Abstract
Amyloid is traditionally viewed as a consequence of protein misfolding and aggregation and is most notorious for its association with debilitating and chronic human diseases. However, a growing list of examples of “functional amyloid” challenges this bad reputation and indicates that many organisms can employ the biophysical properties of amyloid for their benefit. Because of developments in the structural studies of amyloid, a clearer picture is emerging about what defines amyloid structure and the properties that unite functional and pathological amyloids. Here, we review various amyloids and place them within the framework of the latest structural models.
Keywords: Alzheimer Disease, Amyloid, NMR, Prions, Protein Self-assembly
Introduction
The idea that functional proteins can employ an amyloid structure is just the latest step in the evolution of the concept of amyloid. The term amyloid is itself a misnomer arising from its original misidentification as being composed of starch, but even after it was determined to be proteinaceous (reviewed in Ref. 1), its identity was largely defined by its histological properties, such as localization to tissue plaques with specific dye-binding properties. A protein structural view of amyloid began to emerge in the 1960s after the work of Cohen and Calkins (2), who first reported its non-branching fibrillar structure viewed by electron microscopy. A decade later, Eanes and Glenner (3) showed by x-ray diffraction that amyloid is β-sheet-rich, with the β-strands perpendicular to the long axis of the fibrils, the distinctive “cross-β-pattern.” Thus, amyloid came to be viewed as a highly ordered filamentous structure and not simply amorphous deposits in tissue.
The traditional link of amyloid to disease has led some to assert that the term “amyloid-like” should be used for proteins that possess the hallmarks of amyloid but are not associated with pathological plaques (4). Regardless of localization or functionality, there exists a protein biophysical state that is not limited to disease and more broadly represents a low-energy conformation that is common to many polypeptides (5). Knowing which proteins are likely to be amyloidogenic under physiological conditions is difficult because there is no particular defining amino acid sequence motif. Moreover, amyloid-forming proteins display a broad range of sequence composition; however, intrinsic disorder (or destabilized structure) in the amyloid-prone domain is common.
The contemporary definition of amyloid is based primarily on its structural and biophysical properties. Amyloid is a highly ordered protein aggregate with a filamentous morphology that is generally unbranched with indefinite length and diameters of ∼2–20 nm. These fibrils are rich in β-sheet secondary structure and are formed by the noncovalent polymerization of a single protein with the polypeptide chains aligned in the cross-β-configuration. Amyloid is also generally relatively resistant to denaturation and proteolysis and shows yellow-green birefringence on binding Congo red and intense fluorescence on binding thioflavin T. However, these dyes are imperfect reporters, and they are not necessarily specific to a single type of amyloid structure (6). Several algorithms have been developed to predict amyloid propensity based on general sequence characteristics, such as hydrophilic residues, intrinsic disorder, and lack of charge (for example, see Ref. 7).
The “functional amyloid” concept is based on the discovery of several proteins that natively form filamentous aggregates with many of the biophysical qualities of amyloid (Table 1). How functional amyloid may be similar to or different from pathological amyloid is becoming clearer as more is learned about amyloid structure. Several very different amyloid folds have been demonstrated, as outlined below.
TABLE 1.
Examples of proposed functional amyloid and amyloid-like proteins
| Protein | Organism | Amyloid function/characteristics |
|---|---|---|
| Chaplins | S. coelicolor | Spore surface protein (66) |
| Chorion proteins | Antheraea polyphemus (silk moth) | Family of proteins that compose egg shell chorion; protective function (80) |
| Curli | E. coli | Adhesion to surfaces; biofilm component (54) |
| FapC | Pseudomonas fluorescens | Biofilm component (81) |
| Hydrophobins | Neurospora crassa | Form amphipathic surface layer (67, 68) |
| HpaG | Xanthomonas | Plant pathogen virulence factor (39) |
| HET-s | P. anserina | HET-s prion amyloid is determinant of hyphal fusion (82) |
| Fungal adhesins | Candida albicans, Saccharomyces cerevisiae | Cell adhesion (83, 84) |
| Microcin E492 (Mcc) | Klebsiella pneumoniae | Amyloid formation is proposed to regulate Mcc toxicity (85) |
| MSP2 | Plasmodium falciparum | MSP2 (merozoite surface protein 2) has been implicated in erythrocyte invasion (86) |
| Nsp1 | S. cerevisiae | FG repeat forms amyloid-like associations to control nucleocytoplasmic mixing at nuclear pore (87) |
| Peptide hormones | Homo sapiens | Protein hormones in secretory granules are stored in an amyloid-like conformation, which enables controlled release of monomeric functional hormone (88) |
| Pmel17 | H. sapiens | Facilitates melanin synthesis (89) |
| TasA | Bacillus subtilis | Biofilm component (90) |
| Type I antifreeze protein (AFP) | Pseudopleuronectes americanus | May play role in ice inhibition (91) |
Challenges of Structure Elucidation
Determining high-resolution structural information for amyloid has been historically challenging. The standard methods of transmission electron microscopy (TEM)2 and atomic force microscopy have been instrumental in its gross structural characterization, but they do not provide atomic level information. Amyloid is inherently noncrystal-forming, which precludes it from structural elucidation by x-ray crystallography. Also, because of the large size and particulate nature, it is not amenable to solution NMR spectroscopy. Although solution NMR and hydrogen-deuterium exchange can be used in characterizing such proteins in their soluble state and in defining amyloid-forming regions of proteins, they are limited in their ability to provide atomic level details of proteins in the large aggregated state. For these reasons, high-resolution details of amyloid structures have greatly lagged behind other large biological complexes, but during the past decade, advances in several techniques have begun to define some of their atomic level details.
Traditional TEM has served particularly well in the identification of amyloid polymorphisms, in which different structural elements are propagated along the length of the fibril, resulting in different fibril morphologies, such as ribbons or twists. Such morphological heterogeneity is a common characteristic of amyloid and is an indication of atomic level structural heterogeneity (8). EM can also be used to measure the mass per length of fibrils, which gives the number of protein monomers (or fraction of a monomer) that compose a unit length of the fibril, determining important constraints for model building. This method, employed most effectively using a scanning transmission electron microscope (9), can differentiate between categorically distinct fibrillar structures based on their packing densities. A more accessible method using standard TEM has also been developed (10).
Although amyloid fibrils cannot be crystallized, rational approaches using x-ray crystallography have contributed to understanding amyloid structure, particularly with respect to how amino acid side chains and their corresponding β-sheets may pack into fibril cores. Short amyloidogenic segments can form crystals that presumably possess qualities of amyloid structures, which are themselves like one-dimensional crystals. The Eisenberg laboratory determined the atomic architecture of several short peptides (4–10 residues) derived from amyloid proteins, such as Sup35, insulin, amyloid-β peptide (Aβ), Tau, amylin, and prion protein, which they were able to grow as three-dimensional microcrystals (11–13). The structures of these microcrystals all revealed β-sheets with an interlocking of self-complementary surfaces of adjacent β-sheets, a structure termed “steric zipper” (11–13). It is proposed that the presence of short sequences with self-complementary side chains is a determinant of amyloid formation.
Some of the most informative amyloid structure information has resulted from developments in techniques of solid-state NMR (ssNMR) and EPR spectroscopy, which can yield atomic spacing information on full-length proteins in the amyloid state. For ssNMR, uniformly 13C,15N-labeled samples offer the most possible information, but microheterogeneity of amyloid structure in even the best samples and the lower resolution of ssNMR compared with solution NMR often result in spectra that are too crowded to assign signals to specific nuclei. Specifically labeling selected atoms mitigates this problem, but unless the protein is small enough to be synthesized, one is limited to labeling in Escherichia coli using isotopically labeled amino acids in the growth medium. Despite these limitations, ssNMR has produced enough atomic distance constraints in a few cases to build complete high-resolution atomic structures (14–16). Because of such advances, it has become clear that although all amyloids share certain biophysical characteristics, they can be based on very different atomic level structure.
Amyloid Structure
Dozens of amyloid-forming peptides and proteins have been at least partially structurally characterized (Table 2). The structural information indicates that different proteins polymerize into filamentous structures that are rather broadly considered amyloid but based on structurally different arrangements. In some cases, there is the stacking of peptide strands in parallel and antiparallel sheets. In these cases, each contributing polypeptide is part of the greater fold of the fibril but does not necessarily compose a stably folded subunit in itself. In other cases, each subunit stacks to form the fibril, but the subunits are themselves in conformations that are recognizable as distinct protein folds. Both functional and pathological amyloids can be based on either arrangement, but functional amyloids may more frequently employ the stacking of folded subunits.
TABLE 2.
Examples of structurally characterized amyloid-forming proteins
aa, amino acids.
| Protein/peptide | Pathology/function linked to amyloid | Structural information |
|---|---|---|
| α-Synuclein (aa 1–140) | Parkinson disease | Five β-strands within fibril core comprising residues 35–96 (92); parallel in-register β-sheet core region from residues 36–98 (19) |
| Aβ1–40 | Alzheimer disease | Parallel in-register β-sheet (16); two fibril morphologies (twisted and striated ribbons) with 2- and 3-fold symmetry; both are parallel β-sheets, using almost same β-strand segments (8, 24) |
| Aβ1–40 seeded with diseased brain | Alzheimer disease | Parallel in-register β-sheet (75) |
| Aβ1–40(D23N) | Familial Alzheimer disease | Two species: antiparallel (major) and parallel in-register (minor) β-sheets (51) |
| Aβ1–42 | Alzheimer disease | Parallel in-register β-sheet (93); molecules form steric zipper (94) |
| Amylin (aa 1–37) | Type 2 diabetes | Four layers of parallel β-sheets (25) |
| β2-Microglobulin (aa 1–99) | Dialysis-related amyloidosis | Parallel in-register β-sheet (20); fibril core comprising 60–70 residues (95) |
| Curli (CsgA and CsgB) | Proteins secreted by E. coli; biofilm formation, surface colonization | Not parallel in-register, likely β-helix (55) |
| HET-s (aa 218–289) | Regulation of heterokaryon formation in P. anserina | Left-handed β-helix structure (14) |
| Htau40 | Tauopathies | Parallel in-register β-sheet (96, 97) |
| Rnq1 (aa 153–405) | Prion of S. cerevisiae; increased frequency of generation of [URE3] and [PSI+] prions | Parallel in-register β-sheet (36) |
| Sup35 (aa 1–253) | Prion of S. cerevisiae; reduction in fidelity of translation termination | Parallel in-register β-sheet (35); multiple variants employ same basic architecture (34) |
| Ure2 (aa 1–89) | Prion of S. cerevisiae; inappropriate derepression of nitrogen catabolism genes | Parallel in-register β-sheet (98) |
Parallel In-register β-Sheets
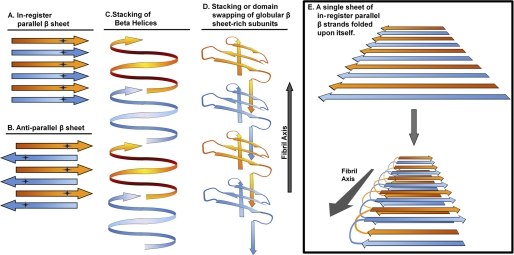
Parallel in-register β-sheet structure is the most common underlying architecture observed for pathological amyloid (Fig. 1, A and E). These fibrils are composed of stacked polypeptide strands that lie perpendicular to the fibril axis and form backbone hydrogen bonds with the adjacent strands aligned in-register and parallel. Each residue thus forms an aligned row along the long axis of the fibrils. As a result, the main chain hydrogen bonds are parallel with the long axis, and the β-sheets run the length of the fibril (Fig. 1E).
FIGURE 1.
Filamentous aggregates can be composed of various arrangements of their constituent proteins. Very different structures are proposed for various amyloids. A, parallel in-register β-sheet structures are composed of individual polypeptides stacking in-register every ∼4.7 Å along the fibril axis (common to many full-length proteins in pathological amyloids). B, antiparallel β-sheet structures are also composed of polypeptides stacking every ∼4.7 Å, but β-strands alternately run in opposite directions (observed primarily in amyloids composed of short polypeptide sequences). C, β-helices are composed of a single polypeptide wrapping around an axis, forming intramolecular parallel β-sheets (likely the structural basis of two functional amyloids). D, some β-sheet-rich proteins can linearly assemble into filamentous structures by other mechanisms, including domain swapping. E, amyloids composed of parallel in-register β-strands form continuous β-sheets that run the length of the fiber. The dimensions and packing densities of such amyloids suggest that the fibrils are composed of sheets folded upon themselves, as has been shown with Aβ (16). Complementary side chains may form steric zippers that stabilize the interlocking sheets. In the case of yeast prion amyloids, multiple and different sheet folds may underlie the variant phenomenon.
EPR is well established in determining parallel in-register amyloid structure (17). The general approach is to introduce a single unpaired electron spin label at a naturally occurring or introduced cysteine residue in the protein (18). Distances between spin labels can yield information about the configuration of the polypeptides. The neuronal aggregation of the protein α-synuclein is associated with neurodegenerative disorders like Parkinson disease. Amyloid fibrils of recombinant α-synuclein were shown by EPR to be based on parallel in-register architecture (19). β2-Microglobulin is a 99-amino acid protein found in amyloid deposits in the joints of patients undergoing long-term hemodialysis. Like α-synuclein, spin-labeled recombinant protein in the amyloid form was determined by EPR to be based on the stacking of polypeptides in parallel in-register β-sheets (20). The spin labels were distributed throughout β2-microglobulin, indicating that most of the protein is involved in the stacking. This is consistent with the findings of Iwata et al. (21), who used a combination of ssNMR and other methods to conclude that a peptide of β2-microglobulin formed the same type of structure.
The best characterized pathological amyloid structure is that of Aβ, which accumulates in brains of patients suffering from Alzheimer disease (reviewed Ref. 22). Aβ1–40, Aβ1–42, and shorter fragments have been studied by ssNMR. The relatively small size of Aβ facilitates structure elucidation because the full-length peptide can be synthesized with the insertion of isotopic labels at specific sites. Like many amyloid proteins, Aβ can form morphologically distinct fibrils, but the underlying structural basis of Aβ fibrils is relatively constant: peptides stacked in-register forming parallel β-sheets (Fig. 2A) (8, 16, 23, 24). The ssNMR data with mass-per-length measurements indicate that morphologically different fibrils can result from different numbers of peptides forming each ∼5-Å layer along the fibril axis. Similar structural models have been proposed for amylin (type 2 diabetes) and human prion protein (25, 26).
FIGURE 2.
Structures of pathological and functional amyloids. A, schematic representation of a parallel in-register β-sheet structure of Aβ1–40 (99). The Protein Data Bank file was kindly provided by Rob Tycko. The fibril is composed of two protofibrils (a single protofibril is shown in the lower half of A), which are each composed of stacked Aβ1–40 peptides in-register with the preceding and following peptides. B, schematic representation of HET-s β-helical amyloid (residues 223–283 from Protein Data Bank code 2KJ3) (100). The monomers alternate between blue and yellow, revealing that each polypeptide provides two β-strands that wrap around the long axis of the fiber.
Prions are infectious proteins that propagate information within the structure of the proteins themselves, as opposed to within nucleic acid. With some exceptions (27), most yeast prions are based on infectious amyloid. The following yeast proteins have been shown to be capable of being amyloid-based prions in yeast: Sup35, Ure2, Rnq1, Swi1, and Mot3 (28–32). The part of each protein responsible for the prion properties (the prion domain) is a large segment rich in hydrophilic amino acids like glutamine and asparagine and poor in charged and hydrophobic amino acids. These regions are presumably natively disordered but capable of aggregating into self-propagating amyloid that is the basis of the respective prion. The prion domains of Ure2, Sup35, and Rnq1 have each been characterized by ssNMR to determine the underlying architecture of their infectious form (33–36). Such experiments employed recoupling methods like PITHIRDS-CT (37), in which the rate of signal decay from selectively labeled nuclei is inversely proportional to the cube of the distance to the next nearest labeled nucleus. The prion domains were labeled at specific residues, and it was determined that, in each case, the nearest labeled neighbor was ∼5 Å distant, essentially the 4.7-Å distance between strands of a β-sheet. That this nearest neighbor was on a different molecule was confirmed by diluting labeled molecules with ∼4-fold unlabeled molecules, showing that the rate of signal decay was dramatically diminished. These results can be explained only by a parallel in-register β-sheet architecture. Mass-per-length measurements of infectious amyloid of various Sup35 and Ure2 constructs show one monomer per 4.7 Å (33, 38), the value predicted for a parallel in-register structure. Based on the similarities among the known yeast prions, it is likely that this is their common structural mechanism.
The parallel in-register architecture places a severe restraint on the possible amyloid structures, but the diameter of fibrils is generally smaller than if they formed a single flat β-sheet. The dimensions suggest that the sheets must be folded along the long axis of the fibrils (Fig. 1E). The locations of these folds could differ in different prion amyloid variants (biologically and structurally different prions that are based on the same prion protein). Thus far, only prions with parallel in-register structure display multiple prion variants determined by the same prion protein. Stacking peptides in-register provides a mechanism by which a single peptide chain can structurally encode and faithfully propagate any of several different prion variants. It is proposed that the same interactions between identical side chains aligned along the long axis of the fibrils that hold the strands in-register also direct a monomer joining the ends of the filaments to adopt the same structure as molecules already in the amyloid fibrils.
Harpins are heat-stable virulence proteins secreted by bacterial plant pathogens and may have a structure similar to that of yeast prions. HpaG, a harpin of Xanthomonas, forms fibrils that are indistinguishable from amyloid by EM and has a functional role during pathogenesis that elicits the plant hypersensitive response (39). HpaG has a repeat sequence nearly identical to a repeat found in the yeast prion protein Rnq1 and may form a functional yet toxic amyloid based on the common parallel in-register stacking of subunits.
Pmel17 forms a functional amyloid for melanin biosynthesis, promoting melanin deposition and possibly protecting cells from reactive intermediates produced during melanin biosynthesis (40). It is processed by a series of proteolytic and glycosylation steps and is assembled into a filamentous form in maturing melanosomes, the organelle where melanin is formed and stored. A fragment of Pmel17, “the repeat domain” (RPT), comprising 10 imperfect repeats, is known to be required for filament formation in vivo (41), is present in the in vivo filaments (42), and forms amyloid in vitro (43). The filaments form only at the mildly acidic pH (∼5) required in melanosomes for melanin synthesis. The RPT domain from mouse and zebrafish, where there is no sequence conservation, also showed selective amyloid formation at acidic pH (44). Both mouse and zebrafish RPT domains were shown by ssNMR to form fibrils based on parallel in-register β-sheet architecture. Moreover, the mouse RPT fibril mass-per-unit length measurement is consistent with stacking of whole polypeptides every 4.7 Å.
Antiparallel β-Sheets
Many small peptides that form amyloid fibrils, including fragments of Aβ, arrange in antiparallel β-sheets (Fig. 1B), which is also the most common β-sheet arrangement in globular proteins. Both Aβ16–22 and Aβ11–25 were shown by ssNMR to form amyloid based on an antiparallel alignment of the peptides (45, 46). The same was shown for Aβ34–42, whose C terminus formed an antiparallel β-sheet (47). Likewise, a 10-residue fragment of amylin, a peptide linked to complications of type 2 diabetes, was found by ssNMR to arrange in antiparallel β-sheets (48). For polyglutamine, often associated with Huntington disease, antiparallel stacking was proposed based on x-ray diffraction data (49), although several other structures have been suggested (reviewed in Ref. 50).
Antiparallel conformations may be overrepresented in the literature because small peptide segments are frequently used in structural studies as a simpler alternative to full-length proteins, especially in cases in which specific isotopic labels are desired and peptide synthesis methods put limits on the possible length of the polypeptide, and yet arguably no full-length amyloid proteins have been observed to preferentially form antiparallel β-sheet structures. A sole exception may be a mutant form of Aβ1–40, associated with early onset familial Alzheimer disease, that was found to form fibrils with antiparallel strands (51).
β-Solenoids/β-Helices
The strands in a β-helix (or solenoid) align to form parallel β-sheets, but the strands wrap around an axis in a helical arrangement, and unlike parallel in-register β-sheets, parallel strands in helices have intramolecular backbone hydrogen bonds. The infectious amyloid of the HET-s prion protein of Podospora anserina yielded good-quality NMR data (52), enabling the elucidation of a high-resolution structure (14). Within this β-helix amyloid, each HET-s monomer makes two helical turns around the filament axis, and the strands form parallel β-sheets (Figs. 1C and 2B). Recently, a HET-s homolog showing only 38% sequence identity to HET-s was found to be able to cross-seed fibril formation (53). This may indicate the importance of three-dimensional structure and not protein sequence, which would make it distinct from in-register stacking of other amyloids.
Curli are extracellular fibrous structures formed by some enterobacteria. They represent a category of adhesin proteins that facilitate the binding to surfaces and the formation of biofilms. The curli of E. coli are essentially filamentous homopolymers composed mostly of the CsgA protein. A β-helix structure of the CsgA monomer was proposed based on sequence alignment (54). SsNMR experiments with isotopically labeled CsgA ruled out a parallel in-register structure for CsgA amyloid (55). Moreover, mass-per-length measurements indicated a packing density that would be consistent with a β-helix structure.
Other β-Folds and Domain Swapping
Some aggregates that have been described as amyloid may be composed of proteins that remain largely in their native fold. Slight misfolding or structural rearrangement may potentiate a linear aggregation phenomenon. Such aggregation as the result of linear domain swapping between globular proteins has been proposed (56, 57). The polymerization of some serpin proteins, a structural class of proteins first identified as inhibitors of proteases, is associated with a family of diseases known as serpinopathies (58). X-ray crystallographic studies of a stable serpin dimer suggested that the β-sheet-rich serpins can form aberrant fibrillar structures through a domain-swapping mechanism in which a β-hairpin from each protomer is inserted into a β-sheet in the next protomer (59). Such polymers are superficially similar to amyloid, as they are disease-associated, fibrillar, and rich in β-sheet structure. Several other proteins have been proposed to polymerize through domain swapping (Fig. 1D). Human cystatin C, which is found in some types of amyloid deposits, was suggested to aggregate via a domain-swapping mechanism (60, 61). Also, the immunoglobulin-binding domain B1 of streptococcal protein G (GB1) forms fibrils through domain swapping (62).
The formation of extracellular amyloid composed of the protein transthyretin is linked to familial amyloid polyneuropathy and senile systemic amyloidosis. EPR and NMR studies suggest that transthyretin remains relatively folded in the amyloid conformation but undergoes structural rearrangements that permit the stacking of β-strands to form filaments (63, 64). Likewise, the superoxide dismutase SOD1, implicated in amyotrophic lateral sclerosis, forms β-sheet-rich filamentous aggregates through a structural rearrangement that enables subunit stacking (65).
Recently, spores of some bacteria and fungi have been proposed to have functional amyloid on their surfaces. The filamentous bacterium Streptomyces coelicolor produces aerial hyphae for spore dispersion. The spores possess surface proteins known as chaplins, which form amyloid-like fibrils (66). In some fungi, hydrophobin proteins form amphipathic monolayers on the surfaces of aerial hyphae and spores. Hydrophobins are cysteine-rich and self-aggregate into small filamentous structures termed rodlets, which have been characterized as amyloid-like because of their dimensions and dye-binding characteristics (67, 68). Like many amyloid-forming proteins, hydrophobins are largely disordered when in the soluble state (67). Multidimensional NMR spectroscopy was used to produce a solution structure of the class I hydrophobin EAS (69). EAS assumes a β-barrel conformation in solution, and it was proposed that the β-barrels stack to form the filamentous rodlet structures.
Non-amyloid Protein Polymers
As new examples of amyloid (both pathological and functional) continue to accumulate, it may become increasingly necessary to draw distinction with what is not amyloid. Many non-amyloid filamentous assemblies of proteins are well characterized and typically involve the linear assembly of folded globular proteins. Cytoskeletal proteins, such as actin and tubulin, assemble into filamentous structures, as do structural proteins, such as keratin. Also, there are many bacterial adhesin proteins that are assembled into extracellular filamentous structures (fimbriae or pili) that are structurally different from amyloid-based adhesins like curli (discussed above). In many cases, these fimbrial structures are composed of proteins with immunoglobulin-like structures that polymerize through donor strand exchange (70), i.e. each subunit donates a β-strand to complete a sheet in the next subunit (Fig. 1D). Such fimbrial assemblies are filamentous and could even have cross-β-structure if the β-strands consistently tended to align perpendicular to the fiber axis, but fimbriae are not described as being amyloid.
Amyloid Intermediate and Oligomeric Structures
The accumulation of protein into an aggregate may produce either a loss or gain of function (if the amyloid has some functional or toxic activity) or even some combination of both. In the case of pathological amyloids, the nature of the actual toxic species is not well understood. There is evidence that distinct toxic oligomeric and annular intermediates may exist on the pathway to amyloid formation (71, 72). It has been observed that soluble oligomers of Aβ correlate better with disease causation than the insoluble fibrillar deposits that are present in amyloid plaques, suggesting that oligomeric forms are the toxic species (73). In such cases, the further conversion to amyloid from toxic oligomer could be considered protective. Because little structural information is available, classifying these oligomeric species has been challenging. However, even off-pathway oligomers could masquerade as intermediates if their formation is reversible, whereas that of the amyloid is more stable. Any pathology or biochemistry that is due to the ends of filaments would be preferentially produced by shorter filaments (even if they have the same basic structure as longer ones). Successive halving of filaments would result in an exponential increase in filament ends, which could explain the much greater toxicity observed for oligomeric species if they are simply the smallest filament units. If toxic oligomeric structures are a common feature of amyloids, functional amyloids would likely require mechanisms to protect cells from their toxicity. Also, because different amyloids are based on very different structures, it is unlikely that each would transition through a similar toxic intermediate.
Functional Versus Pathological Amyloid Structure
The amyloids that are the consequence of misfolding generally appear to be in a low-energy conformation accessible to many polypeptides of very different amino acid sequence and composition. Parallel in-register β-sheet amyloids may represent a common low-energy conformation. Conceivably, many proteins could achieve such a conformation, which may ultimately be more common among pathological amyloids. However, the RPT domain of the functional amyloid Pmel17 adopts this conformation, so there may be cases in which this structure is employed to serve a function. Filamentous structures that result from the linearly assembled folded proteins might be different in this regard because, although they may have a cross-β-configuration as determined by diffraction, they do not necessarily represent a common low-energy structure achievable by divergent polypeptides. The amyloid form of HET-s has an ascribed function, and according to Greenwald and Riek (74), “HET-s has evolved to fold into a cross-β-motif and therefore may be more complex than the disease-related amyloids for which the cross-β-structure is an unfortunate energy minimum on the folding landscape.” The same could be said of CsgA monomers within the curli amyloid, which also has a defined function and is likely based on a β-helical structure. Perhaps the β-helix will prove to be common among functional amyloids; because each subunit is folded, it may avoid issues of “variants” and “strains” that are observed for other amyloids and prions. β-Helices or immunoglobulin folds are most certainly not low-energy conformations common to all members of the diverse family of amyloid proteins. Thus, amyloid is rather loosely defined, including proteins with very different structural folds composing the body of a fibril.
These structures may inform us about their biology. Fibrils composed of parallel in-register β-sheets provide a ready template for polypeptides of the same sequence. This suggests a mechanism of inheritance, which can explain the infectious behavior of yeast prions and their tendency to form multiple variants. This also provides a mechanism for pathological amyloids to propagate in tissue and suggests that non-prion amyloid diseases may also have variants, which was recently demonstrated with extracts from Alzheimer patients seeding distinct Aβ variants in vitro (75).
What Is Amyloid?
There has been a surge in newly proposed functional amyloids, although the list of pathological amyloids also continues to grow. Recently, amyloid adhesins have been proposed to be abundant in natural biofilms produced by a variety of organisms (76, 77). Moreover, bacterial inclusion bodies have been proposed to be composed of amyloid (78), and also there have been amyloid-like inclusions observed in plant chloroplasts (79). As more amyloids and their structures are characterized, it is likely that the concept of amyloid will continue to evolve.
Acknowledgment
We thank Rob Tycko for providing coordinates for a ssNMR Aβ1–40 structure.
This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012.
- TEM
- transmission electron microscopy
- Aβ
- amyloid-β peptide
- ssNMR
- solid-state NMR.
REFERENCES
- 1. Sipe J. D., Cohen A. S. (2000) J. Struct. Biol. 130, 88–98 [DOI] [PubMed] [Google Scholar]
- 2. Cohen A. S., Calkins E. (1959) Nature 183, 1202–1203 [DOI] [PubMed] [Google Scholar]
- 3. Eanes E. D., Glenner G. G. (1968) J. Histochem. Cytochem. 16, 673–677 [DOI] [PubMed] [Google Scholar]
- 4. Sipe J. D., Benson M. D., Buxbaum J. N., Ikeda S., Merlini G., Saraiva M. J., Westermark P. (2010) Amyloid 17, 101–104 [DOI] [PubMed] [Google Scholar]
- 5. Dobson C. M. (1999) Trends Biochem. Sci. 24, 329–332 [DOI] [PubMed] [Google Scholar]
- 6. Groenning M., Olsen L., van de Weert M., Flink J. M., Frokjaer S., Jørgensen F. S. (2007) J. Struct. Biol. 158, 358–369 [DOI] [PubMed] [Google Scholar]
- 7. Toombs J. A., McCarty B. R., Ross E. D. (2010) Mol. Cell. Biol. 30, 319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Petkova A. T., Leapman R. D., Guo Z., Yau W. M., Mattson M. P., Tycko R. (2005) Science 307, 262–265 [DOI] [PubMed] [Google Scholar]
- 9. Goldsbury C., Baxa U., Simon M. N., Steven A. C., Engel A., Wall J. S., Aebi U., Muller S. A. (2011) J. Struct. Biol. 173, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen B., Thurber K. R., Shewmaker F., Wickner R. B., Tycko R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Apostol M. I., Sawaya M. R., Cascio D., Eisenberg D. (2010) J. Biol. Chem. 285, 29671–29675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nelson R., Sawaya M. R., Balbirnie M., Madsen A. Ø., Riekel C., Grothe R., Eisenberg D. (2005) Nature 435, 773–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sawaya M. R., Sambashivan S., Nelson R., Ivanova M. I., Sievers S. A., Apostol M. I., Thompson M. J., Balbirnie M., Wiltzius J. J., McFarlane H. T., Madsen A. Ø., Riekel C., Eisenberg D. (2007) Nature 447, 453–457 [DOI] [PubMed] [Google Scholar]
- 14. Wasmer C., Lange A., Van Melckebeke H., Siemer A. B., Riek R., Meier B. H. (2008) Science 319, 1523–1526 [DOI] [PubMed] [Google Scholar]
- 15. Wasmer C., Schütz A., Loquet A., Buhtz C., Greenwald J., Riek R., Böckmann A., Meier B. H. (2009) J. Mol. Biol. 394, 119–127 [DOI] [PubMed] [Google Scholar]
- 16. Petkova A. T., Ishii Y., Balbach J. J., Antzutkin O. N., Leapman R. D., Delaglio F., Tycko R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16742–16747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Margittai M., Langen R. (2008) Q. Rev. Biophys. 41, 265–297 [DOI] [PubMed] [Google Scholar]
- 18. Margittai M., Langen R. (2006) Methods Enzymol. 413, 122–139 [DOI] [PubMed] [Google Scholar]
- 19. Chen M., Margittai M., Chen J., Langen R. (2007) J. Biol. Chem. 282, 24970–24979 [DOI] [PubMed] [Google Scholar]
- 20. Ladner C. L., Chen M., Smith D. P., Platt G. W., Radford S. E., Langen R. (2010) J. Biol. Chem. 285, 17137–17147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iwata K., Fujiwara T., Matsuki Y., Akutsu H., Takahashi S., Naiki H., Goto Y. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18119–18124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Neet K. E., Thinakaran G. (2008) J. Biol. Chem. 283, 29613–29614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Benzinger T. L., Gregory D. M., Burkoth T. S., Miller-Auer H., Lynn D. G., Botto R. E., Meredith S. C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13407–13412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paravastu A. K., Leapman R. D., Yau W. M., Tycko R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18349–18354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luca S., Yau W. M., Leapman R., Tycko R. (2007) Biochemistry 46, 13505–13522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tycko R., Savtchenko R., Ostapchenko V. G., Makarava N., Baskakov I. V. (2010) Biochemistry 49, 9488–9497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roberts B. T., Wickner R. B. (2003) Genes Dev. 17, 2083–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brachmann A., Baxa U., Wickner R. B. (2005) EMBO J. 24, 3082–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patel B. K., Liebman S. W. (2007) J. Mol. Biol. 365, 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Du Z., Crow E. T., Kang H. S., Li L. (2010) Mol. Cell. Biol. 30, 4644–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tanaka M., Chien P., Naber N., Cooke R., Weissman J. S. (2004) Nature 428, 323–328 [DOI] [PubMed] [Google Scholar]
- 32. Alberti S., Halfmann R., King O., Kapila A., Lindquist S. (2009) Cell 137, 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baxa U., Taylor K. L., Wall J. S., Simon M. N., Cheng N., Wickner R. B., Steven A. C. (2003) J. Biol. Chem. 278, 43717–43727 [DOI] [PubMed] [Google Scholar]
- 34. Shewmaker F., Kryndushkin D., Chen B., Tycko R., Wickner R. B. (2009) Biochemistry 48, 5074–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shewmaker F., Wickner R. B., Tycko R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19754–19759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wickner R. B., Dyda F., Tycko R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2403–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tycko R. (2007) J. Chem. Phys. 126, 064506 [Google Scholar]
- 38. Diaz-Avalos R., King C. Y., Wall J., Simon M., Caspar D. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10165–10170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oh J., Kim J. G., Jeon E., Yoo C. H., Moon J. S., Rhee S., Hwang I. (2007) J. Biol. Chem. 282, 13601–13609 [DOI] [PubMed] [Google Scholar]
- 40. Fowler D. M., Koulov A. V., Balch W. E., Kelly J. W. (2007) Trends Biochem. Sci. 32, 217–224 [DOI] [PubMed] [Google Scholar]
- 41. Hoashi T., Muller J., Vieira W. D., Rouzaud F., Kikuchi K., Tamaki K., Hearing V. J. (2006) J. Biol. Chem. 281, 21198–21208 [DOI] [PubMed] [Google Scholar]
- 42. Theos A. C., Truschel S. T., Tenza D., Hurbain I., Harper D. C., Berson J. F., Thomas P. C., Raposo G., Marks M. S. (2006) Dev. Cell 10, 343–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McGlinchey R. P., Shewmaker F., McPhie P., Monterroso B., Thurber K., Wickner R. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13731–13736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McGlinchey R. P., Shewmaker F., Hu K. N., McPhie P., Tycko R., Wickner R. B. (2011) J. Biol. Chem. 286, 8385–8393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Balbach J. J., Ishii Y., Antzutkin O. N., Leapman R. D., Rizzo N. W., Dyda F., Reed J., Tycko R. (2000) Biochemistry 39, 13748–13759 [DOI] [PubMed] [Google Scholar]
- 46. Petkova A. T., Buntkowsky G., Dyda F., Leapman R. D., Yau W. M., Tycko R. (2004) J. Mol. Biol. 335, 247–260 [DOI] [PubMed] [Google Scholar]
- 47. Lansbury P. T., Jr., Costa P. R., Griffiths J. M., Simon E. J., Auger M., Halverson K. J., Kocisko D. A., Hendsch Z. S., Ashburn T. T., Spencer R. G., Tidor B., Griffin R. G. (1995) Nat. Struct. Biol. 2, 990–998 [DOI] [PubMed] [Google Scholar]
- 48. Nielsen J. T., Bjerring M., Jeppesen M. D., Pedersen R. O., Pedersen J. M., Hein K. L., Vosegaard T., Skrydstrup T., Otzen D. E., Nielsen N. C. (2009) Angew. Chem. Int. Ed. Engl. 48, 2118–2121 [DOI] [PubMed] [Google Scholar]
- 49. Sikorski P., Atkins E. (2005) Biomacromolecules 6, 425–432 [DOI] [PubMed] [Google Scholar]
- 50. Ross C. A., Poirier M. A., Wanker E. E., Amzel M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tycko R., Sciarretta K. L., Orgel J. P., Meredith S. C. (2009) Biochemistry 48, 6072–6084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Siemer A. B., Ritter C., Steinmetz M. O., Ernst M., Riek R., Meier B. H. (2006) J. Biomol. NMR 34, 75–87 [DOI] [PubMed] [Google Scholar]
- 53. Wasmer C., Zimmer A., Sabate R., Soragni A., Saupe S. J., Ritter C., Meier B. H. (2010) J. Mol. Biol. 402, 311–325 [DOI] [PubMed] [Google Scholar]
- 54. Barnhart M. M., Chapman M. R. (2006) Annu. Rev. Microbiol. 60, 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shewmaker F., McGlinchey R. P., Thurber K. R., McPhie P., Dyda F., Tycko R., Wickner R. B. (2009) J. Biol. Chem. 284, 25065–25076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu Y., Gotte G., Libonati M., Eisenberg D. (2001) Nat. Struct. Biol. 8, 211–214 [DOI] [PubMed] [Google Scholar]
- 57. Jaskólski M. (2001) Acta Biochim. Pol. 48, 807–827 [PubMed] [Google Scholar]
- 58. Lomas D. A., Carrell R. W. (2002) Nat. Rev. 3, 759–768 [DOI] [PubMed] [Google Scholar]
- 59. Yamasaki M., Li W., Johnson D. J., Huntington J. A. (2008) Nature 455, 1255–1258 [DOI] [PubMed] [Google Scholar]
- 60. Janowski R., Kozak M., Jankowska E., Grzonka Z., Grubb A., Abrahamson M., Jaskolski M. (2001) Nat. Struct. Biol. 8, 316–320 [DOI] [PubMed] [Google Scholar]
- 61. Staniforth R. A., Giannini S., Higgins L. D., Conroy M. J., Hounslow A. M., Jerala R., Craven C. J., Waltho J. P. (2001) EMBO J. 20, 4774–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Louis J. M., Byeon I. J., Baxa U., Gronenborn A. M. (2005) J. Mol. Biol. 348, 687–698 [DOI] [PubMed] [Google Scholar]
- 63. Serag A. A., Altenbach C., Gingery M., Hubbell W. L., Yeates T. O. (2002) Nat. Struct. Biol. 9, 734–739 [DOI] [PubMed] [Google Scholar]
- 64. Olofsson A., Ippel J. H., Wijmenga S. S., Lundgren E., Ohman A. (2004) J. Biol. Chem. 279, 5699–5707 [DOI] [PubMed] [Google Scholar]
- 65. Elam J. S., Taylor A. B., Strange R., Antonyuk S., Doucette P. A., Rodriguez J. A., Hasnain S. S., Hayward L. J., Valentine J. S., Yeates T. O., Hart P. J. (2003) Nat. Struct. Biol. 10, 461–467 [DOI] [PubMed] [Google Scholar]
- 66. Claessen D., Rink R., de Jong W., Siebring J., de Vreugd P., Boersma F. G., Dijkhuizen L., Wosten H. A. (2003) Genes Dev. 17, 1714–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mackay J. P., Matthews J. M., Winefield R. D., Mackay L. G., Haverkamp R. G., Templeton M. D. (2001) Structure 9, 83–91 [DOI] [PubMed] [Google Scholar]
- 68. Sunde M., Kwan A. H., Templeton M. D., Beever R. E., Mackay J. P. (2008) Micron 39, 773–784 [DOI] [PubMed] [Google Scholar]
- 69. Kwan A. H., Winefield R. D., Sunde M., Matthews J. M., Haverkamp R. G., Templeton M. D., Mackay J. P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3621–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sauer F. G., Fütterer K., Pinkner J. S., Dodson K. W., Hultgren S. J., Waksman G. (1999) Science 285, 1058–1061 [DOI] [PubMed] [Google Scholar]
- 71. Glabe C. G. (2008) J. Biol. Chem. 283, 29639–29643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lashuel H. A., Lansbury P. T., Jr. (2006) Q. Rev. Biophys. 39, 167–201 [DOI] [PubMed] [Google Scholar]
- 73. Lesné S., Koh M. T., Kotilinek L., Kayed R., Glabe C. G., Yang A., Gallagher M., Ashe K. H. (2006) Nature 440, 352–357 [DOI] [PubMed] [Google Scholar]
- 74. Greenwald J., Riek R. (2010) Structure 18, 1244–1260 [DOI] [PubMed] [Google Scholar]
- 75. Paravastu A. K., Qahwash I., Leapman R. D., Meredith S. C., Tycko R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7443–7448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jordal P. B., Dueholm M. S., Larsen P., Petersen S. V., Enghild J. J., Christiansen G., Højrup P., Nielsen P. H., Otzen D. E. (2009) Appl. Environ. Microbiol. 75, 4101–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Larsen P., Nielsen J. L., Dueholm M. S., Wetzel R., Otzen D., Nielsen P. H. (2007) Environ. Microbiol. 9, 3077–3090 [DOI] [PubMed] [Google Scholar]
- 78. Wang L., Maji S. K., Sawaya M. R., Eisenberg D., Riek R. (2008) PLoS Biol. 6, e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Villar-Piqué A., Sabaté R., Lopera O., Gibert J., Torne J. M., Santos M., Ventura S. (2010) PLoS ONE 5, e13625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Iconomidou V. A., Vriend G., Hamodrakas S. J. (2000) FEBS Lett. 479, 141–145 [DOI] [PubMed] [Google Scholar]
- 81. Dueholm M. S., Petersen S. V., Sonderkaer M., Larsen P., Christiansen G., Hein K. L., Enghild J. J., Nielsen J. L., Nielsen K. L., Nielsen P. H., Otzen D. E. (2010) Mol. Microbiol. 77, 1009–1020 [DOI] [PubMed] [Google Scholar]
- 82. Maddelein M. L., Dos Reis S., Duvezin-Caubet S., Coulary-Salin B., Saupe S. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7402–7407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ramsook C. B., Tan C., Garcia M. C., Fung R., Soybelman G., Henry R., Litewka A., O'Meally S., Otoo H. N., Khalaf R. A., Dranginis A. M., Gaur N. K., Klotz S. A., Rauceo J. M., Jue C. K., Lipke P. N. (2010) Eukaryot. Cell 9, 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Otoo H. N., Lee K. G., Qiu W., Lipke P. N. (2008) Eukaryot. Cell 7, 776–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bieler S., Estrada L., Lagos R., Baeza M., Castilla J., Soto C. (2005) J. Biol. Chem. 280, 26880–26885 [DOI] [PubMed] [Google Scholar]
- 86. Adda C. G., Murphy V. J., Sunde M., Waddington L. J., Schloegel J., Talbo G. H., Vingas K., Kienzle V., Masciantonio R., Howlett G. J., Hodder A. N., Foley M., Anders R. F. (2009) Mol. Biochem. Parasitol. 166, 159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ader C., Frey S., Maas W., Schmidt H. B., Görlich D., Baldus M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 6281–6285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Maji S. K., Perrin M. H., Sawaya M. R., Jessberger S., Vadodaria K., Rissman R. A., Singru P. S., Nilsson K. P., Simon R., Schubert D., Eisenberg D., Rivier J., Sawchenko P., Vale W., Riek R. (2009) Science 325, 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Berson J. F., Theos A. C., Harper D. C., Tenza D., Raposo G., Marks M. S. (2003) J. Cell Biol. 161, 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Romero D., Aguilar C., Losick R., Kolter R. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 2230–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Graether S. P., Slupsky C. M., Sykes B. D. (2003) Biophys. J. 84, 552–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Vilar M., Chou H. T., Lührs T., Maji S. K., Riek-Loher D., Verel R., Manning G., Stahlberg H., Riek R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8637–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Balbach J. J., Petkova A. T., Oyler N. A., Antzutkin O. N., Gordon D. J., Meredith S. C., Tycko R. (2002) Biophys. J. 83, 1205–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lührs T., Ritter C., Adrian M., Riek-Loher D., Bohrmann B., Döbeli H., Schubert D., Riek R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17342–17347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Debelouchina G. T., Platt G. W., Bayro M. J., Radford S. E., Griffin R. G. (2010) J. Am. Chem. Soc. 132, 10414–10423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Margittai M., Langen R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10278–10283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Margittai M., Langen R. (2006) J. Biol. Chem. 281, 37820–37827 [DOI] [PubMed] [Google Scholar]
- 98. Baxa U., Wickner R. B., Steven A. C., Anderson D. E., Marekov L. N., Yau W. M., Tycko R. (2007) Biochemistry 46, 13149–13162 [DOI] [PubMed] [Google Scholar]
- 99. Petkova A. T., Yau W. M., Tycko R. (2006) Biochemistry 45, 498–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Van Melckebeke H., Wasmer C., Lange A., Ab E., Loquet A., Böckmann A., Meier B. H. (2010) J. Am. Chem. Soc. 132, 13765–13775 [DOI] [PubMed] [Google Scholar]