Abstract
MicroRNAs (miRNAs) are potent negative regulators of gene expression involved in all aspects of cell biology. They finely modulate virtually all physiological pathways in metazoans, and are deeply implicated in all main pathologies, among which cancer. Mir-221 and miR-222, two closely related miRNAs encoded in cluster from a genomic region on chromosome X, are strongly upregulated in several forms of human tumours. In this work, we report that the ectopic modulation of NF-kB modifies miR-221/222 expression in prostate carcinoma and glioblastoma cell lines, where we had previously shown their oncogenic activity. We identify two separate distal regions upstream of miR-221/222 promoter which are bound by the NF-kB subunit p65 and drive efficient transcription in luciferase reporter assays; consistently, the site-directed mutagenesis disrupting p65 binding sites or the ectopical inhibition of NF-kB activity significantly reduce luciferase activity. In the most distal enhancer region, we also define a binding site for c-Jun, and we show that the binding of this factor cooperates with that of p65, fully accounting for the observed upregulation of miR-221/222. Thus our work uncovers an additional mechanism through which NF-kB and c-Jun, two transcription factors deeply involved in cancer onset and progression, contribute to oncogenesis, by inducing miR-221/222 transcription.
INTRODUCTION
MicroRNAs (miRNAs) are small non-coding RNAs working as post-transcriptional regulators of gene expression through either the degradation or the impairment of the translation of specific target messenger RNAs (mRNAs) (1). A continuously increasing amount of evidence is provided about the involvement of these small regulators in virtually all aspects of cell biology, including physiological modulation and pathological disruption of basic pathways. As an obvious consequence, miRNAs are found to be key players in cancer, where they regulate all main aspects of tumorigenesis and tumor progression, from the first initiating steps to metastasis formation and spreading (2). Those miRNAs whose expression is positively correlated with oncogenesis are often dubbed ‘oncomiRs’. Among these, miR-221 and miR-222 are a pair of miRNAs encoded in cluster on chromosome X, widely overexpressed in a large variety of human cancers, where they were shown to play their oncogenic roles via the downregulation of several tumor suppressors such as p27, p57, PTEN and many others (3,4). In our previous work, we demonstrated that miR-221 and miR-222 are overexpressed in prostate carcinoma and in glioblastoma (5–7), and in both tumors they downregulate the cell cycle inhibitor p27 by impairing the translation of its mRNA (6,7).
To date, the great amount of data describing the tumor-specific modulation of some miRNAs is not yet complemented by studies explaining the basis for the aberrant expression of miRNAs in cancer. There is a general agreement about miRNA gene transcription being performed prevalently by RNA Polymerase II (8,9), which implies that miRNA genes share many common features with protein coding genes, such as basic promoters, polyadenilation of the primary transcripts and specific regulation by transcription factors both at proximal promoters and at distal enhancers.
Two pleiotropic transcription factors deeply involved in almost all aspects of human cancer, NF-kB and AP-1, are good candidates as potent activators of oncogenic miRNAs. NF-kB embraces a family of transcription factors made of hetero- or homo-dimers frequently including one subunit of p65 (relA) and another subunit such as p50, c-rel or relB (10). The dimers are kept in an inactive form in the cytoplasm by one of the members of the IKB family, mainly IKBα. Phosphorylation and proteasome degradation of IKBα sets NF-kB free to enter the nucleus where it controls transcription of a wide variety of genes (11). This activated form of NF-kB is upregulated and functionally correlated with many tumors, including glioblastoma, where it modulates proliferation and invasion (12) and prostate carcinoma, where it marks metastatic cancer (13). AP-1 is another example of dimeric transcription factor containing members of JUN, FOS, ATF and MAF proteins, all sharing a basic leucine zipper motif needed for dimerization and DNA binding (14). FOS and especially JUN are the main members of the AP-1 family, primarily considered as oncogenic activators, even with some interesting exceptions to this rule (15). AP-1 and specifically c-Jun, affects tumor cell proliferation, migration and invasion and influences also the proangiogenic potential of cancer cells by inducing VEGF expression and other angiogenic factors (14). Thus, like NF-kB, AP-1 takes an active part throughout tumorigenesis from the beginning to distal dissemination.
In the present study, we investigated the regulation of miR-221 and miR-222 transcription in prostate carcinoma and glioblastoma cell models. By bioinformatic prediction, we identified two highly conserved genomic regions upstream of miR-221/222, embedding NF-kB and c-Jun responsive elements and we tested them in reporter assays by which we confirmed their transcriptional activation function. The binding of p65 and c-Jun was assessed by electrophoretic mobility shift assays (EMSA) and chromatin immunoprecipitation (ChIP) experiments, while the functional involvement of both NF-kB and c-Jun in miR-221/222 expression was demonstrated by up- and downregulating them via exogenous stimulation or targeted silencing RNAs (siRNAs), respectively.
These results, as a whole, may uncover a new way by which AP-1 and NF-kB contribute to tumorigenesis in prostate carcinoma and glioblastoma, which is by inducing the expression of the oncomiRs miR-221 and miR-222.
MATERIALS AND METHODS
Cell lines, treatments and transfection
All cell lines were maintained in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum. LNCaP cells were starved for 24 h and then treated with 10 ng/ml of TNF-α (SigmaAldrich). Transfections were performed by Lipofectamine 2000 (Invitrogen) using plasmid DNA in Opti-MEM I (Invitrogen), as recommended by the manufacturer. For transient transfections, reporter plasmids were co-transfected with a 1:7 relative amount of the plasmid pEGFP-C3 (Clontech), to monitor transfection efficiency. Cells were analyzed by fluorescence microscopy 48 h after transfection to calculate the transfection efficiency. For each well, the cell number in four random microscopic fields was counted and transfection efficiency for each sample was estimated as the mean value of green fluorescent protein expressing cells over 100 cells per field.
RNA isolation and quantitative real time polymerase chain reaction
Total RNA was isolated from cells by TRIZOL reagent (Invitrogen) as described by manufacturer’s instructions. An amount of 0.05 µg total RNA was reverse-transcribed by using the Taqman MicroRNA Reverse Transcription Kit (Applied Biosystems). Comparative real-time polymerase chain reaction (PCR) was performed in triplicate with the use of the Taqman Universal PCR Master Mix (Applied Biosystems) on the Applied Biosystems 7500 FAST real-time PCR System. Mature miRNA-specific primers and probes were obtained from Applied Biosystems. Normalization was performed by simultaneous quantification of small endogenous nuclear RNA U6 (RNU6B) and relative expression was calculated employing the comparative CT method.
Luciferase reporter constructs and luciferase assay
Regions A and B were amplified by PCR from human genomic DNA (regA primers: fw 5′-agaatgccttagggttttct-3′, rv 5′-tcttatgaacttgattcct-3′; regB primers: fw 5′-ttctgtttggtccttcctct-3′, rv 5′-cattcggttgtgtagagcca-3′). The PCR products were cloned into the pGL3 promoter Vector (Promega) using T4 DNA ligase (Fisher scientific). All constructs were confirmed by sequencing. Mutations were introduced into the NF-kB and AP-1 binding sites using the QuickChange Site-directed Mutagenesis Kit (Stratagene). For luciferase activity measurement, the Dual-Luciferase reporter assay system (Promega) was used. PC3 and U87 cells were transfected with each reporter construct and the TK-Renilla luciferase plasmid that was used as a transfection control. Luciferase detection was performed 48 h after reporter construct transfection. Expression was calculated as the relative Firefly luciferase activity normalized with respect to the activity of transfection control Renilla luciferase. Reporter assays were performed in triplicate.
Chromatin immunoprecipitation assay
Chromatin was prepared by the Millipore/Upstate ChIP assay kit (Millipore) according to the manufacturer’s instruction. For each assay, a total of 107 cells were fixed with 1% of formaldehyde. Cell pellets were resuspended in SDS lysis buffer. Samples were sonicated with a Diagenode Bioruptor (Diagenode SA) for 12 cycles of 30 s at high power, centrifuged and diluted 10-fold in ChIP dilution buffer. After removing an aliquot (whole-cell extract input), the chromatin samples were incubated at 4°C overnight with antibodies against p65 (ab7970 abcam) or c-Jun (sc-8008 Santa Cruz), Polymerase II (sc-55492, Santa Cruz), H3K4me1 (Millipore) or H3K9ac (Millipore). As controls, aliquots of the same chromatins were processed in the same way, but the specific antibodies were omitted from the reaction. The samples were then precipitated by binding to protein A-Agarose/Salmon Sperm DNA beads (Millipore, Billerica, MA, USA) and treated as previously described (16). The PCR primers (RegA primer: fw 5′-ATCTTGTGCCAACCAGTCCCTTCT-3′, rv 5′-AACATTCTCTGGGACGTTCCTGCT-3′; regB primer fw 5′-TGGTCCTTCCTCTAAGATTGGTCC-3′, rv 5′-ACCAAACTGCAGGAACTGACTCAT-3′; transcription start site (TSS) primer: fw 5′-GAAGGTGTAGGTACCCTCAA-3′, rv 5′-TCAACACAACTGCCTACTGC-3′) were designed to amplify 60–120 bp fragments from selected genomic regions. Real-time PCRs of genomic regions containing the putative NF-kB and AP-1-binding site were performed in triplicate by using Power SYBR green master mix (Applied Biosystem). Cycling parameters were 50°C 2 min, 95°C for 10 min, followed by 38–42 cycles of 95°C for 15 s, 60°C 1 min. Control amplifications were performed with oligos 5′-AAACCACCCATCCAGAAGGG-3′ and 5′-CGTGGCAGCACTCGTAAGACT-3′ (chr1:204,366,822–204,366,872) used as the negative control (16). The relative occupancy of the immunoprecipitated factor at a locus was estimated by using the comparative threshold method (17).
Electrophoretic mobility shift assay
32P-labeled double-stranded oligonucleotides of 3.0×104 cpm spanning the NF-kB sites of RegionA (bsA: 5′-GTTCTGGGATTTCCCCCGAT-3′) or regionB (bsB: 5′-CAGCAGGAACGTCCCAGAGAA-3′) were incubated with 5 μg of nuclear extract at room temperature for 20 min with 2 μg of poly(dI-dC) in 50 mM NaCl, 10 mM Tris pH 7.5, 1 mM DTT and 20% glycerol. Competition experiments were performed by co-incubating the probe with 100-fold molar excess of unlabeled double-stranded oligonucleotide or unlabeled double- stranded oligonucleotide mutated in NF-kB sites of regionA (mut-bsA: 5′-GTTCTGAACGGGACCCCGAT-3′) or regionB (mut-bsB: 5′-CAGCAGACCTTGTTCAGAGAA-3′). Supershift assays were performed by incubating nuclear extracts with specific antibodies against p65 (ab7970 abcam) for 30 min at room temperature before the addition of the probe. Band shifts were resolved on non-denaturing 4% polyacrylamide gels.
Immunoblot analysis and c-Jun knockdown
Immunoblot analysis was performed as described previously (7). SMARTpool siRNAs against c-Jun and control siRNAs were obtained from Dharmacon and transfected by Lipofectamine 2000 into PC3 and U87 cells at a final concentration of 100 nM.
Evaluation of NF-kB activity
NF-kB activity was measured in nuclear protein extracts (15 μg) by the TransAMTM NF-kB p65 protein assay (Active Motif, Carlsbad, CA, USA), an ELISA-based method designed to specifically detect and quantify NF-kB p65 subunit activation, with high sensitivity and reproducibility. The assay was performed according to the manufacturer's protocol and analyzed using a microplate absorbance reader Biorad680 (Biorad, CA, USA).
RESULTS
Impaired activation of NF-kB alters miR221/222 expression
We performed a bioinformatic search for transcription factor binding sites (MatInspector software, http://www.genomatix.de) in the human genomic sequence (∼12 000 bp) upstream of miR-222/221 transcriptional unit (18) and identified predicted NF-kB binding sites embedded in two regions highly conserved in mammals (RegA and RegB, Figure 1A), as evidenced from Human Mar. 2006 (NCBI36/hg18) assembly UCSC Genome Browser (http://genome.ucsc.edu/). To verify the involvement of NF-kB in miR-222/221 expression, we performed experiments of induction/repression of the NF-kB pathway in cell lines endogenously characterized by either low/high NF-kB activity or low/high levels of miR222/221 (5,7). First, we analyzed LNCaP prostate carcinoma cells that have low NF-kB activity (19) and low levels of miR221/222 (7). After incubating these cells with TNFα for 3 h, we verified NF-kB activation (Figure 1B) by TransAM NF-kB p65 kit (Active Motif) and we analyzed mir-222 and miR-221 by qRT-PCR. In these conditions of NF-kB stimulation, we found an upregulation of about 2-fold of both miRNAs (Figure 1C, left panel). We then analyzed PC3 prostate carcinoma cells and U87 glioblastoma cells, both characterized by high NF-kB activity (12,19) and high levels of miR221/222 (5,7). To inhibit NF-kB pathway, we transfected these cells with pCMV-IkBαM plasmid that encodes a dominant-negative mutant of IκBα, thus blocking the phosphorylation events that trigger NF-kB activation (Figure 1B). As shown in the central and right panels of Figure 1C, the blockade of NF-kB after pCMV-IkBαM transfection, decreased miR-221/miR-222 expression of 50% in both cell lines. Similar results were obtained when PC3 cells were incubated in the presence of acetyl-β-boswellic acid (data not shown) that in these cells inhibits the constitutively activated NF-kB signaling (20). These results demonstrate that the modulation of NF-kB pathway affects miR-221/222 expression in prostate carcinoma and glioblastoma cells, suggesting that NF-kB is involved in miR-221/222 expression regulation. We also showed the effects of NF-kB inhibition on p27Kip1 expression in PC3 and U87 cells, where we had previously demonstrated that miR-221/222 target p27 (7). We decided to measure the effects of NF-kB in a reporter assay with a luciferase construct under the control of p27 3′-UTR (p3′-UTR-p27). In this way, we highlighted the effects on p27 3′-UTR, where miR-221/222 work. The results we obtained (Figure 1D) show that the downregulation of NF-kB is able to consistently upregulate luciferase activity driven by p27 3′-UTR, strongly indicating that this effect is directly mediated by miR-221/222.
Figure 1.
NF-kB modulates miR-221/222 expression through two genomic sites upstream of miR-221/222 transcriptional unit. (A) Schematic diagram of Regions A (RegA) and B (RegB) genomic loci on human chromosome X derived from Human Mar. 2006 (NCBI36/hg18) assembly UCSC Genome Browser. The direction of miR-221 and miR-222 transcription is indicated by an arrow. (B) NF-kB activity assay. LNCaP cells were starved for 24 h and then stimulated with TNFα for 3 h. PC3 and U87 cells were transfected for 48 h with the pCMV-IκBαM (+IκBαM) or pCMV (−IκBαM) plus pEFGP, to assess transfection efficiency. Nuclear protein extracts were harvested and NF-κB activity was measured by transAM NFκB p65 protein kit. (C) Total RNA from LNCaP, PC3 and U87 cells treated as in (B) was extracted for quantitative real time polymerase chain reaction (qRT-PCR) analysis, miRNAs were normalized to snoRNA U6. For each protocol, three experiments were performed independently and the mean values (±SE) among experiments are presented. (D) p3′-UTR-p27 construct harboring the 3′-UTR of p27kip1, was transfected into PC3 and U87 cells after 24 h of control (p3′-UTR-p27) or pCMV-IkBαM (p3′-UTR-p27 + IκBαM) plasmids transfection. (E) Left panel: empty reporter plasmid (c) or pRegA and pRegA-mp65 luciferase constructs containing, respectively, a wild-type and a mutated Region A were transfected into PC3 and U87 cells after 24 h of control (−) or pCMV-IkBαM (+IκBαM) plasmids transfection. Another complete set of samples was treated with TNFα for 3 h (+TNFα) in each cell line. Luciferase activity was determined 48 h after reporter plasmid transfection in all cases. The ratio of normalized sensor to control luciferase activity is shown. Right panel: the same set of experiments was performed for constructs pRegB and pRegB-mp65 harboring a wild-type or a mutated Region B, respectively. Data are presented as mean ± SE from three separate experiments with at least n = 3 for each experiment.
Two regions upstream of miR-221/222 encoding sequence embed NF-kB binding sites and are endowed with NF-kB-dependent enhancer activity
In order to better investigate the role of NF-kB and to test a putative enhancer activity of RegA and RegB, we constructed reporter plasmids by inserting the genomic fragments corresponding to each region into the pGL3-promoter vector that contains a minimal SV40 promoter upstream of luciferase cDNA. We transfected the two plasmids named pRegA and pRegB for Regions A and B, respectively, into PC3 and U87 cells (Figure 1E). Luciferase assays showed that both regions actively enhance transcription in both cell lines: pRegA yields a ∼2-fold induction of luciferase activity compared with the empty vector (Figure 1E, left panel), while pRegB is even more effective, producing a 6-fold enhancement of luciferase activity (Figure 1E, right panel). The introduction of point mutations disrupting the predicted NF-kB binding site (pRegA-mp65, Figure 1E, left panel) restored the luciferase levels to those of the empty vector in Region A, while in Region B (pRegB-mp65, Figure 1E, right panel) luciferase activity, even if significantly reduced, was still above the levels of the negative control, indicating the presence of other relevant activation sites in that region. A reduction of luciferase activity was achieved even when the two cell lines were transfected with pCMV-IkBαM plasmid 24 h before pRegA and pRegB transfection (+IkBαM, Figure 1E, in both panels, in each cell line). As expected, IkBαM transfection did not affect luciferase activity driven by the mutated constructs. A further confirmation of the involvement of the NF-kB pathway in the transcriptional activity of RegA and RegB was provided by treating PC3 and U87 cells with TNFα, that in fact enhanced luciferase activity of both wild-type constructs, but not of the mutated ones (+TNFα, Figure 1E, left and right panel). Altogether, these assays indicate that Regions A and B own an intrinsic transcriptional activation potential working in two different cancer cell contexts and this likely relies on the predicted NF-kB sites present in each region.
NF-kB is recruited at two regulatory regions upstream of mir-222/221 locus
The reduction in transcriptional activity of both Regions A and B observed when NF-kB sites were mutated as well as in the presence of a dominant-negative mutant of IkBα suggested that NF-kB directly binds those sites in Regions A and B. To assess the ability of the two binding sites to actually bind NF-kB nuclear protein, we performed EMSA. As probes, we used sequences bsA and bsB corresponding to the NF-kB binding sites, respectively, in the Regions A and B and the reaction was carried out in the presence of either PC3 or U87 nuclear extracts. EMSA revealed DNA–NF-kB-containing complexes on both sequences in both cell lines (Figure 2A and B). A specific competition of the gel retardation was induced by the co-incubation of labeled probes together with a 100-fold molar excess of unlabeled bsA and bsB probes (Figure 2A, lane 4 and Figure 2B, lane 5). Accordingly with our hypothesis that NF-kB binds Regions A and B sites, we did not detect any competition when the ‘cold’ competitors used were the mutagenized sequences bsA or bsB, where NF-kB sites were disrupted (Figure 2A, lanes 3 and Figure 2B, lanes 3). The specificity was confirmed by supershift with antibody anti-RelA/p65 (Figure 2A, lane 5 and Figure 2B, lane 4).
Figure 2.
NF-kB binding to Regions A and B. (A) Electrophoretic mobility shift assay. Nuclear extracts were prepared as described in the text, and assayed for protein binding to sequence bsA, corresponding to NF-kB binding site in the Region A. An unlabeled bsA probe (lane 4) or mut-bsA probe mutated in NF-kB site (lane 3) were used as competitors. Where indicated, an antibody recognizing the NFk-B p65 subunit was included (lane 5). Products of binding reactions were resolved by electrophoresis on 4% polyacrylamide gels and detected by autoradiography. (B) The same set of experiments was performed for Region B; competition with unlabeled wt probe (bsB) and mutated probe (mut-bsB) are indicated in lanes 5 and 3, respectively. Supershift with p65 antibody is evidenced in lane 4. (C) ChIP assay of chromatin isolated from PC3 and U87 cell lines transfected with control (bsA and bsB) or pCMV-IkBαm (bsA+ IkBαm and bsB+ IkBαm) plasmids, and immunoprecipitated by anti-p65 or control IgG, followed by qPCR analysis with primers targeted to sequences bsA and bsB and to a region on chromosome 1 used as the negative control (16). The data show occupancy relative to control IgG and represent mean ± SE of three independent experiments.
Finally, the direct interaction between these putative cis-regulatory elements and NF-kB was investigated by ChIP assays. Isolated chromatin from PC3 and U87 cell lines was immunoprecipitated by anti-RelA/p65 or control IgG, followed by qPCR analysis with primers targeted to sequences bsA and bsB and to a region on chromosome 1 (chr1:204,366,822–204,366,872), used as the negative control (16). As shown in Figure 2C, p65 binds both elements in both cell lines, with binding to the highly conserved B region being always stronger. The specific involvement of p65 was further confirmed by experiments carried out in pCMV-IkBαM transfected cells: the inhibition of NF-kB pathway significantly impaired p65 binding to both Regions A and B (Figure 2C). These results demonstrate that NF-kB binds directly to the identified sites in Regions A and B upstream of miR-221/222 genomic region.
NF-kB and c-Jun bind together to one of the regulatory regions upstream of miR-221/222 and cooperate to induce transcription
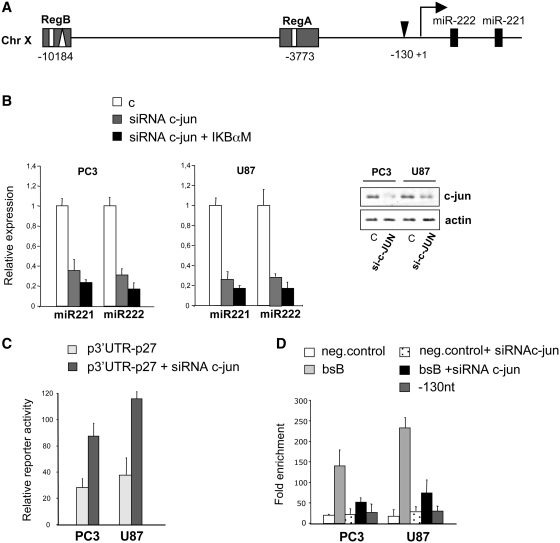
MiRNA transcription, just like that of mRNAs, can be co-regulated by more than one transcription factor, working either in cooperation or independently. In fact, our bioinformatic search for relevant transcription factors revealed that Region B harbors a canonical AP-1 binding site 115 bp downstream the NF-kB bsB (Figure 3A). AP-1 is a dimeric basic leucine zipper protein that belongs to the Jun and Fos subfamilies; c-Jun is the most potent transcriptional activator in its group (21). Interestingly, a recent paper demonstrated that c-Jun is involved in miR-222/221 activation in lung and liver cancer (3). To address the involvement of c-Jun in miR-221/222 activation in prostate carcinoma and in glioblastoma, we performed a knockdown of c-Jun by siRNAs in U87 and PC3 cells and measured miR-221/222 expression by qRT-PCR (Figure 3B). c-Jun depletion resulted in a 60–70% decrease in miR-221 and miR-222 levels in both cell lines (Figure 3B). In agreement with what we found by inhibiting NF-kB, we observed that c-Jun depletion too was able, per se, to increase the luciferase activity driven by p27kip1 3′-UTR reporter construct (Figure 3C), again showing that c-Jun can indirectly modulate p27 through miR-221/222. Moreover, the concomitant downregulation of c-Jun by siRNAs and of NF-kB by pCMV-IkBαM plasmid transfection yielded a miR-221/222 reduction stronger than any one obtained in either single knockdown condition (i.e. c-Jun siRNAs or NF-kB inhibition), thus indicating that NF-kB and AP-1 may cooperate to the induction of miR-221/222 through binding Region B in prostate carcinoma and glioblastoma. To further assess the functional role of c-Jun in miR-221/222 expression, we carried out ChIP assays measuring the binding of c-Jun to Region B in PC3 and U87 cell lines treated with control (bsB) or anti-c-Jun (bsB +siRNA c-jun) siRNAs. Indeed, c-Jun binding was remarkably enriched in Region B as compared to the negative control region (Figure 3D) and no relevant chromatin enrichment by c-Jun ChIP was observed in c-Jun depleted cells, testifying the specificity of the ChIP assay. We also checked whether a previously identified AP-1 site (3) found 130 nt upstream of the 5′-end of miR221/222 (Figure 3A and D, −130 nt) is bound by c-Jun in PC3 and U87 cells, but we could not find any significant enrichment in these contexts.
Figure 3.
C-Jun binding to Region B. (A) Schematic representation of NF-kB (white rectangles) and c-Jun (white triangle) binding sites, inside Regions A and B. (B) PC3 and U87 cells were transfected with control siRNAs or anti-c-JUN siRNAs and after 48 h total RNA and proteins were extracted. Levels of miR-221 and miR-222 were analyzed by qRT-PCR (left and central panels). The efficiency of siRNA-mediated depletion of c-Jun was assessed by western blot (right panel). Western blot analysis of β-actin is shown as a loading control. (C) p3′-UTR-p27 construct harboring the 3′-UTR of p27kip1, was transfected into PC3 and U87 cells after 24 h of control siRNA (p3′-UTR-p27) or anti-c-JUN siRNAs (p3′UTR-p27 + siRNA c-jun) transfection. (D) ChIP assay of chromatin isolated from PC3 and U87 cell lines transfected with control (bsB) or anti-c-Jun (bsB +siRNA c-jun) siRNAs and immunoprecipitated by anti-c-Jun or control IgG, followed by qPCR analysis with primers targeted to sequence bsB or to a sequence 130 nt upstream of the 5′-end of miR221/222 (−130 nt, black triangle in A) that represents a previously identified c-Jun site (3). The same region shown in Figure 2C was used as the negative control. The data show occupancy relative to control IgG and represent mean ± SE of three independent experiments.
In order to directly link our results about c-Jun binding to Region B with those showing that c-Jun is an activator of miR-221/222 expression, we designed experiments to prove that Region B works as an enhancer through both its NF-kB and its AP-1 binding sites. As a first approach, we performed luciferase assays using pRegB plasmid, pRegB mutagenized in c-Jun binding site (pRegB mc-Jun) and pRegB mutagenized in both sites (pRegB mp65/c-jun) (Figure 4A). Interestingly, the enhancer activity of RegB was almost abolished when point mutations disrupting the predicted c-Jun binding site were introduced or when pRegB plasmid was transfected 24 h after siRNAs against c-Jun. Moreover, when we transfected pRegB mp65/cjun, the effect of the mutagenesis in both p65 and c-Jun sites was comparable with that obtained by mutating the c-Jun site alone, suggesting that c-Jun is necessary for the recruitment of NF-kB p65 to Region B. To better understand the relationship between p65 and c-Jun in the binding of Region B, we performed a ChIP assay in cells transfected with c-Jun siRNAs, using an antibody against p65. As shown in Figure 4B, in both PC3 and U87 cells, we confirmed that p65 is significantly bound to Region B, but its binding is strongly impaired by the siRNA-mediated depletion of c-Jun. To further confirm the specificity of this c-Jun/p65 interaction for Region B, we assayed also the binding of p65 to Region A in c-Jun depleted cells: p65 recruitment to Region A, where no predicted AP-1 site is present, was not affected by c-Jun downregulation. Finally, we performed a sequential ChIP experiment where we first immunoprecipitated chromatin with anti-c-Jun antibodies, and then with anti-p65 ones; in this way we were able to show that Region B simultaneously binds c-Jun and p65 (Figure 4C). As a whole, these results show that p65 and c-Jun induce miR-221/222 expression by binding to Region B, and strongly indicate that c-Jun is the main activator in this region, being necessary for p65 activation function.
Figure 4.
C-Jun is the main activator of Region B. (A) Empty reporter plasmid (c) or pRegB, pRegB-mc-jun and pRegB mp65/c-jun luciferase constructs containing, respectively, a wild-type, a mutated Region B in c-Jun site or a mutated Region B in both p65 and c-Jun sites were transfected into PC3 and U87 cells. pRegB was also transfected into cells previously treated with anti-c-Jun siRNAs (pRegB +siRNA c-jun). Luciferase activity was determined 48 h after transfection. The ratio of normalized sensor to control luciferase activity is shown and data are represented as mean ± SE from three separate experiments with at least n = 3 for each experiment. (B) ChIP assay was performed as described in Figure 3C but the chromatin was immunoprecipitated by anti-p65 antibody and the data regarding bsA (bsA) or bsA in the presence of anti-c-Jun siRNAs (bsA + siRNA c-jun) were included. (C) Sequential ChIP assay performed in U87 cells on bsA (bsA) or bsB (bsB), by a preliminary immunoprecipitation with anti-c-Jun antibodies, and a subsequent one with anti-p65 antibodies. The data show occupancy relative to control IgG and represent mean ± SE of three independent experiments.
P65 and c-Jun binding to Regions A and B modifies the local epigenetic state of the chromatin and induces RNA Polymerase II recruitment to miR-221/222 transcriptional start site
To check if the binding of p65 and c-Jun to Regions A and B can induce the recruitment of RNA Polymerase II to miR-221/222 TSS, we performed ChIP assays for the binding of RNA Polymerase II to the previously defined TSS (18) and compared the results with those obtained in cells transfected with pCMV-IkBαM plasmid or with anti-c-jun siRNAs. As depicted in Figure 5A, RNA Polymerase II binding to TSS is significantly impaired by NF-kB inhibition (TSS + IkBαM), and even more by c-Jun depletion (TSS + siRNA c-jun). We then analyzed some relevant epigenetic markers, such as the monomethylated form of histone H3 lysine 4 (H3K4me1) and the acetylated form of histone H3 lysine 9 (H3K9ac), at Regions A and B, as well as in TSS. Figure 5B shows that H3K4me1 enrichment, typically associated with enhancers (22), is reduced, in both Regions A and B, when NF-kB is inhibited by IkBαM, whereas the siRNA-mediated depletion of c-Jun influences only H3K4me1 enrichment on Region B, as expected. Comparable results are shown in Figure 5C concerning H3K9ac enrichment, except for TSS: in this case H3K9ac is reduced by NF-kB inhibition and almost totally knocked down by anti-c-jun siRNAs. This is in agreement with the role of H3K9ac as a general marker of open chromatin structure, while H3K4me1 preferentially marks enhancers (22). As a whole, these data show that alterations of NF-kB or c-Jun activity induce epigenetic modifications at Regions A and B which are consistent with their role as enhancers, which in turn can modulate RNA Polymerase II recruitment to miR-221/222 TSS.
Figure 5.
Epigenetic modifications and RNA Polymerase II recruitment at miR-221/222 regulatory regions. (A) ChIP assay of chromatin isolated from U87 cells transfected with control (TSS), pCMV-IkBαM (TSS+IkBαm) or anti-c-Jun siRNAs (TSS+siRNA c-jun) and immunoprecipitated by anti-POLII or control IgGs, followed by qPCR analysis with primers targeted to miR-221/222 TSS or a negative control region (16). The same chromatin as in (A) was immunoprecipitated by anti-H3K4me1 (B) or anti-H3K9ac (C) antibodies and qPCR analysis was performed with primers targeted to the sequences bsA, bsB, TSS and negative control. The data show occupancy relative to control IgGs and represent mean ± SE of three independent experiments.
DISCUSSION
MiRNA genes, like protein-coding genes, are predominantly transcribed by RNA Polymerase II, and own many of the common features typical of RNA Pol II genes, such as the regulation by core promoters and distal enhancers, or the 5′ capping and the polyadenilation of primary transcripts (8,9). In this frame, a number of papers have been published describing transcription factors that regulate miRNA expression by binding to promoter regions, in either physiological or pathological contexts (23,24). Fewer works have recently described the role of enhancers in the specific induction of miRNA expression (25,26).
In this study, we show that the transcription of miR-221/222, whose promoter region had been previously identified (3,18,27), is under the positive control of two distal enhancer regions, binding NF-kB p65 and c-Jun, which are known as ‘master regulators’ in cancer. We identified these two regions, that we dubbed Regions A and B, on the basis of their striking evolutionary conservation (Figure 1A). We noticed that, among all the well conserved regions located upstream of miR-221/222 genomic sequence, those two were unique. In fact, they are conserved in all species where miR-221/222 are present and when we assayed each of them in transcription reporter experiments, both of them behaved as good transcriptional enhancers (Figure 1E). Our study is performed in prostate carcinoma and glioblastoma cell lines, where we and others demonstrated the oncogenic role of miR-221/222 via the downregulation of p27, p57, PTEN and other targets (3,4,6,7). In prostate carcinoma, NF-kB is constitutively activated and represents a survival signal sustaining tumor growth (28). Interestingly, the level of NF-kB activation is directly proportional to tumor aggressiveness; this is well represented also in vitro, by the differential activation of NF-kB that is detectable in the highly aggressive PC3 cell line as compared with LNCaP cells (19). Very recently, a prominent involvement of NF-kB as a driver of metastasis has been uncovered in prostate carcinoma (13). Glioblastoma provides a further relevant example of tumor-specific activation of NF-kB, which works as a signaling event mediating the effects of PDGF (12). Here, we show that the ectopical modulation of NF-kB in prostate carcinoma and glioblastoma cells caused consistent changes in miR-221/222 transcription (Figure 1C) through the binding to Regions A and B, as confirmed by mutagenis experiments where we disrupted each of the p65 predicted sequences in Region A or in Region B (Figure 1E). Consistently, when we impaired NF-kB activation by the transfection of a dominant-negative form of IkBα, we obtained comparable results, again strongly suggesting that NF-kB works in the activation of miR-221/222 likely by binding to the two regions. These results were further complemented by our EMSA and ChIP experiments. However, in both tumor cell types, Region B appeared to be more potent than Region A in luciferase assays and, importantly, while the abolishment of NF-kB site in Region A completely cancelled its transcriptional activity, the mutation in Region B produced a strong, but not total, reduction (Figure 1E). The presence of a totally conserved c-Jun predicted site in Region B prompted us to investigate c-Jun as an additional important candidate for miR-221/222 regulation operated through Region B.
Indeed, a broad potential in tumor control is owned by the members of AP-1 family, among which c-Jun is a prominent representative (15): in human prostate cancer, c-Jun is considered a marker of high risk tumors and elevated levels of this transcription factor are associated with disease recurrence (29). In glioblastoma cells, the activated form of c-Jun favors cell survival and protects tumor cells against the cytotoxic effects of DNA-damaging agents (30). In fact, we demonstrated that c-Jun is strongly involved in miR-221/222 transcription enhancement, as indicated by the great reduction of miR-221/222 expression in cells where we had depleted c-Jun via siRNAs (Figure 3B). Accordingly, we showed that the site-directed mutagenesis of c-Jun site completely eliminated Region B activity in luciferase assays (Figure 4A), and in fact Region B is bound by c-Jun, as demonstrated by our ChIP experiments (Figure 3D).
Thus, our findings correlating the high expression of miR-221/222 in prostate and brain tumors with NF-kB and c-Jun represent a novel case of concomitant and possibly cooperative regulation of an oncogenic cluster by these two factors. To our knowledge, only one work has been published so far describing the concurrent action of NF-kB p65 and AP-1 in the induction of a miRNA, miR-155, via the recognition of close sites in the promoter region (31). Like miR-221/222, miR-155 is one of the miRNAs most frequently upregulated in cancer (32). In mRNA genes, however, the functional interaction of NF-kB and AP-1 is not uncommon, especially in the regulation of tumor- or inflammation-related genes. In breast cancer cells, the urokinase-type plasminogen activator (uPA) gene, driving cancer cell motility and metastasis, is induced by the conjunct work of AP-1 and NF-kB (33). Another relevant example of a gene concomitantly regulated by NF-kB and c-Jun is ccl2, a highly regulated inflammatory gene strongly overexpressed in both prostate carcinoma and glioblastoma (34,35), whose promoter harbors c-Jun and NF-kB functional sites. Notably, in that case the stronger enhancing effect was demonstrated for c-Jun, needed for the recruitment of NF-kB p65 (36). The performance of Regulatory Region B that we are describing in our work shows some relevant similarities with that pattern. In fact, luciferase reporter and ChIP experiments have shown that the disruption of the c-Jun recognition site has a much stronger effect than that yielded by mutating the NF-kB one (Figures 1E and 4A). Most importantly, abolishing c-Jun binding by anti-c-Jun-siRNAs affects p65 binding to its own site too (Figure 4B). This implies that c-Jun is the major player in Region B, where it helps also p65 binding. A recent paper described that the transcriptional repression of p27Kip1 gene is mediated by AP-1 through the binding of a site in p27Kip1 promoter (37). A very intriguing hypothesis is that AP-1 exerts an important and pleiotropic role in the knockdown of p27 in cancer, both directly, by repressing its transcription, and indirectly, by inducing its negative regulators miR-221/222. In fact, we showed that the siRNA-mediated inhibition of c-Jun induced an upregulation of p27Kip1 expression that was at least in part mediated by miR-221/222 action on p27Kip1 3′-UTR (Figure 3C). The transcription activation potential of Region A, on the contrary, seems to be mostly due to the NF-kB site that we have identified. In fact, both in prostate carcinoma and in glioblastoma cells, the site-directed mutagenesis of that site completely flattens down luciferase activity in reporter assays (Figure 2A). NF-kB p65 involvement in the functional role of miR-221/222 as repressors of p27Kip1 was confirmed when we inhibited p65 activation by IkBαM and observed an upregulation of the reporter activity yielded by constructs harboring the 3′-UTR of p27Kip1 (Figure 1D). In 2008, Ozsolak et al. (38) demonstrated that an MITF binding site is present in Region A, at a short distance upstream of the NF-kB site identified by us. This supports our definition of Region A as a relevant regulatory region for miR-221/222, even if in our tumor models a different factor, p65, is playing its role in place of the melanoma-related MITF.
Previous studies on the transcriptional regulation of miR-221/222 cluster were carried out in other human cancers (3,18,27) and always led to the identification of promoter regions proximal to pre-miR-222 5′-end. In one paper, the MET oncogene was shown to activate miR-221/222 in non-small cell lung cancer and in hepatocellular carcinoma, through c-Jun recruitment to a promoter region at −130 bp upstream of pre-miR-222 5′-end (3). This observation is in accordance with our finding of c-Jun involvement in miR-221/222 upregulation in cancer. However, in our tumor models, the c-Jun site identified by Garofalo et al. (3) is not bound in ChIP experiments, suggesting that in different tumor types the same factor may use different sites to drive miR-221/222 transcription.
Our experiments describing the epigenetic state of Regions A and B strongly indicate that they likely work as enhancers when they are bound by NF-kB or c-Jun (Figure 5), acting as long-distance inducers of RNA Polymerase II recruitment at miR-221/222 TSS. Moreover, our results about H3K4me1 and H3K9ac enrichment at Regions A and B are in agreement with the data presented in the Human Mar. 2006 (NCBI36/hg18) assembly UCSC Genome Browser (http://genome.ucsc.edu/) that indicate a clear gathering of H3K4me1 and H3K9ac modifications at the same regions, even if in different cell contexts.
In conclusion, we think that our work opens a new perspective on miR-221/222 transcriptional regulation, by linking these two proven oncomiRs with master transcriptional regulators involved in cancer.
FUNDING
Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR) (PRIN 2007 no. 20077YZTL8_002); Ministero della Salute, Italy–USA collaborative programme (to M.G.F.). Funding for open access charge: Ministero della Salute, Italy-USA collaborative programme (to M.G.F.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Beatrice Salvatori for advice in setting up ChIP experiments, Gianluca Canettieri for providing anti-c-Jun antibodies and Cristina Buè for technical help.
REFERENCES
- 1.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 2.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J. Clin. Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl Acad. Sci. USA. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciafrè SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 6.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafrè SA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafrè SA, Farace MG. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J. Biol. Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 8.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baeuerle PA, Baltimore D. NF-kappa B: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 11.Cortés Sempere M, Rodríguez Fanjul V, Sánchez Pérez I, Perona R. The role of the NFkappaB signalling pathway in cancer. Clin. Transl. Oncol. 2008;10:143–147. doi: 10.1007/s12094-008-0171-3. [DOI] [PubMed] [Google Scholar]
- 12.Smith D, Shimamura T, Barbera S, Bejcek BE. NF-kappaB controls growth of glioblastomas/astrocytomas. Mol. Cell Biochem. 2008;307:141–147. doi: 10.1007/s11010-007-9593-4. [DOI] [PubMed] [Google Scholar]
- 13.Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T, Guney I, Strochlic DE, Macconaill LE, Beroukhim R, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat. Med. 2010;16:286–294. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 15.Shaulian E. AP-1–The Jun proteins: Oncogenes or tumor suppressors in disguise? Cell Signal. 2010;22:894–899. doi: 10.1016/j.cellsig.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakrabarti SK, James JC, Mirmira RG. Quantitative assessment of gene targeting in vitro and in vivo by the pancreatic transcription factor, Pdx1. Importance of chromatin structure in directing promoter binding. J. Biol. Chem. 2002;277:13286–13293. doi: 10.1074/jbc.M111857200. [DOI] [PubMed] [Google Scholar]
- 18.Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C, Iorio MV, Li M, Volinia S, Alder H, et al. MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J. Natl. Cancer Inst. 2010;102:706–721. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasparian AV, Yao YJ, Kowalczyk D, Lyakh LA, Karseladze A, Slaga TJ, Budunova IV. The role of IKK in constitutive activation of NF-kappaB transcription factor in prostate carcinoma cells. J. Cell Sci. 2002;115(Pt 1):141–151. doi: 10.1242/jcs.115.1.141. [DOI] [PubMed] [Google Scholar]
- 20.Syrovets T, Gschwend JE, Büchele B, Laumonnier Y, Zugmaier W, Genze F, Simmet T. Inhibition of IkappaB kinase activity by acetyl-boswellic acids promotes apoptosis in androgen-independent PC-3 prostate cancer cells in vitro and in vivo. J. Biol. Chem. 2005;280:6170–6180. doi: 10.1074/jbc.M409477200. [DOI] [PubMed] [Google Scholar]
- 21.Ryseck RP, Bravo R. c-JUN, JUN B, and JUN D differ in their binding affinities to AP-1 and CRE consensus sequences: effect of FOS proteins. Oncogene. 1991;6:533–542. [PubMed] [Google Scholar]
- 22.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 23.Zhou R, Hu G, Gong AY, Chen XM. Binding of NF-kappaB p65 subunit to the promoter elements is involved in LPS-induced transactivation of miRNA genes in human biliary epithelial cells. Nucleic Acids Res. 2010;38:3222–3232. doi: 10.1093/nar/gkq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song G, Wang L. A conserved gene structure and expression regulation of miR-433 and miR-127 in mammals. PLoS One. 2009;4:e7829. doi: 10.1371/journal.pone.0007829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc. Natl Acad. Sci. USA. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Löffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermüller J, Kretzschmar AK, Burger R, Gramatzki M, Blumert C, Bauer K, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 27.Felicetti F, Errico MC, Bottero L, Segnalini P, Stoppacciaro A, Biffoni M, Felli N, Mattia G, Petrini M, Colombo MP, et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;68:2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 28.Domingo-Domenech J, Mellado B, Ferrer B, Truan D, Codony-Servat J, Sauleda S, Alcover J, Campo E, Gascon P, Rovina A, et al. Activation of nuclear factor-kappaB in human prostate carcinogenesis and association to biochemical relapse. Br. J. Cancer. 2005;93:1285–1294. doi: 10.1038/sj.bjc.6602851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouyang X, Jessen WJ, Al-Ahmadie H, Serio AM, Lin Y, Shih WJ, Reuter VE, Scardino PT, Shen MM, Aronow BJ, et al. Activator protein-1 transcription factors are associated with progression and recurrence of prostate cancer. Cancer Res. 2008;68:2132–2144. doi: 10.1158/0008-5472.CAN-07-6055. [DOI] [PubMed] [Google Scholar]
- 30.Potapova O, Basu S, Mercola D, Holbrook NJ. Protective role for c-Jun in the cellular response to DNA damage. J. Biol. Chem. 2001;276:28546–28553. doi: 10.1074/jbc.M102075200. [DOI] [PubMed] [Google Scholar]
- 31.Gatto G, Rossi A, Rossi D, Kroening S, Bonatti S, Mallardo M. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 2008;36:6608–6619. doi: 10.1093/nar/gkn666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sliva D, English D, Lyons D, Lloyd FP., Jr Protein kinase C induces motility of breast cancers by upregulating secretion of urokinase-type plasminogen activator through activation of AP-1 and NF-kappaB. Biochem. Biophys. Res. Commun. 2002;290:552–557. doi: 10.1006/bbrc.2001.6225. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Lu Y, Pienta KJ. Multiple roles of chemokine (C-C motif) ligand 2 in promoting prostate cancer growth. J. Natl. Cancer Inst. 2010;102:522–528. doi: 10.1093/jnci/djq044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Desbaillets I, Tada M, de Tribolet N, Diserens AC, Hamou MF, Van Meir EG. Human astrocytomas and glioblastomas express monocyte chemoattractant protein-1 (MCP-1) in vivo and in vitro. Int. J. Cancer. 1994;58:240–247. doi: 10.1002/ijc.2910580216. [DOI] [PubMed] [Google Scholar]
- 36.Wolter S, Doerrie A, Weber A, Schneider H, Hoffmann E, von der Ohe J, Bakiri L, Wagner EF, Resch K, Kracht M. c-Jun controls histone modifications, NF-kappaB recruitment, and RNA polymerase II function to activate the ccl2 gene. Mol. Cell. Biol. 2008;28:4407–4423. doi: 10.1128/MCB.00535-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khattar E, Kumar V. Mitogenic regulation of p27(Kip1) gene is mediated by AP-1 transcription factors. J. Biol. Chem. 2010;285:4554–4561. doi: 10.1074/jbc.M109.029280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]