Abstract
A mutant of Streptococcus uberis carrying a single copy of ISS1 within pauA was unable to activate bovine plasminogen. Contrary to a hypothesis postulated previously, this mutation did not alter the ability of the bacterium to grow in milk or to infect the lactating bovine mammary gland.
Clinical bovine mastitis due to intramammary infection with Streptococcus uberis is common throughout the world. One review (11) cited four surveys conducted in Ontario, Canada; Ohio; The Netherlands; and the United Kingdom which indicated that S. uberis was responsible for about 26% of all clinical cases. Another study estimated that the level in the United Kingdom may be as high as 33% (6).
Following experimentally induced infection of the lactating mammary gland, S. uberis is found predominantly in the lumenal areas of secretory alveoli and ductular tissue (21), indicating that much of the bacterial growth occurs in residual and newly synthesized milk. This environment is likely to be deficient in free and peptide-associated amino acids (1). Streptococci, because of the nature of their metabolic biochemistry, are nutritionally fastidious, and members of the genus Streptococcus that cluster with the pyogenic streptococci (3) are auxotrophic for a number of amino acids. S. uberis is no exception. A survey of 12 strains revealed that this species required between 10 and 13 amino acids to grow in a chemically defined medium and that 8 amino acids were required by all strains (8). It has been suggested that early in pathogenesis, prior to the induction of an inflammatory response, the growth of S. uberis is facilitated by the ability of the organism to hydrolyze host proteins (9).
S. uberis has not been shown to hydrolyze protein directly (8, 9). In a chemically defined medium in which a single essential amino acid was omitted, the inclusion of intact alpha, beta, or kappa bovine caseins failed to restore growth (8). However, S. uberis has been shown to activate bovine, ovine, and, to a lesser extent, equine plasminogens (9) to the serine protease plasmin. Plasminogen occurs naturally in bovine milk (2). In the absence of certain essential amino acids, the growth of S. uberis can be restored by the inclusion of plasmin-hydrolyzed caseins (8), demonstrating that the acquisition of some essential amino acids as peptides may be achieved by this route by using the oligopeptide ABC transporter encoded within the opp operon (19).
A number of plasminogen activators have been found in streptococci isolated from humans and animal species, and it appears that in many cases these activities are specifically directed at plasminogen from the host (4, 14, 17). Although these findings do not provide evidence that plasminogen activators play a central role in infection and pathogenesis, the observation that different bacteria, even biotypes of the same species that are capable of infection in different animal species, activate plasminogen specifically from their respective hosts is compelling.
In addition to activating bovine plasminogen, S. uberis is also able to bind the active protease plasmin on its surface (13). This phenomenon has also been reported for other streptococci, for which it has been postulated that acquisition of plasmin may promote invasion, although no direct evidence for this in S. uberis has yet been reported. Plasmin bound on the surface of S. uberis may also allow the production of nutritionally beneficial peptides in close proximity to the bacterial cell. The bound enzyme is also more resistant to the inhibitory effects of molecules such as α2-antiplasmin (15).
There is no direct evidence to demonstrate the roles of any of these molecules or activities in vivo. However, data on the use of the plasminogen activator PauA as a vaccine antigen have been presented (12), and these data have supported the potential role of PauA in the colonization of the bovine mammary gland. Animals vaccinated with preparations containing PauA produced immunoglobulin G capable of inhibiting plasminogen activation by this protein. Following experimental challenge, vaccinated animals shed 104-fold fewer bacteria in their milk and displayed a substantially reduced inflammatory response to infection than nonvaccinated animals (12). This finding led to the hypothesis that the activation of plasminogen by PauA plays a significant role in disease and that its neutralization by immunoglobulin may afford significant protection from disease. In order to generate strains that would allow investigation of this hypothesis in vivo, random insertional mutagenesis (16) was used to generate strains lacking the ability to activate plasminogen.
Approximately 1,600 colonies from a bank of random insertional mutants of S. uberis (strain 0140J [19]) (each containing a single copy of pGhost9 integrated into the chromosome between two parallel copies of ISS1) were screened for the ability to produce zones of clearing on Todd-Hewitt agar containing skimmed milk (1%, vol/vol) and bovine plasminogen (1 U/ml) as described previously (9). Mutants unable to activate plasminogen were analyzed further. Amplification of pauA from boiled culture extracts by PCR (5) was attempted with the primers P044 (5′-GACGACGACAAGATAACCGGTTATGATTCCGAC) and P045 (5′-GGAACAAGACCCGTATTTAATGGATACTTCCTTTA).
The plasmid was cured from PauA mutants by repeated subculturing at 28°C and analysis for loss of erythromycin resistance at 37°C, as described previously (5, 19) for other mutants of S. uberis. All subsequent analyses were performed with a cured derivative of a single pauA mutant, PNW-001.
Whole-cell lysates and concentrated culture supernatants were produced as described previously (11, 20), and the presence of PauA was detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting (20) using the PauA-specific monoclonal antibody EC3 (10). In the clearing assay (10), the level of plasminogen activation was determined by incubation of 20 μl of each extract in skimmed milk agarose. In addition, the ability of concentrated culture supernatant to activate plasminogen was determined by a plasmin-specific chromogenic assay (14).
The ability of the mutant and wild-type strains to bind plasmin was determined directly by using washed bacterial suspensions to which plasmin had been added and following overnight culture in Todd-Hewitt broth containing plasminogen in a manner that had been used previously to analyze this interaction in streptococci (14).
The ability of bacteria to grow in bovine skim milk was determined as described previously (18, 19). Milk was obtained from disease-free cattle from the dairy herds at the Institute for Animal Health (Compton, Berks, United Kingdom), and skim milk was produced following centrifugation (18, 19). Bacteria from overnight cultures were inoculated into milk at a density of 103 CFU/ml and incubated at 37°C for 24 h. Samples were removed and bacteria were enumerated (19, 20) at various time intervals.
Both strains were used in an intramammary challenge experiment to determine their relative virulence levels. The protocol used was that reported previously for analysis of a hasA mutant of S. uberis (5). In brief, approximately 103 CFU of each strain in 1 ml of pyrogen-free saline was infused into the mammary gland after the evening milking through the teat duct. Strain PNW-001 was used to challenge eight quarters of four dairy cows (two contralateral quarters per animal), and the wild-type strain was similarly infused simultaneously into four quarters of two other dairy cows. Samples of milk were collected at each milking (at 0700 and 1600 h) for enumeration of bacteria and somatic cells (5). Clinical evaluation of the condition of milk and udders was undertaken at each milking, and animals showing clinical signs of mastitis were treated with commercially available intramammary antibiotic preparations.
Of 27 mutants that were unable to activate plasminogen, pauA was shown to be present and intact in 26. Despite repeated attempts, it was not possible to amplify pauA from one mutant (PNW-001), indicating the possible presence of the 5.4-kb ISS1-pGhost9-ISS1 sequence in this location. PCR amplification of pauA from the cured derivative of this strain was subsequently shown to yield a product of around 1.8 kb, consistent with the presence of a single, remnant, integrated copy of ISS1 within this open reading frame (ORF). This finding was later confirmed by analysis of the sequence of this product, which revealed the integration of ISS1 between codons 195 and 196 of pauA. As is common following the integration of IS6 family insertion sequence elements, an 8-nucleotide direct repeat was duplicated on either side of the insertion. Southern hybridization of HindIII-digested chromosomal DNA revealed only a single copy of ISS1 in strain PNW-001 and no such element in the wild-type strain, 0140J (data not shown).
The sequence of the mutated ORF revealed direct translational read-through from pauA into ISS1 for 18 bases before a termination codon was encountered. Consequently, pauA::ISS1 of PNW-001 was potentially able to produce a protein encoding the first 195 amino acids of PauA, lacking 66 amino acids from the C terminus and incorporating a further 6 amino acids encoded by ISS1. The potential product was determined to have a molecular massof 23.4 kDa and a theoretical pI of 4.80.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting of culture supernatants from the wild-type and pauA mutant strains revealed that the mutant strain was able to secrete a protein of a size consistent with that of the truncated ORF (pauA::ISS1) but that the wild-type strain produced a PauA protein with a size of 30 kDa, consistent with the size previously reported (7). PauA or truncated PauA was detected only in culture supernatant and not in whole-cell fractions, suggesting that the truncated protein was processed (cotranslationally synthesized, cleaved, and secreted) by S. uberis in a manner similar to that for the full-length molecule. However, the precise sequences at the N and/or C terminus of the truncated product were not determined to confirm this possibility.
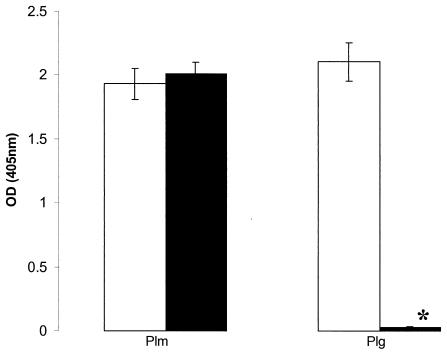
In contrast to what occurred with the wild-type strain, culture supernatant from PNW-001 was unable to activate bovine plasminogen to a detectable level in either the clearing assay with skimmed milk agarose or the chromogenic assay. Also, following overnight growth of PNW-001 in Todd-Hewitt broth containing plasminogen, no plasmin activity was detected in association with the bacterial cells (Fig. 1). This finding contrasts with the observations made for the wild-type strain (0140J), on which plasmin was readily detected following similar cultivation (Fig. 1). The inability of PNW-001 to bind plasmin following growth in plasminogen was most likely due to its inability to activate plasminogen, as this strain was shown to be able to bind the enzyme at levels similar to those seen when the wild-type strain was presented with this molecule directly (Fig. 1). This observation is consistent with those made previously during the investigation of a natural isolate that lacked the ability to produce PauA (13), further indicating that activation of plasminogen precedes binding of plasmin in this organism. The failure of the truncated protein to activate bovine plasminogen is also consistent with previous observations made in this and other laboratories indicating that recombinant PauA proteins with C-terminal deletions are similarly inactive (P. N. Ward et al., unpublished data).
FIG. 1.
Association of plasmin activity with S. uberis 0140J and the pauA mutant PNW-001. The data are the arithmetic means (± standard errors) of levels of plasmin activity (towards the plasmin-specific substrate H-d-val-leu-lys-pNA detected at an optical density [OD] at 405 nm) associated with S. uberis 0140J (open bars) (n = 9) and PNW-001 (filled bars) (n = 9) from overnight cultures incubated in the presence of plasmin (Plm) or grown overnight in the presence of plasminogen (Plg) (14). The asterisk denotes a value significantly different from the corresponding value for strain 0140J (P < 0.0001).
Following inoculation of bovine skim milk, the mutant and wild-type strains grew at similar rates and achieved similar final cell numbers, an indication that activation of plasminogen was not required to release essential amino acids from milk proteins during growth in this medium in vitro. Growth rates and final cell densities of the two strains in Todd-Hewitt broth were also similar.
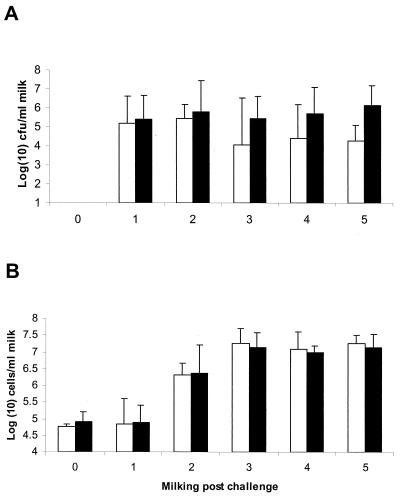
In order to assess the collective role of plasminogen activation on other processes affecting pathogenesis, the virulence of strain PNW-001 was compared to that of the wild-type strain following intramammary challenge in a well-established model for bovine mastitis (5, 12). Both strains were detected in similar numbers in milk obtained from the first milking postchallenge, and no significant difference in the bacterial numbers obtained from both groups was detected for the duration of the experiment (Fig. 2). The genotypes of isolates from each quarter were confirmed by PCR amplification of pauA (in the case of strain 0140J) or pauA::ISS1 (in the case of strain PNW-001). In each case, the level of colonization was consistent with that reported on several previous occasions for the wild-type strain (5, 12) and was also consistent with the earlier observation that the growth rate of S. uberis in milk was unaffected by the inability of the organism to activate plasminogen. No differences in levels of inflammation, measured by the speed and magnitude of neutrophil infiltration into milk, were detected following challenge with either strain (Fig. 2). Similarly, there were no discernible differences in the clinical signs associated with infection with either strain. All challenged quarters showed clinical signs of mastitis (clotted milk and swollen and tender udder quarters), and all quarters of all animals required antibiotic therapy to eliminate the infection.
FIG. 2.
Bacterial recovery and inflammatory response following challenge with S. uberis 0140J and the pauA mutant PNW-001. The data are geometric means (± standard errors) of the numbers of bacteria (A) and somatic cells (B) detected in milk samples obtained before challenge (milking 0) and after challenge (milkings 1 to 5) with S. uberis 0140J (open bars) (n = 4) or PNW-001 (filled bars) (n = 8).
It appears that the ability to activate plasminogen through the action of PauA does not play a major role in the ability of S. uberis to either grow in milk or infect the bovine mammary gland. This series of observations is, therefore, at odds with the hypothesis that plasminogen activation is required for growth in milk (9) and that activation of plasminogen is central to the pathogenesis of bovine mastitis (12). However, data supporting the role of PauA as an effective vaccine against mastitis due to S. uberis have been presented (12); furthermore, these data indicated not only that vaccination was effective but, more specifically, that responses which resulted in a neutralizing effect were the most protective (12). As the published studies showing the protective effect of vaccination with PauA used partially purified fractions of PauA-enriched extracellular proteins, it is possible that protection resulted from additional responses to molecules other than PauA. However, similar preparations lacking PauA showed no protective effect (12). Alternately, the data presented here may indicate that the truncated form of PauA retains some activities of the full-length molecule (other than the ability to activate plasminogen, which is clearly absent) that are important for pathogenesis and that these activities are sufficient for full virulence.
Given the high frequency with which PauA can be detected in S. uberis (7), a role for this molecule during interaction with its host is implicated. Indeed, only one strain of S. uberis has been shown to lack pauA, and this strain was shown to carry a gene for an alternate plasminogen activator, PauB (7, 22). Also, given the ability of many species of pyogeneic streptococci to activate plasminogen from their natural host species (4, 13, 16) through molecules with differing sequences and from ORFs located in different regions of the respective chromosomes (18), a role for such activities in host-pathogen interaction appears likely. However, this investigation, in which the virulence of a mutant strain of S. uberis was shown to be unaffected by the absence of bacterially induced plasminogen activation, indicates that in this species the role of the plasminogen activator PauA remains compelling but unproved.
Editor: A. D. O'Brien
REFERENCES
- 1.Aston, J. W. 1975. Amino acids in milk. Their determination by gas-liquid chromotography and their variation due to mastitic infection. Aust. J. Dairy Technol. 30:55-59. [Google Scholar]
- 2.Benslimane, S., M. J. Dogin-Bergeret, J. L. Bergadue, and Y. Gaudemer. 1990. Variation with season and lacatation of plasmin and plasminogen in Montebeliard cows milk. J. Dairy Res. 57:423-435. [Google Scholar]
- 3.Bentley, R. W., J. A. Leigh, and M. D. Collins. 1991. Intrageneric structure of Streptococcus based on comparative analysis of small-subunit rRNA sequences. Int. J. Syst. Bacteriol. 41:487-494. [DOI] [PubMed] [Google Scholar]
- 4.Caballero, A. R., R. Lottenberg, and K. H. Johnston. 1999. Cloning, expression, sequence analysis, and characterization of streptokinases secreted by porcine and equine isolates of Streptococcus equisimilis. Infect. Immun. 67:6478-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Field, T. R., P. N. Ward, L. H. Pedersen, and J. A. Leigh. 2003. The hyaluronic acid capsule of Streptococcus uberis is not required for the development of infection and clinical mastitis. Infect. Immun. 71:132-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hillerton, J. E., M. F. S. Shearn, R. M. Teverson, S. Langridge, and J. M. Booth. 1993. Effect of pre-milking teat dipping on clinical mastitis on dairy farms in England. J. Dairy Res. 60:31-41. [DOI] [PubMed] [Google Scholar]
- 7.Johnsen, L. B., K. Poulsen, M. Kilian, and T. E. Petersen. 1999. Purification and cloning of a streptokinase from Streptococcus uberis. Infect. Immun. 67:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitt, A. J., and J. A. Leigh. 1997. The auxotrophic nature of Streptococcus uberis: the acquisition of essential amino acids from plasmin derived casein peptides. Adv. Exp. Med. Biol. 418:647-650. [PubMed] [Google Scholar]
- 9.Leigh, J. A. 1993. Activation of bovine plasminogen by Streptococcus uberis. FEMS Microbiol. Lett. 114:67-72. [DOI] [PubMed] [Google Scholar]
- 10.Leigh, J. A. 1994. Purification of a plasminogen activator from Streptococcus uberis. FEMS Microbiol. Lett. 118:153-158. [DOI] [PubMed] [Google Scholar]
- 11.Leigh, J. A. 1999. Streptococcus uberis: a permanent barrier to the control of bovine mastitis? Vet. J. 157:225-238. [DOI] [PubMed] [Google Scholar]
- 12.Leigh, J. A., J. M. Finch, T. R. Field, N. C. Real, A. Winter, A. W. Walton, and S. M. Hodgkinson. 1999. Vaccination with the plasminogen activator from Streptococcus uberis induces an inhibitory response and protects against experimental infection in the dairy cow. Vaccine 17:851-857. [DOI] [PubMed] [Google Scholar]
- 13.Leigh, J. A., and R. A. Lincoln. 1997. Streptococcus uberis acquires plasmin activity following growth in the presence of bovine plasminogen through the action of its specific plasminogen activator. FEMS Microbiol. Lett. 154:123-129. [DOI] [PubMed] [Google Scholar]
- 14.Leigh, J. A., S. M. Hodgkinson, and R. A. Lincoln. 1998. The interaction of Streptococcus dysgalactiae with plasmin and plasminogen. Vet. Microbiol. 61:121-135. [DOI] [PubMed] [Google Scholar]
- 15.Lincoln, R. A., and J. A. Leigh. 1998. Characterisation of the interaction of bovine plasmin with Streptococcus uberis. J. Appl. Microbiol. 84:1104-1110. [DOI] [PubMed] [Google Scholar]
- 16.Maguin, E., H. Prévost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCoy, H. E., C. C. Broder, and R. Lottenberg. 1991. Streptokinases produced by pathogenic group C streptococci demonstrate species-specific plasminogen activation. J. Infect. Dis. 164:515-521. [DOI] [PubMed] [Google Scholar]
- 18.Rosey, E. L., R. A. Lincoln, P. N. Ward, R. J. Yancey, Jr., and J. A. Leigh. 1999. PauA: a novel plasminogen activator from Streptococcus uberis. FEMS Microbiol. Lett. 178:27-33. [DOI] [PubMed] [Google Scholar]
- 19.Smith, A. J., A. J. Kitt, P. N. Ward, and J. A. Leigh. 2002. Isolation and characterization of a mutant strain of Streptococcus uberis, which fails to utilize a plasmin derived beta-casein peptide for the acquisition of methionine. J. Appl. Microbiol. 93:631-639. [DOI] [PubMed] [Google Scholar]
- 20.Smith, A. J., P. N. Ward, T. R. Field, C. L. Jones, R. A. Lincoln, and J. A. Leigh. 2003. MtuA, a lipoprotein receptor antigen from Streptococcus uberis, is responsible for acquisition of manganese during growth in milk and is essential for infection of the lactating bovine mammary gland. Infect. Immun. 71:4842-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas, L. H., W. Haider, A. W. Hill, and R. S. Cook. 1994. Pathologic findings of experimentally induced Streptococcus uberis infection in the mammary gland of cows. Am. J. Vet. Res. 55:1723-1728. [PubMed] [Google Scholar]
- 22.Ward, P. N., and J. A. Leigh. 2002. Characterization of PauB, a novel broad-spectrum plasminogen activator from Streptococcus uberis. J. Bacteriol. 184:119-125. [DOI] [PMC free article] [PubMed] [Google Scholar]