijrers I, de Wit E, Teunissen H, Talhout W, van Steensel B, van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 2006;38:599–727. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz YB, Kahn TG, Nix DA, Li X-Y, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 2006;38:599–727. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 19.Negre N, Hennetin J, Lavrov S, Bellis M, White KP, Cavalli G. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006;4:e170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orlando V. Polycomb, epigenomes, and control of cell identity. Cell. 2003;112:599–606. doi: 10.1016/s0092-8674(03)00157-0. [DOI] [PubMed] [Google Scholar]
- 21.Bantignies F, Cavalli G. Cellular memory and dynamic regulation of polycomb group proteins. Curr. Opin. Cell Biol. 2006;18:275–283. doi: 10.1016/j.ceb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Kwong C, Adryan B, Bell I, Meadows L, Russel S, Manak R, White R. Stability and dynamics of Polycomb target sites in Drosophila development. PLoS Genet. 2008;4:e1000178. doi: 10.1371/journal.pgen.1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ringrose L, Rehmsmeier M, Dura JM, Paro R. Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Dev. Cell. 2003;5:759–771. doi: 10.1016/s1534-5807(03)00337-x. [DOI] [PubMed] [Google Scholar]
- 24.Grosby MA, Miller C, Alon T, Watson KL, Verrijzer CB, Goldman-Levi R, Zak NB. The trithorax group gene moira encodes a brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol. Cell. Biol. 1999;19:1159–1170. doi: 10.1128/mcb.19.2.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon JA, Tamkun JW. Programming off and on states in chromatin: mechanisms of Polycomb and trithorax group complexes. Curr. Opin Genet. Dev. 2002;12:210–218. doi: 10.1016/s0959-437x(02)00288-5. [DOI] [PubMed] [Google Scholar]
- 26.Tchurikov NA, Pavlova GV, Korochkin LI, Krasnov AN, Shostak NG. Detection of an extended protein-binding domain in the ct6-enhancer region of the Drosophila cut locus. Dokl. Akad. Nauk. 1998;363:835–838. (Russian) [PubMed] [Google Scholar]
- 27.Fritsch C, Brown JL, Kassis JA, Müller J. The DNA-binding polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development. 1999;126:3905–3913. doi: 10.1242/dev.126.17.3905. [DOI] [PubMed] [Google Scholar]
- 28.Belyakin SN, Christophides GK, Alekseyenko AA, Kriventseva EV, Belyaeva ES, Nanayev RA, Makunin IV, Kafatos FC, Zhimulev IF. Genomic analysis of Drosophila chromosome underreplication reveals a link between replication control and transcriptional territories. Proc. Natl Acad. Sci. USA. 2005;102:8269–8274. doi: 10.1073/pnas.0502702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandmann T, Jensen LJ, Jakobsen JS, Karzynski MM, Eichenlaub MP, Bork P, Furlong EE. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev. Cell. 2006;10:797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 30.Bergman CM, Carlson JW, Celniker SE. Drosophila DNase I footprint database: a systematic genome annotation of transcription factor binding sites in the fruitfly, Drosophila melanogaster. Bioinformatics. 2005;21:1747–1749. doi: 10.1093/bioinformatics/bti173. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jone RS. Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Kwong C, Adryan B, Bel I, Meadows L, Russell S, Manak JR, White R. Stability and dynamics of polycomb target sites in Drosophila development. PLoS Genet. 2008;4:e1000178. doi: 10.1371/journal.pgen.1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 34.King IF, Emmons RB, Francis NJ, Wild B, Müller J, Kingston RE, Wu CT. Analysis of a polycomb group protein defines regions that link repressive activity on nucleosomal templates to in vivo function. Mol. Cell. Biol. 2005;25:6578–6691. doi: 10.1128/MCB.25.15.6578-6591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ananiev EV, Gvozdev VA, Ilyin YV, Tchurikov NA, Georgiev GP. Reiterated genes with varying location in intercalary heterochromatin regions of Drosophila melanogaster polytene chromosomes. Chromosoma. 1978;70:1–17. doi: 10.1007/BF00292211. [DOI] [PubMed] [Google Scholar]
- 36.Spradling AC, Rubin GM. Drosophila genome organization: conserved and dynamic aspects. Annu. Rev. Genet. 1981;15:219–264. doi: 10.1146/annurev.ge.15.120181.001251. [DOI] [PubMed] [Google Scholar]
- 37.Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM, et al. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 2002;3:RESEARCH0084. doi: 10.1186/gb-2002-3-12-research0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tchurikov NA, Zelentsova ES, Georgiev GP. Clusters containing different mobile dispersed genes in the genome of Drosophila melanogaster. Nucleic Acids Res. 1980;8:1243–1258. doi: 10.1093/nar/8.6.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 40.Gasser SM, Laemmli UK. The organisation of chromatin loops: characterization of a scaffold attachment site. EMBO J. 1986;5:511–518. doi: 10.1002/j.1460-2075.1986.tb04240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tchurikov NA, Krasnov AN, Ponomarenko NA, Golova YB, Chernov BK. Forum domains in Drosophila melanogaster cut locus possesses looped domains inside. Nucleic Acids Res. 1998;26:3221–3227. doi: 10.1093/nar/26.13.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mirkovitch J, Mirault M-E, Laemmli UK. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- 43.Surdej P, Brandli D, Miassod R. Scaffold-associated regions and repeated or cross-hybridizing sequences on an 800 kilobase DNA stretch of the Drosophila X chromosome. Biol. Cell. 1991;73:111–120. doi: 10.1016/0248-4900(91)90093-3. [DOI] [PubMed] [Google Scholar]
- 44.Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009;23:1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moshkin YM, Belyakin SN, Rubtsov NB, Kokoza EB, Alekseyenko AA, Volkova EI, Belyaeva ES, Makunin IV, Spierer P, Zhimulev IF. Microdissection and sequence analysis of pericentric heterochromatin from the Drosophila melanogaster mutant Suppressor of Underreplication. Chromosoma. 2002;111:114–125. doi: 10.1007/s00412-002-0190-8. [DOI] [PubMed] [Google Scholar]
- 46.Zhimulev IF, Belyaeva ES, Makunin IV, Pirrotta V, Volkova EI, Alekseyenko AA, Andreyeva EN, Makarevich GF, Boldyreva LV, Nanayev RA, et al. Influence of the SuUR gene on intercalary heterochromatin in Drosophila melanogaster polytene chromosomes. Chromosoma. 2003;111:377–398. doi: 10.1007/s00412-002-0218-0. [DOI] [PubMed] [Google Scholar]
- 47.Hannah A. Localization and function of heterochromatin in Drosophila melanogaster. Adv. Genet. 1951;4:87–125. doi: 10.1016/s0065-2660(08)60232-1. [DOI] [PubMed] [Google Scholar]
- 48.Kaufmann B, Iddles MK. Ectopic pairing in salivary gland chromosomes of Localization and function of heterochromatin in Drosophila melanogaster. Portugaliae Acta Biol. 1963;7:225–248. [Google Scholar]
- 49.Spofford JB. Position-effect variegation in Drosophila. Genet. Biol. Drosophila. 1976;1:955–1018. [Google Scholar]
- 50.Wakimoto BT. Beynd the nucleosome: epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- 51.Recillas-Targa F, Valadez-Graham V, Farrell CM. Prospects and implications of using chromatin insulators in gene therapy and transgenesis. Bioessays. 2004;26:796–807. doi: 10.1002/bies.20059. [DOI] [PubMed] [Google Scholar]
- 52.Perrod S, Gasser SM. Long-range silencing and position effects at telomeres and centromeres: parallels and differences. Cell Mol. Life. 2003;60:2303–2318. doi: 10.1007/s00018-003-3246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rincón-Arano H, Furlan-Magaril M, Recillas-Targa F. Protection against telomeric position effects by the chicken cHS4 β-globin insulator. Proc. Natl Acad. Sci. USA. 2007;104:14044–14049. doi: 10.1073/pnas.0704999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Majumder P, Roy S, Belozerov VE, Bosu D, Puppali M, Cai HN. Diverse transcription influences can be insulated by the Drosophila SF1 chromatin boundary. Nucleic Acids Res. 2009;37:4227–4233. doi: 10.1093/nar/gkp362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belyaeva ES, Zhimulev IF, Volkova EI, Alekseyenko AA, Moshkin YM, Koryakov DE. Su(UR)ES: a gene suppressing DNA underreplication in intercalary and pericentric heterochromatin of Drosophila melanogaster polytene chromosomes. Proc. Natl Acad. Sci. USA. 1998;95:7532–7537. doi: 10.1073/pnas.95.13.7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moshkin YM, Alekseyenko AA, Semeshin VF, Spierer A, Spierer P, Makarevich GF, Belyaeva ES, Zhimulev IF. The bithorax complex of Drosophila melanogaster: underreplication and morphology in polytene chromosomes. Proc. Natl Acad. Sci. USA. 2001;98:570–574. doi: 10.1073/pnas.021353598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laird CD, Lamb MM. Intercalary heterochromatin of Drosophila as a potential model for human fragile sites. Am. J. Med. Genet. 1988;30:689–691. doi: 10.1002/ajmg.1320300170. [DOI] [PubMed] [Google Scholar]
- 59.Hansen RS, Canfield TK, Lamb MM, Gartler SM, Laird CD. Association of fragile X syndrome with delayed replication of tha FMR1 gene. Cell. 1993;73:1403–1409. doi: 10.1016/0092-8674(93)90365-w. [DOI] [PubMed] [Google Scholar]
- 60.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 61.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 62.Sander M, Hsieh T. Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985;13:1057–1072. doi: 10.1093/nar/13.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tchurikov NA. Molecular mechanisms of epigenetics. Biochemistry. 2005;70:406–423. doi: 10.1007/s10541-005-0131-2. [DOI] [PubMed] [Google Scholar]
- 64.Gao X, Hou Y, Ebina H, Levin HL, Voytas DF. Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res. 2008;18:359–369. doi: 10.1101/gr.7146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez-Pulido L, Hoe KL, Kim DU, Park HO, Ponting CP, Rappsilber J, et al. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010;140:666–677. doi: 10.1016/j.cell.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.WuL., Zhou H, Zhang Q, Zhang J, Ni F, Chang Liu C, Qi Y. DNA Methylation mediated by a microRNA pathway. Mol. Cell. 2010;38:465–475. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 67.Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr. Opin. Genet. Dev. 2010;20:134–141. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moshkovich N, Lei EP. HP1 Recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet. 2010;6:e1000880. doi: 10.1371/journal.pgen.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.