Abstract
A recent report (Zhong, D., Xiong, L., Liu, T., Liu, X., Liu, X., Chen, J., Sun, S. Y., Khuri, F. R., Zong, Y., Zhou, Q., and Zhou, W. (2009) J. Biol. Chem. 284, 23225–23233) details that 2-deoxy-d-glucose (2-DG), a well known inhibitor of glycolysis and a candidate antineoplastic agent, also induces insulin-like growth factor 1 receptor (IGF-1R) signaling through the inhibition of insulin-like growth factor 1-insulin-like growth factor-binding protein 3 (IGF-1-IGFBP-3) complex formation. Zhong et al. hypothesized that disrupted IGF-1/IGFBP-3 binding by 2-DG led to increased free IGF-1 concentrations and, consequently, activation of IGF-1R downstream pathways. Because their report suggests unprecedented off-target effects of 2-DG, this has profound implications for the fields of metabolism and oncology. Using ELISA, surface plasmon resonance, and novel “intensity-fading” mass spectrometry, we now provide a detailed characterization of complex formation between IGF-1 and IGFBP-3. All three of these independent methods demonstrated that there was no effect of glucose or 2-DG on the interaction between IGF-1 and IGFBP-3. Furthermore, we show examples of 2-DG exposure associated with reduced rather than increased IGF-1R and AKT activation, providing further evidence against a 2-DG increase in IGF-1R activation by IGF-1-IGFBP-3 complex disruption.
Keywords: Glucose, Insulin-like Growth Factor (IGF), Mass Spectrometry (MS), Protein-Protein Interactions, Surface Plasmon Resonance (SPR), 2-DG, IGFBP-3
Introduction
Insulin-like growth factors 1 and 2 (IGF-1 and IGF-2) are peptide hormones similar in molecular structure to insulin, and they regulate a variety of cellular activities, including metabolism, proliferation, and growth. Both IGFs2 bind to the IGF-1 receptor (IGF-1R) at the cell membrane and initiate a signaling cascade that results in the activation of the phosphatidylinositol 3-kinase/AKT/mammalian target of rapamycin pathway (1–5). The IGF-2 receptor, which binds to IGF-2, lacks an intracellular signaling domain and is therefore considered to only act as a sink for excess IGF-2 (6).
IGF actions are tightly modulated by a family of proteins called insulin-like growth factor-binding proteins (IGFBPs), of which IGFBP-1–6 have been characterized. Most IGFBPs in the blood originate from the liver, but they are also expressed in many other tissues. IGFBP-3 and -5 are the most abundant IGFBPs in the circulation. They form a ternary complex with IGFs and a third protein termed acid-labile subunit (7–9). Insulin-like growth factor-binding proteins are known to modulate actions of IGFs both in vitro and in vivo (7). Because only free IGFs are considered ligands for the IGF-1R, significant research efforts have focused on small molecules capable of interfering with IGF-IGFBP binding.
A recent report showed that 2-deoxy-d-glucose (2-DG), a close derivative of glucose, promoted dissociation of IGF-1 from IGFBP-3 and consequently contributed to elevated phosphoserine 473 AKT levels in a variety of cancer cell lines (10). 2-DG has been proposed as a potential therapeutic agent in the treatment of cancer because it interferes with glycolysis (11). Entering the cell through glucose transporters, 2-DG inhibits enzymes of the glycolytic pathway both competitively (phosphoglucose isomerase) (12) and noncompetitively (hexokinase) (13). Because tumor cells depend more heavily on glycolysis compared with normal cells, 2-DG is under investigation for cancer treatment (14).
The recent study by Zhong et al. (10) showed that 2-DG, apart from its classic activities as an inhibitor of glycolysis, can also increase IGF-1R signaling by disrupting IGF-1/IGFBP-3 binding. As increased IGF-1R signaling is associated with greater proliferation of tumor cells, this report has shed doubt on the efficacy of 2-DG as a cancer therapeutic. To examine this issue further, this study characterizes the effect of glucose and 2-DG on binding between IGF-1 and IGFBP-3. Using ELISA, surface plasmon resonance (SPR), and novel “intensity-fading” mass spectrometry (MS), we report that glucose and 2-DG have no effect on IGF-1-IGFBP-3 complex formation.
EXPERIMENTAL PROCEDURES
Materials
Human recombinant IGF-1 was purchased from Feldan Biosciences (Montreal, Canada), and human recombinant IGFBP-3 was obtained from Insmed Inc. (Glen Allen, VA). Glucose, 2-DG, and fatty acid-free bovine serum albumin (BSA) were purchased from Sigma. MCF-7, T47D, and HeLa cell lines were obtained from American Type Culture Collection (ATCC) (Manassas, VA) and cultured in standard DMEM m supplemented with 10% fetal bovine serum (FBS) and 20 μg/ml gentamycin. Phosphate buffered saline (PBS; 10 mm Sodium Phosphate, 2.7 mm KCl, 137 mm NaCl) and Hepes buffered saline (HBS-EP; 10 mm Hepes pH 7.4, 150 mm NaCl, 3 mm EDTA, 0.005% (v/v) Tween-20) were prepared using analytical grade chemicals.
ELISA
Microtiter plates (Costar, Lowell, MA) were coated with either human recombinant IGF-1 (5 μg/ml) or human recombinant IGFBP-3 (2 μg/ml) in PBS overnight at room temperature. Plates were washed three times with sample buffer (PBS or HBS-EP; 100 μl/well) and then blocked for 3 h at room temperature using 5% (w/v) casein (in sample buffer; 300 μl/well). Plates were washed again in a similar manner before 30-min incubations with IGFBP-3 or 2.5-h incubations with IGF-1 at room temperature. After washing, primary antibodies (anti-IGFBP-3 or anti IGF-1; Santa Cruz Biotechnology, Santa Cruz, CA) were added to each well (100 μl of 1:50 dilution in sample buffer containing 5% (w/v) casein) for 1 h at room temperature. After washing, secondary antibody (Santa Cruz Biotechnology) was added to each well in the similar manner. After washing, the color developing solution (R&D Systems, Minneapolis, MN) was added to the wells, and absorbance readings were monitored at 450 nm.
SPR
Label-free, real time binding between IGF-1 (7.6 kDa) and IGFBP-3 (29 kDa) was examined using a Biacore 3000 system (GE Healthcare) at 25 °C with filtered (0.2 μm) and degassed HBS-EP running buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, 3 mm EDTA, 0.005% (v/v) Tween 20). IGFBP-3 (9 μg/ml in 10 mm sodium acetate, pH 5.5) was immobilized to CM4 sensor chips using the Biacore amine coupling kit (∼1000 RU final); corresponding reference surfaces were prepared in the absence of IGFBP-3. IGF-1 (0–25 nm) (or BSA as negative control) was injected over reference and IGFBP-3-immobilized surfaces, in the absence or presence of competitor (25–100 mm glucose or 2-DG), using variable flow rates (10–75 μl/min) and contact times (3–5 min association, 5–10 min dissociation). Between sample injections, sensor chip surfaces were regenerated using Pierce gentle elution buffer (Thermo Scientific) containing 0.1% (v/v) Empigen (Affymetrix-Anatrace).
All SPR data were double-referenced (15) and are representative of duplicate injections acquired from two independent trials. For each titration series, a buffer blank (± glucose or 2-DG) was injected first, the highest IGF-1 concentration second, and serial dilutions then followed (from the lowest to the highest concentration repeated). Comparing binding responses between the highest IGF-1 injections verified consistent IGFBP-3-immobilized surface activity throughout each assay. Apparent equilibrium dissociation constants (KD), as well as individual association (ka) and dissociation (kd) rate constants, were determined by global fitting of the data to a “1:1 kinetic” model with or without mass transport (BIAevaluation version 4.1 software). The kinetic estimates represent fits to the experimental data where χ2 values were <1.5.
MS
Eppendorf tubes containing IGF-1 (13 μm) or IGF-1 plus IGFBP-3 (13 or 26 μm) samples were incubated for 3 h on a rocking platform at room temperature. A normal phase (NP-20) protein chip (Bio-Rad) was washed three times with 5 μl of HPLC-grade water before the addition of 5 μl of sample per spot and air drying. Subsequently, each spot was washed three times with 5 μl of a low stringency buffer (Bio-Rad) and then left to air dry. 5 mg of α-cyano-4-hydroxycinnamic acid (Bio-Rad) was dissolved in 200 μl of solution A (50% (v/v) HPLC-grade water, 49.5% (v/v) acetonitrile, and 0.5% (v/v) trifluoroacetic acid); the mixture was vortexed for 5 min and then centrifuged for 10 min before the supernatant was collected and further diluted with 200 μl of solution A. The diluted supernatant was then added to each spot (1 μl per spot) and left to air dry; this was repeated twice to create two layers. The chip was then analyzed using a Ciphergen Protein Chip Series 4000 instrument (Ciphergen Biosystems, Fremont, CA).
Western Blots
Total cell lysates were obtained using RIPA buffer as described earlier (16). 50 μg of total protein were loaded per lane, and membranes were immunoblotted with the following antibodies: phosphoserine 473 AKT, AKT, phosphotyrosine 1135/1136 IGF-1R β chain, IGF-1R β chain, and β-actin (Cell Signaling, Danvers, MA).
RESULTS
Qualitative Binding of IGF-1 to IGFBP-3
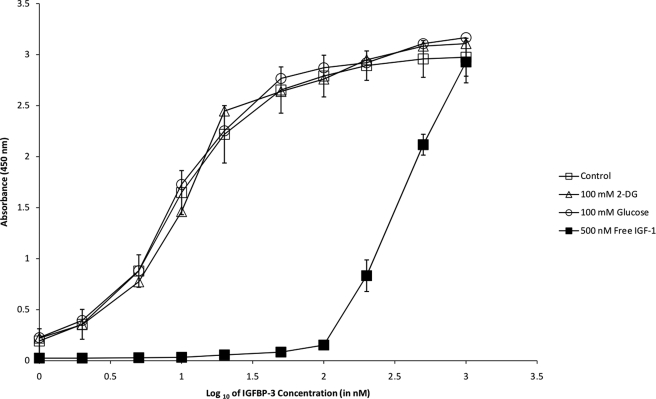
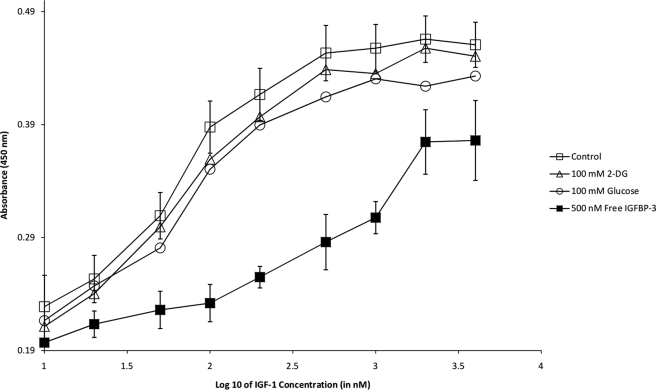
Initially, ELISAs were performed in which IGF-1 was immobilized in the microtiter wells (Fig. 1). In an HBS-EP buffer system, saturable, dose-dependent binding of IGFBP-3 was detected and yielded sigmoidal curves (log scale) as expected for typical protein interactions. Binding specificity was evidenced by decreased colorimetric responses in the presence of free IGF-1 (Fig. 1), as well as high sodium chloride and high Tween 20 detergent concentrations (data not shown). In the presence of excess glucose or 2-DG (25–100 mm), however, the binding of IGFBP-3 to immobilized IGF-1 was unaltered in all cases. As negative controls, microtiter wells lacking immobilized protein or primary/secondary antibodies failed to generate any colorimetric response (data not shown). In the reversed orientation, ELISAs were also performed in which IGFBP-3 was immobilized (Fig. 2). Saturable, dose-dependent binding of IGF-1 was detected, and similar to above, the interaction could be competed with free IGFBP-3 but was unaltered by glucose or 2-DG. ELISAs were performed in other buffer settings (PBS, Krebs-Ringer) with identical conclusions (data not shown).
FIGURE 1.
ELISA to monitor binding of IGFBP3 (0–1000 nm in HBS-EP) to immobilized IGF-1 in the absence (open squares) or presence of 100 mm 2-DG (open triangles), 100 mm glucose (open circles), and 500 nm IGF-1 (closed squares). Error bars (standard deviation of triplicate measurements) are only depicted for the “control” and “500 nm IGF-1” series for simplicity.
FIGURE 2.
Reverse ELISA to monitor binding of IGF-1 (0–5000 nm in HBS-EP) to immobilized IGFBP-3 in the absence (open squares) or presence of 100 mm 2-DG (open triangles), 100 mm glucose (open circles), and 500 nm IGFBP-3 (closed squares). Error bars (standard deviation of triplicate measurements) are only depicted for the control and 500 nm IGFBP-3 series for simplicity.
Quantitative Binding of IGF-1 to IGFBP-3
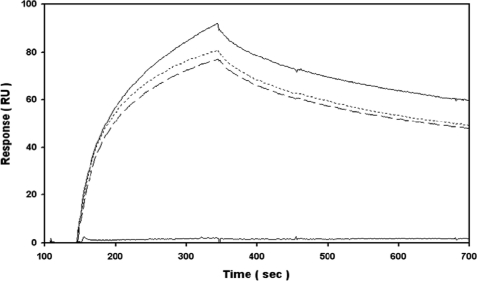
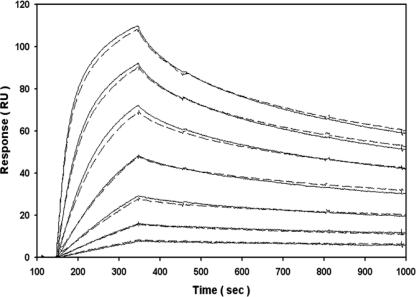
To validate the ELISA data, binding between IGF-1 and IGFBP-3 was then examined using label-free, real time SPR and the identical HBS-EP buffer system. A single low nanomolar concentration of IGF-1 specifically bound to immobilized IGFBP-3 under high flow rate conditions (Fig. 3), whereas an equimolar concentration of BSA failed to interact (i.e. negative binding control). The overall amounts of IGF-1 bound, and the individual association and dissociation kinetics were not significantly altered in the presence of excess glucose or 2-DG (25–100 mm). Expanding upon the fixed concentration specificity tests, IGF-1 was then titrated in the absence and presence of excess 2-DG (Fig. 4). Visually, the overall dose dependence and individual kinetics of IGF-1 binding to immobilized IGFBP-3 were not significantly altered in the presence of excess glucose or 2-DG. Likewise, global fitting of each titration series to a 1:1 kinetic model indicated that there was no significant difference in the subnanomolar affinity for this interaction over replicate trials (Table 1). In the absence or presence of an additional “kt” parameter, we also confirmed that the curve fitting was not significantly altered by mass transport limitations (i.e. fitted kt coefficient was ∼108 RU m−1 s−1 as expected).
FIGURE 3.
SPR analysis to monitor specificity of IGF-1 (25 nm in HBS-EP) binding to IGFBP-3 (1000 RU amine-coupled) in the absence (upper solid line) or presence of 100 mm 2-DG (dashed line) and 100 mm glucose (dotted line) at 75 μl/min. As a negative binding control, no significant response was observed with 25 nm BSA (bottom solid line). Double-referenced data are representative of duplicate injections acquired from two independent trials.
FIGURE 4.
SPR analysis to monitor real time kinetics of IGF-1 (0–25 nm in HBS-EP; 2-fold dilution series) binding to IGFBP-3 (1000 RU amine-coupled) in the absence (solid lines) or presence of 100 mm 2-DG (dashed lines) at 75 μl/min (3. 5 min association + 10 min dissociation). Double-referenced data are representative of duplicate injections acquired from two independent trials.
TABLE 1.
Apparent kinetics of IGF-1 binding to immobilized IGFBP-3 in the absence and presence of 2-DG, as assessed by SPR
Estimates (means ± S.E.; n = 4) represent global analysis of titration series to a 1:1 kinetic model where goodness-of-fit (χ2) values were <1.5; association rate (ka), dissociation rate (kd), and equilibrium dissociation (KD = kd/ka) constants were not significantly altered in the presence of an additional mass transport (kt ∼108 RU m −1 s−1) fitting parameter.
| 100 mm 2-DG | ka × 106 | kd × 10−4 | KD |
|---|---|---|---|
| m−1s−1 | s−1 | pm | |
| − | 1.40 ± 0.01 | 8.40 ± 0.04 | 604 ± 5 |
| + | 1.33 ± 0.01 | 7.93 ± 0.05 | 599 ± 5 |
Complex Formation between IGF-1 and IGFBP3
To validate the ELISA and SPR data, we also used MS to demonstrate that 2-DG and glucose have no effect on IGF-1/IGFBP-3 binding. In recent years, surface-enhanced laser desorption/ionization and matrix-enhanced laser desorption/ionization (MALDI)-time of flight (TOF) MS have become powerful tools for the investigation of noncovalent protein complexes (17). Intensity fading experiments are relatively novel techniques used to directly visualize protein-protein binding without the use of cross-linkers (18). In these experiments, a fixed concentration of protein A (ideally between 5 and 16 kDa) is incubated with different concentrations of its binding partner B; as the concentration of protein B is increased, the intensity of the peak corresponding to protein A slowly diminishes or “fades” (19).
In our MS experiments, we incubated IGF-1 at 13 μm alone, with an equimolar concentration of IGFBP-3 (13 μm), or with excess IGFBP-3 (26 μm) in PBS. Fig. 5A shows that the intensity of IGF-1 (7600 Da) fades with increasing concentrations of IGFBP-3 (29,000 Da), decreasing from 1000 intensity units to under 50 at the highest IGFBP-3 concentration. Epidermal growth factor (EGF) (9 μm) was included as an internal nonbinding control, and its intensity (6400 Da) remained unaltered across all IGFBP-3 concentrations. As commonly encountered in intensity fading experiments (18, 19), detection of the resultant IGF-1-IGFBP-3 complex (36,000 Da) was not proportional to theoretical expectations. The same experiments were performed in PBS containing 100 mm 2-DG (Fig. 5B) or 100 mm glucose (data not shown), and similar spectral outcomes were observed. Because neither glucose nor 2-DG was able to reverse the intensity fading of the IGF-1 peak, this assay indicates that they do not influence IGF-1/IGFBP-3 binding.
FIGURE 5.
MS analysis to monitor binding between IGF-1 (∼7600 Da) and IGFBP-3 (∼29,000 Da) in the absence or presence of 100 mm 2-DG (and internal EGF nonbinding control, ∼6400 Da). A, upper boxes represent 13 nm IGF-1 alone; middle boxes represent 13 nm IGF-1 with 13 nm IGFBP-3, and lower boxes represent 13 nm IGF-1 with 26 nm IGFBP-3. B, identical intensity fading experiments performed in the presence of 100 mm 2-DG. The data presented are representative of three independent experiments. y axes are representative of signal response (microampere (uA)), and x axes are representative of molecular mass (Da).
Effects of 2-DG on AKT and IGF-1R
Furthermore, we investigated the effect of 2-DG on IGF-1R and AKT activation in two breast cancer cell lines (MCF-7 and T47D) and one cervical cancer cell line (HeLa). We observed that addition of 2-DG at 25 mm, a concentration previously reported to enhance AKT phosphorylation in these cell lines (20), resulted in a consistent decrease in IGF-1R activation. As shown in Fig. 6, exposure to 2-DG actually reduced IGF-1R β chain tyrosine 1135/1136 phosphorylation in both basal and IGF-1 (130 nm)-stimulated contexts and across all three cell lines. We also observed that AKT activation is not universally induced by 2-DG. Fig. 6 shows that phosphoserine 473 AKT is increased in T47D cells and decreased in MCF-7 cells in basal 10% FBS conditions. In IGF-1 (130 nm)-stimulated conditions, 2-DG does not appear to have a considerable effect on AKT activation in either MCF-7 or T47D cells. In HeLa cells, AKT phosphorylation is basally high and is not substantially altered in the presence of IGF-1 or 2-DG. These results indicate that 2-DG-induced AKT activation is not universal and not IGF-1R-dependent, as implied by Zhong et al. (20). Moreover, 2-DG-induced activation of 5′-adenosine monophosphate-activated protein kinase was consistently high across all cell lines (data not shown), which is in agreement with previously published results by Zhong et al. (20) suggesting that 2-DG-induced changes in AKT activation are 5′-adenosine monophosphate-activated protein kinase-independent.
FIGURE 6.
Western blot assay of IGF-1R signaling. MCF-7, T47D, and HeLa cell lines were plated at 106 cells/well in 6-well plates in DMEM containing 10% FBS. The following day, cells were treated with 10% FBS DMEM containing 2-DG (25 mm), IGF-1 (130 nm), or both. Four hours later, cells were harvested, and levels of signaling proteins were assayed by Western blot.
DISCUSSION
Our main objective was to investigate the effects of 2-DG and glucose on IGF-1/IGFBP-3 binding, as a follow-up to a recent publication by Zhong et al. (10). They showed that a concentration of 25 mm 2-DG was sufficient to disrupt more than 60% of IGF-1-IGFBP-3 complexes, a finding that, if confirmed, would have considerable impact both on IGF and cancer research. The reported finding that 2-DG exposure is associated with higher free IGF-1 concentrations challenges its utility as an antineoplastic agent and even its utility as an experimental strategy to selectively inhibit glycolysis without affecting other aspects of cellular physiology. It became important to determine whether its relatively well characterized inhibition of glycolysis would predominate over this newly described stimulation in IGF signaling.
We sought to extend the findings of Zhong et al. (10) by identifying changes in affinity constants and by evaluating the potential effects of glucose, obviously structurally related to 2-DG, on IGF-1-IGFBP-3 complex formation. Using ELISA, we have shown that IGF-1/IGFBP-3 binding is unaltered by either 2-DG or glucose at various concentrations both within and exceeding physiological range. The assay was performed both in different orientations and in different buffer systems with identical conclusions.
To date, several groups have used SPR to examine the interaction between IGF-1 and its binding proteins (21, 22). Similar to the assay design detailed in previous reports, our SPR results are based upon IGF-1 injected over immobilized IGFBP-3 at a high flow rate (75 μl/min) and low signal range (<150 RU) to minimize any potential mass transport effects. Global fitting of the titration series to a 1:1 kinetic model with or without mass transport effects yielded similar outcomes. Our kinetic estimates were biologically relevant (e.g. ka ∼103–107 m−1 s−1 and kd ∼10−1–10−6 s−1 for typical protein interactions) and correlated well with values reported previously (21, 22) (i.e. overall affinity between IGF-1 and IGFBP-3 is subnanomolar). Ultimately, the presence of excess glucose or 2-DG failed to significantly alter the binding interaction between IGF-1 and immobilized IGFBP-3 in our SPR assay.
Using mass spectrometry, we demonstrated for the first time that a newer intensity fading approach (19) is appropriate for the study of IGF-IGFBP complexes. Although the resultant signal for the complexes did not match the anticipated theoretical predictions, this issue has been encountered by others (18, 19) and may be the result of higher ionization energies required for complexes compared with the individual protein and its binding partner alone. Nevertheless, our intensity fading experiments clearly show that the signal corresponding to IGF-1 was decreased in a dose-dependent manner upon IGFBP-3 addition and that 2-DG or glucose had no effect compared with the control MS spectra.
Upon examination of 2-DG-induced changes in IGF-1R signaling in MCF-7, T47D, and HeLa cell lines, we were unable to confirm the previously published finding that exposure to 2-DG induces IGF-1R activation (10). In fact, we observed that 2-DG treatment reduces phosphorylation of IGF-1R in all three cell lines tested. This finding provides further evidence against the hypothesis that 2-DG disrupts IGF-1-IGFBP-3 complex formation, which leads to activation of the IGF-1R. Moreover, the fact that phosphoserine 473 AKT levels changed in a cell-specific manner and were uncorrelated with phospho-IGF-1R levels supports the hypothesis that 2-DG-induced changes in AKT activation are independent of IGF-1R in these cell lines.
In conclusion, we have utilized three unique experimental strategies to demonstrate that 2-DG does not alter the binding interaction between IGF-1 and IGFBP-3. Although our direct measures of IGF-1-IGFBP-3 complex formation contrast with the experimental findings reported by Zhong et al. (10), we suspect that the commercial assay (which was designed to measure free IGF-1 in serum samples) utilized in their study may have been limited by its ability to provide only an indirect measure of IGF-1-IGFBP-3 complex formation. The mechanism responsible for the 2-DG-induced increase in IGF-1R signaling observed by Zhong et al. (10, 20) in some cell lines remains unknown. However, our results using several independent methods do not support the hypothesis of a universal mechanism involving IGF-1-IGFBP-3 complex disruption but rather suggest a mechanism involving cell-specific intracellular signaling differences.
Acknowledgments
We thank the Genome Quebec and Lady Davis Institute Clinical Proteomics Facility for their assistance with mass spectrometry experiments. Sheldon Biotechnology Centre is supported by a research resource grant from the Canadian Institutes of Health Research.
Footnotes
- IGF
- insulin-like growth factor
- IGF-1R
- IGF-1 receptor
- 2-DG
- 2-deoxy-d-glucose
- RU
- resonance unit
- SPR
- surface plasmon resonance
- IGFBP
- insulin-like growth factor-binding protein.
REFERENCES
- 1. Pollak M. (2008) Nat. Rev. Cancer 8, 915–928 [DOI] [PubMed] [Google Scholar]
- 2. Laviola L., Natalicchio A., Giorgino F. (2007) Curr. Pharm. Des. 13, 663–669 [DOI] [PubMed] [Google Scholar]
- 3. Collett-Solberg P. F., Cohen P. (2000) Endocrine 12, 121–136 [DOI] [PubMed] [Google Scholar]
- 4. Vincent A. M., Feldman E. L. (2002) Growth Horm. IGF Res. 12, 193–197 [DOI] [PubMed] [Google Scholar]
- 5. LeRoith D., Roberts C. T., Jr. (2003) Cancer Lett. 195, 127–137 [DOI] [PubMed] [Google Scholar]
- 6. Ghosh P., Dahms N. M., Kornfeld S. (2003) Nat. Rev. Mol. Cell Biol. 4, 202–212 [DOI] [PubMed] [Google Scholar]
- 7. Firth S. M., Baxter R. C. (2002) Endocr. Rev. 23, 824–854 [DOI] [PubMed] [Google Scholar]
- 8. Hwa V., Oh Y., Rosenfeld R. G. (1999) Endocr. Rev. 20, 761–787 [DOI] [PubMed] [Google Scholar]
- 9. Jones J. I., Clemmons D. R. (1995) Endocr. Rev. 16, 3–34 [DOI] [PubMed] [Google Scholar]
- 10. Zhong D., Xiong L., Liu T., Liu X., Liu X., Chen J., Sun S. Y., Khuri F. R., Zong Y., Zhou Q., Zhou W. (2009) J. Biol. Chem. 284, 23225–23233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wick A. N., Drury D. R., Morita T. N. (1955) Proc. Soc. Exp. Biol. Med. 89, 579–582 [DOI] [PubMed] [Google Scholar]
- 12. Wick A. N., Drury D. R., Nakada H. I., Wolfe J. B. (1957) J. Biol. Chem. 224, 963–969 [PubMed] [Google Scholar]
- 13. Chen W., Guéron M. (1992) Biochimie 74, 867–873 [DOI] [PubMed] [Google Scholar]
- 14. Singh D., Banerji A. K., Dwarakanath B. S., Tripathi R. P., Gupta J. P., Mathew T. L., Ravindranath T., Jain V. (2005) Strahlenther Onkol 181, 507–514 [DOI] [PubMed] [Google Scholar]
- 15. Myszka D. G. (1999) J. Mol. Recognit. 12, 279–284 [DOI] [PubMed] [Google Scholar]
- 16. Levitt R. J., Georgescu M. M., Pollak M. (2005) Biochem. Biophys. Res. Commun. 336, 1056–1061 [DOI] [PubMed] [Google Scholar]
- 17. Bolbach G. (2005) Curr. Pharm. Des. 11, 2535–2557 [DOI] [PubMed] [Google Scholar]
- 18. Yanes O., Nazabal A., Wenzel R., Zenobi R., Aviles F. X. (2006) J. Proteome Res. 5, 2711–2719 [DOI] [PubMed] [Google Scholar]
- 19. Yanes O., Villanueva J., Querol E., Aviles F. X. (2007) Nat. Protoc. 2, 119–130 [DOI] [PubMed] [Google Scholar]
- 20. Zhong D., Liu X., Schafer-Hales K., Marcus A. I., Khuri F. R., Sun S. Y., Zhou W. (2008) Mol. Cancer Ther. 7, 809–817 [DOI] [PubMed] [Google Scholar]
- 21. Beattie J., Phillips K., Shand J. H., Szymanowska M., Flint D. J., Allan G. J. (2008) Mol. Cell. Biochem. 307, 221–236 [DOI] [PubMed] [Google Scholar]
- 22. Vorwerk P., Hohmann B., Oh Y., Rosenfeld R. G., Shymko R. M. (2002) Endocrinology 143, 1677–1685 [DOI] [PubMed] [Google Scholar]