Abstract
The application of isothermal amplification technologies is rapidly expanding and currently covers different areas such as infectious disease, genetic disorder and drug dosage adjustment. Meanwhile, many of such technologies have complex reaction processes and often require a fine-tuned primer set where existing primer design tools are not sufficient. We have developed a primer selection system for one important primer, the turn-back primer (TP), which is commonly used in loop-mediated amplification (LAMP) and smart amplification process (SmartAmp). We chose 78 parameters related to the primer and target sequence, and explored their relationship to amplification speed using experimental data for 1344 primer combinations. We employed the least absolute shrinkage and selection operator (LASSO) method for parameter selection and estimation of their numerical coefficients. We subsequently evaluated our prediction model using additional independent experiments and compared to the LAMP primer design tool, Primer Explorer version4 (PE4). The evaluation showed that our approach yields a superior primer design in isothermal amplification and is robust against variations in the experimental setup. Our LASSO regression analysis revealed that availability of the 3′- and 5′-end of the primer are particularly important factors for efficient isothermal amplification. Our computer script is freely available at: http://gerg.gsc.riken.jp/TP_optimization/.
INTRODUCTION
Isothermal amplification enables amplification of a small quantity of nucleic acid in a short time without a thermocycling apparatus. Its popularity is rapidly growing and gradually covering different applications such as pathogen detection and SNP genotyping (1–4). This is because of several advantages of isothermal amplification, such as easy operation, quick results, cost and energy efficiency, and modest equipment requirements. To achieve DNA amplification at a constant temperature, strand separation is the crucial limiting step. In most cases, the DNA polymerases used in isothermal amplification reactions provide such functionality by their strong strand displacement activity. For this reason, the Pfu, Bst and Aac DNA polymerases are used, for example in rolling circle amplification (RCA) (5), loop-mediated amplification (LAMP, Supplementary Figure S1A) (6,7) and smart amplification process (SmartAmp, Supplementary Figure S1C) (4) DNA amplification reactions. In other cases, additional enzymes such as helicase are added to enable or accelerate the isothermal amplification reactions (8). While only one primer is sufficient for the RCA reaction, the LAMP and SmartAmp systems are more complicated, utilizing 3–6 primers at once. RCA amplifies a circular DNA at room temperature, and has been applied for a variety of purposes (9–11). LAMP is mainly used for pathogen detection with its rapid and accurate detection (3). SmartAmp is a particularly useful method for SNP and mutation detection (4). The method enables detection of genetic polymorphisms or somatic mutations in ∼30 min from whole blood without any DNA purification. SmartAmp uses asymmetric primers, a turn-back primer (TP) and a folding primer (FP) (Supplementary Figure S1C), which play key roles in the acceleration (TP) and control (FP) of the reaction (12). In contrast, LAMP has a symmetric primer design (Supplementary Figure S1A), where the most important primers, the inner primers (IP), have the same structure as the TP in SmartAmp. For LAMP as well as SmartAmp, TPs are the drivers of the amplification reactions, and therefore an optimal TP design is essential for an efficient amplification reaction.
An effective primer design is required for all primer-based methods. Accumulated experience on the behavior of different primers has revealed several important physical properties of the primer sequence, such as its GC content, melting temperature, self-complementarity and free energy for hybridization at the 3′-end, as well as parameters related to secondary-structure formation. Many PCR primer design tools have been developed over the years (13–21) and are widely used. However, many isothermal DNA amplification technologies have more complex reaction processes and require a fine-tuned primer design. Often the available primer design tools are not sufficient, with a large and complicated experimental primer screening procedure becoming an essential step to obtain a final primer set.
TPs are prominent examples for primers preferably used for isothermal amplification reactions, and they are in principle able to maintain an amplification reaction without support from other primers (TP–TP system shown in Supplementary Figure S1B). Therefore we focused on the analysis of TPs and their mode of action in SmartAmp reactions to find the optimal conditions for the use of TPs in isothermal amplification. Optimized TP designs using our parameters were proven effective for LAMP and SmartAmp reactions, having both a higher reaction speed and reduced background amplification from primer dimers.
MATERIALS AND METHODS
Amplification reactions
In this study, we examined two isothermal amplification systems, TP–TP system and TP–FP–BP system, under two different reaction conditions, Aac reaction condition and Bst reaction condition. In all the setups, 2400 copies of plasmids or 8 ng of human genomic DNA were used as DNA templates. All the reactions were performed in 10 μl total volume and assayed in triplicate.
In the Aac reaction condition, each reaction volume contained 1.4 mM of dNTPs, 5% DMSO, 20 mM of Tris–HCl (pH 8.0), 10 mM of KCl, 10 mM of (NH4)2SO4, 8 mM of MgSO4, 0.1% Tween-20, 1/100 000 diluted original SYBR Green I (Invitrogen), 0.24 U/μl of Aac DNA polymerase (DNAFORM K.K.). In the Bst reaction condition, 0.8 M Betaine instead of DMSO and 0.32 U/μl of Bst DNA polymerase (New England Biolabs) instead of Aac DNA polymerase were used.
For the TP–TP system, 3.3 μM each of forward and reverse TP were used. For the TP–FP–BP system, 2.66 μM each of TP and FP and 1.33 μM of BP (TP:FP:BP = 2:2:1) were used.
All reactions were assembled on ice. The DNA templates were heated at 98°C for denaturation prior to setting up the reactions. All amplification reactions were incubated at 60°C for 90 min and monitored in real time with 1-min interval using a Mx3000 (STRATAGENE) or LightCycler480 (Roche).
Calculation of parameters
The free energy of the most stable DNA secondary structure was calculated by hybrid-ss-min in the UNAfold package (22).
The free energy of the most stable hybridization between two DNA sequences was calculated by hybrid-2s.pl in the UNAfold package (22).
The probability of non-paired state, which can be an indicator of single-stranded state of a particular base in a given DNA sequence, was calculated by the following procedure: first, the probabilities of all possible base pairs within a given DNA sequence were calculated by hybrid-ss in the UNAfold package (22), which employs the partition function of statistical thermodynamics. For a particular base, the sum of the probabilities of base pairing with any other base was calculated to find the total base pairing probability Ptotal. 1–Ptotal was then defined as the probability of a particular base not being paired.
The free energy of the 3′-anchored primer binding were calculated by Fastagrep (20).
Calculation of the reaction speed
Generation of double-stranded DNA was monitored in real-time by addition of SYBR Green I. The amplification was measured by the emitted fluorescence at 1-min intervals. We defined the reaction speed as the inverse of the reaction time, which is defined as the time at which the signal exceeds a certain threshold. The signals from 1 to 10 min were omitted because they tend to be unstable. First, the signal amplitude at 11 min relative to the maximum signal for each well in a reaction set was calculated. Then the maximum amplitude across all the wells in the reaction set was used as the normalization factor NF. Next, the reaction time T was obtained as the time where the signal exceeded one-third of the NF. If the signal did not exceed the threshold during the reaction, the reaction time T was set as ‘undefined’. The reaction speed S was then obtained as 60/T (if T was defined) or 0 (if T was undefined). The mean reaction speed <S> of a particular primer set was finally obtained as the average of S over the replicates.
Prediction of reaction speed with LASSO
We created a linear regression model for the reaction speed with the least absolute shrinkage and selection operator (LASSO) method (23), a popular method for estimation in linear models. The LASSO procedure enabled us to avoid over-fitting by selecting a subset of the predictors setting the coefficients of the other predictors to zero. We used this method for parameter selection and estimation of the numerical values of the coefficients. We examined 68 parameters for target amplification and 28 parameters for background amplification (Supplementary Table S1) using a training data set comprising of 24 target regions, 420 primers and 1344 primer combinations under the Aac reaction conditions.
To choose the shrinkage factor in the LASSO, we performed 5-fold cross-validation 100 times, in which we selected four or five out of 24 targets for validation and used the remaining targets for model fitting. The overall mean absolute difference between prediction and observation was calculated as the cross-validation error (CVE). The CVE was calculated for all shrinkage factors, choosing the shrinkage factor which gave the minimum CVE as the optimal shrinkage factor. With the optimal shrinkage factor, the LASSO determined the numerical values for coefficients of the parameters. We then calculated the t-value of each parameter by dividing the numerical coefficient by the standard error, taking the LASSO structure into consideration (24), and evaluated its statistical significance assuming a t-distribution with N–p–1 degrees of freedom (N being the number of observations and p being the number of parameters), under the null hypothesis that the numerical coefficient is zero. For statistical analysis and modeling, R version 2.8 was used. We employed the LAR package (25) for the LASSO procedure.
Primer selection for comparison with Primer explorer version 4
We compared our predictions with primer explorer version 4 (PE4), a tool to assist designing LAMP primer sets (https://primerexplorer.jp/lamp4.0.0/index.html). We used the default condition and the standard selection steps in the primer selection with PE4. To perform the comparison in the same region, we chose the center position, for which eight forward and eight reverse TPs could be selected on each side. We then selected eight forward and eight reverse TPs randomly with different 3′-end positions on each side. We calculated our prediction scores for PE4 selections and confirmed that our prediction scores for the random selection had the same distribution as all selections (data not shown).
Our prediction was performed on the DNA region around the center position of the same DNA sequence used in primer design by PE4. The criteria for the selection of TPs were higher predicted target amplification speed and lower predicted background amplification speed with different 3′-end positions. The difference between the observed target and the observed background amplification speed was calculated as the evaluation score for the comparison between PE4 and our method.
RESULTS
Important parameters for TP
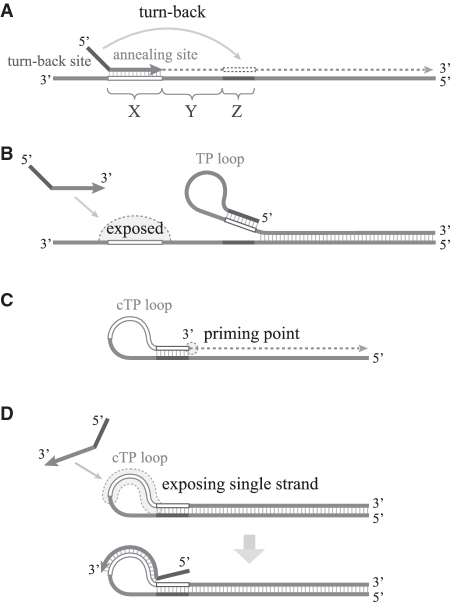
To concentrate on the amplification efficacy of the TP as a major primer in SmartAmp and LAMP, we considered an amplification system driven by only two TPs (TP–TP system, Supplementary Figure S1B). The principal primer reactions in the TP–TP system are illustrated in Supplementary Figure S2. In our designs for the TP–TP system, the amplified region between a forward TP and a reverse TP does not exceed 200 nt. An exponential amplification using TP is achieved by combining a turn-back site with an annealing site. The turn-back site is intentionally designed to bind a sequence downstream of the annealing site to form a loop structure (Figure 1A). The loop can be formed by either the sense or anti-sense strand of TP, and is termed the TP loop or cTP loop (complementary TP loop), respectively. Sense TP loop formation allows the annealing site to be exposed for hybridization of new TP (Figure 1B), where an extension from this new TP can displace the existing strand. Anti-sense cTP loop formation creates a priming site for DNA extension at the 3′-end (Figure 1C). The cTP loop also functions as a structure to keep a single-strand DNA exposed in amplified products, to which a new TP can hybridize and initiate additional primer extension reactions (Figure 1D). TP–TP mediated amplification yields a specific amplification product commonly seen after 40 min. However, in most TP–TP-mediated amplification reactions, a template-free amplification is seen after longer incubation times. This ‘background amplification’ can lead to false positive results, if there is no clear time difference between the specific and the unspecific amplification reactions. To avoid such false positive results, we measured two types of amplification for each primer set: ‘target amplification’ and ‘background amplification’, where the target amplification contained a DNA template but the background amplification did not.
Figure 1.
Conceptual figures of TP roles. (A) The turn-back site is intentionally designed to bind the sequence downstream of the annealing site to form a loop structure. (B) The sense TP loop formation allows the annealing site to be exposed for the next hybridization. (C) The anti-sense cTP loop formation creates a priming site for DNA extension at the 3′-end and (D) the cTP loop works as a structure to keep single-stranded DNA exposed in amplified products, where a new TP can hybridize very easily (D).
For creating prediction models for TP, we assumed that the following factors are important for efficient amplification using TP: accessibility for priming, specificity of priming, stability of priming, strand extension efficacy by DNA polymerase, and efficacy of the TP loop formation. These factors were then represented as parameters using the following physical features: length, free energy and probability of non-paired state (‘Materials and Methods’ section). The accessibility for priming, which is mainly determined by the secondary structure of both primer and template sequences, was estimated by the probability of non-paired state. This probability was calculated for the annealing and turn-back regions as well as for their template sequences. The specificity of priming was evaluated as the number of 3′-anchored primer binding sites on the whole template whose free energies exceeded a certain threshold. The stability of priming was evaluated as the free energy of hybridization, which was calculated for the annealing and turn-back regions. The strand extension efficacy by DNA polymerase was evaluated as the length of the amplified product. The efficacy of the TP loop formation was assessed by the ratio of lengths of the annealing, turn-back and TP loop regions. For the background amplification, template-related features were not considered, such as those related to the turn-back region and the TP loop. In total 78 unique parameters were considered: 68 parameters for target amplification and 28 parameters for background amplification (refer to Supplementary Table S1 for more details).
We employed LASSO (‘Materials and Methods’ section) to select significant parameters and to calculate the coefficients that maximize the amplification speed (shown in Table 1), using a training data set comprising of 24 target regions, 420 TPs, and 1344 TP–TP combinations assayed under the Aac reaction conditions (Supplementary Figure S3). The most significant parameter for the target amplification was ‘dG_3end_complementarity_TP’, which is the propensity for the 3′-end of two TPs of the same type to bind to each other. Its coefficient being negative indicates that this type of hybridization is wasteful and reduces the reaction speed. The second most significant parameter was ‘Probability_non-paired_TP_5end_1_3’, which is the probability of non-paired state in the first three bases of the TP 5′-end. Its coefficient being positive suggests that a non-paired structure in the TP 5′-end, which is important in the turn-back event, accelerates the reaction. Two parameters relating to free energy for hybridization to the target template, which is well known to be important in PCR amplification, were in the third and sixth place.
Table 1.
Significant parameters (P < 0.2) derived from LASSO
| Parameter_name | Description | Coefficient | P-value |
|---|---|---|---|
| Parameters significant for target amplification | |||
| dG_3end_complementarity_TP | −ΔG of 3′-anchored binding of two TPs within the same TP type | −9.73E–02 | 5.10E–26 |
| Probability_non-paired_TP_5end_1_3 | The probability for non-paired state in 1–3 bases of TP 5′-end | 1.75E–01 | 4.52E–03 |
| dG_cTP_turnback_5end_1_6 | −ΔG of binding of 1–6 base of cTP turn-back 5′-end to its complementary sequence | 7.97E–02 | 3.66E–02 |
| Probability_non-paired_TP_3end_1_3 | The probability for non-paired state in 1–3 bases of TP 3′-end | 1.41E–01 | 5.62E–02 |
| |(X–Y)/X| | |(X–Y)/X| X: length of the annealing region Y: distance between the annealing and the turn-back region (excluding binding region) Very long Y makes TP loop formation hard. Very short Y does not make enough space in the TP loop for coming new TP. | –7.91E–02 | 1.22E–01 |
| dG_TP_anneal_3end_1_3 | −ΔG of binding of 1–3 base of TP annealing 3′-end | 7.80E–02 | 1.35E–01 |
| Parameters significant for background amplification | |||
| dG_3end_binding_TP_hetero_TP | −ΔG of 3′-anchored binding of two TPs between forward and reverse TP | 2.40E–02 | 2.69E–10 |
| dG_3end_complementarity_TP | −ΔG of 3′-anchored binding of two TPs within the same TP type | −3.01E–02 | 2.78E–07 |
| dG_TP_3end_1_6 | −ΔG of binding of 1–6 base of TP 3′-end to its complementary sequence | 9.58E–02 | 2.61E–05 |
| dG_TP_5end_4_9 | −ΔG of binding of 4–9 base of TP 5′-end to its complementary sequence | −7.44E–02 | 1.20E–04 |
| dG_TP_3end_7_12 | −ΔG of binding of 7–12 base of TP 3′-end to its complementary sequence | 5.67E–02 | 3.49E–03 |
| dG_TP_heterodimer | −ΔG of dimer formation between forward and reverse TP | 2.03E–02 | 8.90E–03 |
| dG_TP_homodimer | −ΔG of dimer formation within the same TP type | 1.87E–02 | 9.41E–03 |
| dG_TP_5end_1_6 | −ΔG of binding of 1–6 base of TP 5′-end to its complementary sequence | 4.05E–02 | 1.29E–02 |
| Probability_non-paired_TP_3end_1_3 | The probability for non-paired state in 1–3 bases of TP 3′-end | 1.33E–01 | 1.92E–02 |
| dG_TP_self-folding | −ΔG of self-folding of TP | −3.52E–02 | 5.07E–02 |
| dG_TP_5end_7_12 | −ΔG of binding of 7–12 base of TP 5′-end to its complementary sequence | −4.08E–02 | 5.43E–02 |
| Probability_non-paired_TP_5end_1_3 | The probability for non-paired state in 1–3 bases of TP 5′-end | 5.73E–02 | 1.34E–01 |
| dG_TP_3end_4_9 | −ΔG of binding of 4–9 base of TP 3′-end to its complementary sequence | 2.03E–02 | 1.35E–01 |
The most significant parameter for the background amplification was ‘dG_3end_binding_TP_hetero_TP’, which is the free energy of hybridization between the 3′-ends of the forward and reverse TP. Its positive coefficient indicates that this hetero interaction contributes to the background amplification. The second most important parameter was again ‘dG_3end_complementarity_TP’.
Evaluation in TP–TP system
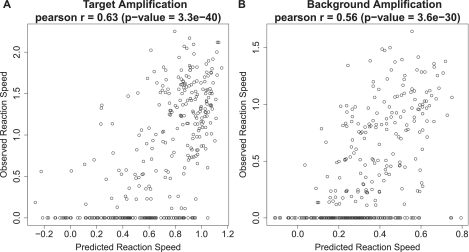
To evaluate the obtained linear prediction model, we applied it to an independent test set consisting of six target regions with 100 TPs, and 352 TP–TP combinations assayed under the Aac reaction conditions (Supplementary Figure S4). Predicted and observed reaction speeds were compared for target amplification and background amplification in the absence of a template. The resultant Pearson correlation coefficients (r) were 0.63 (P < 10−39) and 0.56 (P < 10−29) for target and background amplification, respectively (Figure 2A and B). These values did not vary significantly when trying different settings of training and test data set. Considering the fact that the observed reaction speeds showed some variation among replicates, with Pearson correlations coefficients around 0.9 and 0.8 for target and background amplification, respectively (Supplementary Figure S5), we conclude that the prediction performs well.
Figure 2.
Evaluation in the TP–TP system. The predicted reaction speeds were compared with the observed reaction speeds, both in target amplification (A) and in background amplification (B).
Whereas many reactions had a reaction speed predicted to be high but observed to be low (false positives, right bottom side of the Figure 2A and B), almost no reactions had a low predicted speed but a high observed reaction speed (false negatives, left top side of the Figure 2A and B). We concluded that our prediction always assigned a high speed for the primer that subsequently yielded the fastest amplification experimentally.
Evaluation in TP–FP–BP system
Since our predictions showed satisfying results for a simple TP–TP system, we evaluated predicted TPs next in the TP–FP–BP system, a simplified version of the SmartAmp system. The full set of the SmartAmp system consists of five primers: one TP along with a FP, a boost primer (BP) and two additional outer primers (OP) (4) (Supplementary Figure S1C). The TP–FP–BP experiments performed for our evaluation used only the major primers TP, FP and BP. Beside the already described TP, a reverse FP has the unique feature that an annealing site is combined with a tail that can form a hairpin structure. BP is a linear primer which can anneal fully to the template and is designed to hybridize to the TP loop (Supplementary Figure S1D.). For our comparison, TPs were selected with our prediction model whereas FPs and BPs were selected manually and stayed the same in each target region.
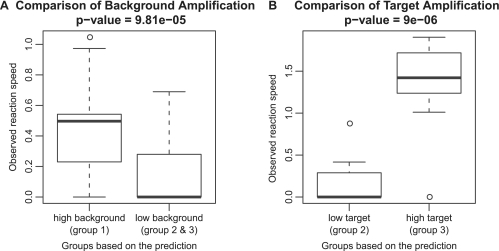
In the TP–FP–BP reactions, the background and target amplification were monitored. TPs were grouped according to predicted reaction speed, with group 1 having a high predicted background amplification, group 2 having a low background and a low target amplification, and group 3 having low predicted background but high predicted target amplification. For each group, two target regions and 16 primers with different 3′-end positions were selected (Supplementary Figure S6). The reactions were performed in the Aac reaction conditions.
First we compared group 1 against group 2 and 3 to look at high versus low background amplification, respectively. The observed background speed showed a clear reduction in the low background group [P < 10−4 (Wilcoxon rank sum test)] (Figure 3A). The low background TPs (groups 2 and 3) were then further assessed in terms of their target amplification, where the observed target speed showed a significant correlation in the high target amplification group, group 3 (P < 10−5, Wilcoxon rank sum test) (Figure 3B). The results suggested that our prediction method is also applicable for the TP in the SmartAmp system.
Figure 3.
Evaluation in the TP–FP–BP system. Three groups based on the predicted reaction speed, group 1: high background, group 2: low background and low target amplification, and group 3: low background and high target amplification, were compared in terms of their observed reaction speed. (A) Comparison of the background amplification: high background (group 1) versus low background TPs (groups 2 and 3). (B) The low background TPs (groups 2 and 3) were then further assessed by the target amplification: low target amplification (group 2) versus high target amplification (group 3).
Comparison with existing tool Primer explorer version 4
The two major primers in LAMP, the IP, have the same features as the TP in SmartAmp. Primer explorer version 4 (PE4), a tool to assist designing LAMP primer sets, is available online (https://primerexplorer.jp/lamp4.0.0/index.html). Although PE4 does not use scores to predict primers, it allows for selection of primers based on user-defined conditions. To compare our predictions with PE4 predictions, we used the default settings in PE4. We examined two target regions, in which eight forward and eight reverse TPs were selected per target (in total two times 64 assays).
Since SmartAmp and LAMP originally employ different DNA polymerases and buffer conditions, we compared them in both the Aac (SmartAmp) and the Bst (LAMP) experimental setups.
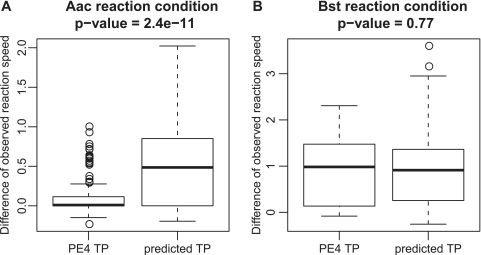
The difference between the observed target and the observed background amplification speed was calculated as an evaluation score in this comparison. Our predictions performed significantly better under the Aac conditions (P < 10−10, Wilcoxon rank sum test) (Figure 4A) than TP designed by PE4. Most of the PE4-based reactions showed either no amplification within 90 min, or equal amplification for the background and the target amplification signals. Under the Bst conditions, the overall performance of our prediction and PE4 were virtually the same (P = 0.77, Wilcoxon rank sum test; Figure 4B). Additionally, again we found that the best primer set, which had the largest difference in reaction speed, was included in our predicted primer sets for all targets. Hence, our prediction performed well regardless of the DNA polymerase and buffer conditions used. This implies that the performance of our prediction method is robust with respect to changes in the experimental setup, suggesting that the importance of TP features is independent of the specific amplification conditions. Hence our model may be widely used where TP are applied for isothermal DNA amplification.
Figure 4.
Comparison with an existing primer design tool, Primer explorer version 4 (PE4) (https://primerexplorer.jp/lamp4.0.0/index.html). PE4 is a tool to assist designing LAMP primer sets, where one of the major primers in LAMP, inner primer (IP), has the same features as TP in SmartAmp. Since SmartAmp and LAMP employ different experiment conditions, the Aac and Bst condition, respectively, the comparison was made under both the Aac (A) and Bst (B) experiment conditions. The difference between the observed target and the observed background amplification speed was calculated as an evaluation score in this comparison.
DISCUSSION
We have developed a primer selection system of the TP for use in SmartAmp and LAMP by predicting the amplification speed for both the target and background amplification. As the development of fine-tuned primers used in the complicated isothermal amplification processes often needs extensive experimental screening, this in silico approach facilitates rapid development of primer sets.
We examined various parameters to establish a model for predicting the reaction speed. Availability of the 3′-end region of TP, which is calculated as free energy of 3′-anchored binding of two TPs within the same primer type, was commonly found to be very important for both the target and background amplification. We speculate that it is because the availability of 3′-end which involves many kinds of priming events, including primer dimer formation. On the other hand, to differentiate the target amplification from the background amplification, other parameters that appeared exclusively in either the target or background amplification model are important. One such parameter in the target amplification was the probability of non-paired state at the 5′-end region of TP. This is an indicator of the availability of the 5′-end region of TP, which is important for the turn-back event followed by loop formation (Figure 1). In the background amplification, one such parameter was free energy of hybridization between the 3′-ends of forward and reverse TP, which is an indicator of hetero dimer formation.
We tested our prediction models using independent test data. The overall correlations between predicted and observed reaction speed for the target and background amplification were around 0.6. Some reactions were predicted to be rapid but observed to be slow (Figure 2A and B). This suggests that there may be additional parameters affecting the reaction speed that have not yet been identified. Considering reaction spans longer than 90 min may also improve the correlation. On the other hand, reaction speeds observed as high were always predicted as high. Using this method, it is thus possible to avoid the unnecessary experimental examinations of primers likely to have a low efficiency as predicted by our method.
A comparison with the LAMP primer design tool PE4 showed that some primer sets exhibit large differences in reaction speed under different experimental conditions. However, importantly our LASSO prediction methodology performed well for both the Aac and Bst reaction conditions, despite having been optimized using the Aac conditions alone. Conversely, most of the primers selected by PE4 performed poorly under the Aac reaction conditions. PE4 selection relies on conventional parameters such as length, GC content, melting temperature and free energy of hybridization, while the availability of the 3′- and 5′-ends of a primer, which we found to be important, are not included. This suggests that those availabilities in a primer are important for isothermal amplification, and improves the robustness of the computational prediction in the face of variations in the experimental procedure.
Through the exploration of important physical properties of the TP and target template, we have identified parameters that affect amplification significantly. This knowledge may be applicable also for other primers in other isothermal amplification methods, and for conventional PCR. With prediction models based on these significant parameters, our prediction system offers an efficient selection of the TP for both SmartAmp and LAMP, and potentially other isothermal amplification systems, regardless of their experimental setups.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Research Grant for RIKEN Omics Science Center from Ministry of Education, Culture, Sports, Science and Technology (MEXT) (to Y.H.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to acknowledge Matthias Harbers, Takeshi Hanami, Yuki Tanaka and Kengo Usui for helpful comments on the article; and also Takefumi Ishidao, Shiro Fukuda, Atsuko Oguchi-Katayam, Jean-Etienne Morlighem, Timo Lassmann and Thierry Sengstag for scientific discussions.
REFERENCES
- 1.Gill P, Ghaemi A. Nucleic acid isothermal amplification technologies - a review. Nucleosides Nucleotides Nucleic Acids. 2008;27:224–243. doi: 10.1080/15257770701845204. [DOI] [PubMed] [Google Scholar]
- 2.Jeong YJ, Park K, Kim DE. Isothermal DNA amplification in vitro: the helicase-dependent amplification system. Cell. Mol. Life Sci. 2009;66:3325–3336. doi: 10.1007/s00018-009-0094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mori Y, Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Inf. Chemother. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitani Y, Lezhava A, Kawai Y, Kikuchi T, Oguchi-Katayama A, Kogo Y, Itoh M, Miyagi T, Takakura H, Hoshi K, et al. Rapid SNP diagnostics using asymmetric isothermal amplification and a new mismatch-suppression technology. Nature Methods. 2007;4:257–262. doi: 10.1038/nmeth1007. [DOI] [PubMed] [Google Scholar]
- 5.Liu DY, Daubendiek SL, Zillman MA, Ryan K, Kool ET. Rolling circle DNA synthesis: Small circular oligonucleotides as efficient templates for DNA polymerases. J. Am. Chem. Soc. 1996;118:1587–1594. doi: 10.1021/ja952786k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 7.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vincent M, Xu Y, Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004;5:795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silander K, Saarela J. Whole genome amplification with Phi29 DNA polymerase to enable genetic or genomic analysis of samples of low DNA yield. Methods Mol. Biol. 2008;439:1–18. doi: 10.1007/978-1-59745-188-8_1. [DOI] [PubMed] [Google Scholar]
- 10.Zhao W, Ali MM, Brook MA, Li Y. Rolling circle amplification: applications in nanotechnology and biodetection with functional nucleic acids. Angew. Chem. Int. Ed. Engl. 2008;47:6330–6337. doi: 10.1002/anie.200705982. [DOI] [PubMed] [Google Scholar]
- 11.Johne R, Muller H, Rector A, van Ranst M, Stevens H. Rolling-circle amplification of viral DNA genomes using phi29 polymerase. Trends Microbiol. 2009;17:205–211. doi: 10.1016/j.tim.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Kimura Y, Oguchi-Katayama A, Kawai Y, Naito K, Mitani Y, Morlighem JE, Hayashizaki Y, Lezhava A. Tail variation of the folding primer affects the SmartAmp2 process differently. Biochem. Biophys. Res. Commun. 2009;383:455–459. doi: 10.1016/j.bbrc.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 13.Rychlik W, Rhoads RE. A computer program for choosing optimal oligonucleotides for filter hybridization, sequencing and in vitro amplification of DNA. Nucleic Acids Res. 1989;17:8543–8551. doi: 10.1093/nar/17.21.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowe T, Sharefkin J, Yang SQ, Dieffenbach CW. A computer program for selection of oligonucleotide primers for polymerase chain reactions. Nucleic Acids Res. 1990;18:1757–1761. doi: 10.1093/nar/18.7.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brownie J, Shawcross S, Theaker J, Whitcombe D, Ferrie R, Newton C, Little S. The elimination of primer-dimer accumulation in PCR. Nucleic Acids Res. 1997;25:3235–3241. doi: 10.1093/nar/25.16.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rozen S, Skaletsky HJ. Primer3 on the WWW for General Users and for Biologist Programmers. Totowa, NJ: Humana Press; 2000. [DOI] [PubMed] [Google Scholar]
- 17.Yuryev A, Huang JP, Pohl M, Patch R, Watson F, Bell P, Donaldson M, Phillips MS, Boyce-Jacino MT. Predicting the success of primer extension genotyping assays using statistical modeling. Nucleic Acids Res. 2002;30:e131. doi: 10.1093/nar/gnf131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miura F, Uematsu C, Sakaki Y, Ito T. A novel strategy to design highly specific PCR primers based on the stability and uniqueness of 3′-end subsequences. Bioinformatics. 2005;21:4363–4370. doi: 10.1093/bioinformatics/bti716. [DOI] [PubMed] [Google Scholar]
- 19.SantaLucia J. Physical Principles and Visual-OMP Software for Optimal PCR Design. Totowa, NJ: Humana Press; 2007. [DOI] [PubMed] [Google Scholar]
- 20.Andreson R, Mols T, Remm M. Predicting failure rate of PCR in large genomes. Nucleic Acids Res. 2008;36:e66. doi: 10.1093/nar/gkn290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mann T, Humbert R, Dorschner M, Stamatoyannopoulos J, Noble WS. A thermodynamic approach to PCR primer design. Nucleic Acids Res. 2009;37:e95. doi: 10.1093/nar/gkp443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markham NR, Zuker M. In: Bioinformatics, Volume II. Structure, Function and Applications. Keith JM, editor. Vol. 453. Totowa, NJ: Humana Press; 2008. pp. 3–31. [Google Scholar]
- 23.Tibshirani R. Regression shrinkage and selection via the Lasso. J. Roy. Stat. Soc. B. 1994;58:267–288. [Google Scholar]
- 24.Osborne MR, Presnell B, Turlach BA. On the LASSO and its dual. J. Comput. Graph. Stat. 2000;9:319–337. [Google Scholar]
- 25.Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. Ann. Stat. 2004;32:407–451. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.