Abstract
AS160 (TBC1D4) is a known Akt substrate that is phosphorylated downstream of insulin action and that leads to regulated traffic of GLUT4. As GLUT4 vesicle fusion with the plasma membrane is a highly regulated step in GLUT4 traffic, we investigated whether AS160 and 14-3-3 interactions are involved in this process. Fusion was inhibited by a human truncated AS160 variant that encompasses the first N-terminal phosphotyrosine-binding (PTB) domain, by either of the two N-terminal PTB domains, and by a tandem construct of both PTB domains of rat AS160. We also found that in vitro GLUT4 vesicle fusion was strongly inhibited by the 14-3-3-quenching inhibitors R18 and fusicoccin. To investigate the mode of interaction of AS160 and 14-3-3, we examined insulin-dependent increases in the levels of these proteins on GLUT4 vesicles. 14-3-3γ was enriched on insulin-stimulated vesicles, and its binding to AS160 on GLUT4 vesicles was inhibited by the AS160 tandem PTB domain construct. These data suggest a model for PTB domain action on GLUT4 vesicle fusion in which these constructs inhibit insulin-stimulated 14-3-3γ interaction with AS160 rather than AS160 phosphorylation.
Keywords: Glucose Transport, Insulin, Membrane Fusion, Membrane Trafficking, Signal Transduction, AS160, GLUT4
Introduction
AS160 (also called TBC1D4) is a Rab GTPase-activating protein (GAP)3 that is part of a subfamily of Rab GAPs that contain a conserved TBC1 (Tre-2/Bub2/Cdc16) domain. AS160 was first implicated as a regulator of insulin-stimulated GLUT4 traffic by Lienhard and co-workers (1–3), who discovered that AS160 (which has a mass of 160 kDa) is a downstream substrate of the kinase Akt. Insulin action, through activation of PI3K and phosphatidylinositol 3,4,5-triphosphate generation, leads to activation of phosphatidylinositol-dependent kinases PDK1 and PDK2, and both these activities are required for Akt activation. These activations of Akt then lead to phosphorylation of AS160 on serine and tyrosine residues, most importantly Ser-588 and Thr-642 (1, 4).
Evidence that AS160 is involved in regulation of GLUT4 traffic includes use of a construct coding for a form of AS160 with 4 putative phosphorylatable serine and threonine residues mutated to alanine (1). This AS160–4P construct inhibits GLUT4 traffic in insulin target cells, including fat and muscle (5, 6). The implication from these studies is that AS160 is a negative regulator of vesicle traffic of GLUT4 and that its conversion to a 4P mutant prevents insulin action from switching off the GAP activity. AS160–4P acts as a fully functional GAP, and this activity leads to termination of the action of a downstream Rab that is also closely associated with GLUT4 traffic (7, 8).
A mouse model in which a similar mutant (AS160 T649A) has been expressed using a knock-in approach has been described. This mutation leads to a glucose-intolerant phenotype with reduced insulin sensitivity and altered GLUT4 traffic (9). Human insulin resistance has also been found to be associated with AS160 mutations. A human truncated variant of AS160 leads to insulin resistance and postprandial hyperinsulinemia (10). The effects of this truncation are associated with impaired GLUT4 translocation in insulin target cells. As this N-terminal truncation produces AS160 with an intact phosphotyrosine-binding (PTB) domain, we have examined here the functional and mechanistic consequences of AS160 PTB domain expression by focusing primarily on the fusion of GLUT4 vesicles with the plasma membrane.
Extensive work by James and co-workers (11–13) and MacKintosh and co-workers (14, 15) has provided evidence that AS160 action is highly dependent on association with 14-3-3 proteins. The investigation by Larance et al. (11) led to the hypothesis that AS160 is bound to GLUT4 vesicles in the basal state and dissociates from the vesicles when phosphorylated and when bound to 14-3-3. Furthermore, although dissociation is evident, this dissociation is not necessary for insulin-stimulated inactivation of the function of AS160 (12, 13). Support for the latter notion is the observation that an AS160-GLUT4 construct, directly tied to vesicles by virtue of the transmembrane-associated GLUT4 component, is functionally inactivated by insulin action without dissociation from the vesicles (13).
The site of action of AS160 in GLUT4 vesicle traffic has been partially resolved, and it has been found that the AS160–4P inhibits GLUT4 exocytosis but not its endocytosis (16). Further resolution of its site of action within the exocytosis pathway has been problematic. Some evidence supports a role in release of GLUT4 vesicles from an intracellular reservoir compartment (16), whereas other data suggest a role in docking of vesicles in close proximity of the plasma membrane (16–19). The fusion of GLUT4 vesicles at the plasma membrane is known to be a highly insulin signaling-dependent step in GLUT4 traffic and can be resolved in temporal detail using total internal reflection fluorescence microcopy (18, 20–22). We have shown that these final steps in insulin action on GLUT4 traffic can be reconstituted using a cell-free in vitro approach (23). We have used this reductionist in vitro approach here to examine the role of AS160 in fusion of GLUT4 vesicles. An advantage of the in vitro fusion assay is that it reduces the complexity of the possible component involvement in fusion and allows separation of this step from cellular vesicle release that may influence vesicle docking.
In view of the importance of AS160 in glucose metabolism and human insulin resistance, we have used cell-free in vitro assays to study the association of AS160 with 14-3-3 and the role of this interaction in GLUT4 vesicle fusion with the plasma membrane. In particular, we have investigated how protein constructs of the PTB domains of AS160 act as fusion inhibitors and have evaluated the extent to which these inhibitory effects occur through 14-3-3- and AS160-dependent generation of fusion-competent GLUT4 vesicles.
EXPERIMENTAL PROCEDURES
DNA Constructs and Recombinant Protein Expression and Purification
His-tagged N-terminal AS160(1–290), AS160(230–532), and AS160(1–532) constructs were amplified from a rat adipocyte cDNA library with primers containing BamHI and HindIII restriction sites and cloned into the pET28a(+) vector (Novagen). His-tagged N-terminal constructs were all expressed in Escherichia coli strain Rosetta(DE3)pLysS by incubation with 0.1 mm isopropyl β-d-thiogalactopyranoside for 4 h at room temperature. Recombinant proteins were purified on HisTrap columns using an ÅKTA chromatography system (GE Healthcare) with a linear imidazole gradient from 50 to 300 mm. The purified proteins were dialyzed against PBS. The HA-AS160(1–532) construct was obtained by PCR amplification with primers containing KpnI and EcoRI restriction sites and cloned into the pHM6 vector (Roche Applied Science). The FLAG-AS160(1–532) construct was obtained by PCR amplification with primers containing NotI and BamHI restriction sites and cloned into the p3×FLAG-CMV-10 expression vector (Sigma). The pCis2-HA-GLUT4 construct was a gift from Dr. Samuel Cushman (24).
The human p3×FLAG-AS160 R363XTr construct was a kind gift from Dr. David Savage and has been described previously (10). The human p3×FLAG-AS160 (full-length) construct was a kind gift from Dr. Gustav Lienhard and has been described previously (25). FLAG-tagged AS160 R363X or full-length AS160 was transfected into the human embryonic kidney HEK293T cell line using a calcium phosphate transfection method, and after 48 h of expression, the cells were lysed, and the recombinant protein was purified by immunoprecipitation with anti-FLAG antibody-agarose conjugate (Sigma). The recombinant protein was eluted from the beads with excess 3×FLAG peptide.
GST-14-3-3β, -γ, and -ϵ cDNA constructs were purchased from Addgene (plasmids 13276, 13280, and 13279, respectively). GST fusion proteins were expressed in E. coli strain DH5α by induction with 0.3 mm isopropyl β-d-thiogalactopyranoside for 2 h at 37 °C. The recombinant proteins were purified on a glutathione-Sepharose column, and the eluted proteins were dialyzed against PBS.
Quantification of 14-3-3 Isoform mRNA Levels
Total RNA was extracted from rat brain and epididymal adipose tissue with TriPure isolation reagent (Roche Applied Science) according to the manufacturer's instructions. The RNA was treated with DNase I to remove trace genomic DNA. 500 ng of total RNA, treated with DNase I, was reverse-transcribed to cDNA using the Superscript III First-Strand Synthesis Supermix kit for qRT-PCR (Invitrogen). Quantitative real-time PCR was performed with the StepOnePlusTM real-time PCR system (Applied Biosystems) using iTaqTM SYBR® Green Supermix with ROX (Bio-Rad) according to the manufacturer's instructions. Primers were designed using Primer3 software (26) and synthesized by Sigma. Primers were validated against a standard curve of varying amounts of brain cDNA, a tissue in which all isoforms are known to be present. The comparative CT method was used to quantify the relative expression of 14-3-3 in rat adipose tissue. 2.5 ng of adipose cDNA and 500 nm primers (final concentration) were used in the final quantitative real-time PCR. The following primer sequences were used: 14-3-3β, TAATGTTTGCTTCCGTGGTG (forward) and GGAGGGGGTCTTTTTCTTTTT (reverse); 14-3-3ϵ, AGAGGCTATTGCGCTGTCAT (forward) and AAACTCCCCCAAAACACCTC (reverse); 14-3-3η, CGAAATCAGCAAAGAGCACA (forward) and TGGTGCATTCTGGATCTCAT (reverse); 14-3-3γ, GCGCTCAACTACTCCGTTTT (forward) and GAGTCCTCGTTCAGAGTGTCG (reverse); 14-3-3θ, TGACAACAGGGCGTTTCC (forward) and ACGAAGGCAGGAGGTGTAAGT (reverse); 14-3-3ζ, GCGGGGAATAAACAGGATAA (forward) and AGCCTCACAAGTGCTCCAAG (reverse); and 14-3-3σ-like, CCACTTCCTACCACCACTCC (forward) and CAGTCCAGTTCTCAGCCACA (reverse).
Assay of in Vitro GLUT4 Vesicle Fusion
The individual components of the in vitro fusion assay were prepared as described previously (23). When recombinant proteins were preincubated with the GLUT4 vesicles for 1 h at 4 °C prior to the fusion assay, the control vesicles were preincubated with an equivalent amount of BSA. The in vitro fusion assay was performed as described previously (23), and the extent of fusion at 5 min was compared with the maximum fusion obtained by incubation at 37 °C for 30 min.
Phosphoprotein Pulldown Assays with GST-14-3-3
Equal amounts (7 μg) of GST-14-3-3β, -γ, and -ϵ fusion proteins were prebound to glutathione-Sepharose. Fresh lysates from basal or insulin-stimulated rat adipocytes were obtained by maintenance for 20 min at 18 °C in phosphate lysis buffer (50 mm Na2HPO4 (pH 7.4), 150 mm NaCl, 2% octaethylene glycol dodecyl ether, and protease and phosphatase inhibitors). 500 μg of cell lysate was incubated with immobilized 14-3-3 for 2 h at 4 °C. After extensive washing, bound protein was eluted by incubating in elution buffer (20 mm reduced glutathione in 50 mm Na2HPO4 (pH 8.5) and 150 mm NaCl). Eluted proteins were analyzed by SDS-PAGE, followed by electrotransfer and immunoblotting with anti-AS160 antibody (Millipore) and anti-phospho-Akt substrate (PAS) antibody (Cell Signaling Technology).
Quantification of AS160 and 14-3-3 Isoform Levels on GLUT4 Vesicles
Rat adipocytes at 40% cytocrit were maintained in the basal state or stimulated with 20 nm insulin for 20 min at 37 °C. The cells were then homogenized and processed as described previously (27) to obtain a post-high density microsome supernatant containing the GLUT4 vesicles. The GLUT4 vesicles were purified by immunoisolation using a rabbit anti-GLUT4 antibody (28) prebound to a maltose-binding protein-protein A construct (23) attached to amylose resin. After 2 h of incubation, the amylose column was washed with HES buffer (20 mm HEPES (pH 7.0), 0.5 mm EGTA, and 250 mm sucrose), and the GLUT4 vesicles were eluted in HES buffer containing 40 mm maltose. Eluted proteins were resolved by SDS-PAGE and analyzed by Western blotting with the following antibodies: anti-GLUT4 antibody (custom-made sheep anti-GLUT4 antibody raised against the same peptide as described previously (28)), anti-AS160 antibody, anti-PAS antibody, and anti-14-3-3β/ϵ (Santa Cruz Biotechnology) or anti-14-3-3γ (Millipore) antibody. For quantification purposes and when required, a standard curve of the respective recombinant protein was run on the same gel.
Rat Adipocyte Transfection
Rat adipocytes were electroporated with FLAG-AS160 (full-length) and HA-AS160(1–532) according to the method described by Al-Hasani et al. (29). Briefly, 200 μl of 50% cytocrit rat adipocytes was electroporated with 2 μg of pHM6-HA-AS160(1–532) and 0.1 μg of p3×FLAG-AS160 (full-length). Cells were incubated for 12 h at 37 °C in DMEM supplemented with 3.5% BSA. After washing with Krebs-Ringer HEPES buffer supplemented with 1% BSA and 200 nm adenosine, cells were left unstimulated or stimulated with 60 nm insulin for 20 min at 37 °C. The adipocytes were homogenized in HES buffer, and GLUT4 vesicles were isolated and analyzed as described above.
Detection of Cell-surface HA-tagged GLUT4
HA-GLUT4 levels at the surface of transfected rat adipocytes were measured using an adaptation of previously described methods (29, 30). 5 h after electroporation with 0.1 μg of pCis2-HA-GLUT4 and 0.4 μg of p3×FLAG-AS160(1–532), adipose cells were washed with Krebs-Ringer HEPES buffer supplemented with 1% BSA and 200 nm adenosine. Cells were maintained in the basal state or stimulated with 60 nm insulin for 20 min at 37 °C. To stop further GLUT4 translocation, the cells were then treated with 2 mm KCN (final concentration) for 3 min at 37 °C. The adipocytes were incubated for 1 h at 25 °C in the presence of 1 μg/ml anti-HA antibody (Covance). After two washes with Krebs-Ringer HEPES buffer, the cells were incubated for an additional 1 h at 25 °C in the presence of 1 μg/ml β-galactosidase-conjugated anti-mouse IgG (Southern Biotech). Aliquots of cells at 40% cytocrit were then transferred into 96-well plates, and the fluorogenic β-galactosidase substrate fluorescein digalactoside (Invitrogen) was added to give a final concentration of 0.1 mm. The resulting fluorescence was measured every 15 s over a 60-min period in a PHERAstar FS multiwell plate reader (BMG Labtech). The level of HA-GLUT4 present at the cell surface was then calculated from the fluorescence generated (rate of increase/mg of protein).
Interaction of Full-length AS160 and N-terminal AS160(1–532) in HEK293 Cells
HEK293 cells were transfected with 0.5 μg of p3×FLAG-AS160 (full-length) and 2 μg of pHM6-HA-AS160(1–532) with Lipofectamine LTX (Invitrogen) according to the manufacturer's instructions. After expression for 18 h, cells were incubated with serum-free medium for 16 h and left untreated or stimulated with 50 ng/ml IGF1 for 20 min at 37 °C. At the end of the incubation, cells were washed with PBS and lysed in Tris lysis buffer (50 mm Tris-HCl (pH 7.4), 1 mm EDTA, 150 mm NaCl, and 0.5% Triton X-100, supplemented with protease and phosphatase inhibitors). Lysates were cleared by centrifugation at 17,000 × g for 20 min, and portions of the cleared lysates were immunoprecipitated with anti-FLAG antibody-agarose conjugate or anti-HA tag antibody (Abcam) conjugated to protein G-agarose. Non-transfected cells were used as a control. Immunoprecipitated proteins were eluted with SDS sample buffer and analyzed by immunoblotting with anti-FLAG antibody, anti-HA antibody, anti-pan-14-3-3 protein antibody (Millipore), or anti-PAS antibody.
RESULTS
In Vitro GLUT4 Vesicle Fusion with the Plasma Membrane
We have developed an in vitro fusion assay that is reconstituted from separately isolated fractions from rat adipose cells that are maintained in the basal state or stimulated with insulin (23). The separate isolation of fractions allows them to be mixed such that GLUT4 vesicles from basal cells can be mixed with the plasma membrane and cytosol of either basal cells or insulin-stimulated cells. With this approach, we found that GLUT4 vesicles from basal cells can be activated to become fusion-competent using the plasma membrane from insulin-stimulated but not basal cells (23). To examine whether AS160 has a role in facilitating vesicle fusion and to investigate the mechanism for the inhibitory effects of human AS160 truncation on GLUT4 traffic (10), we examined the inhibitory effects of protein constructs generated from AS160 N-terminal PTB domains (Fig. 1A).
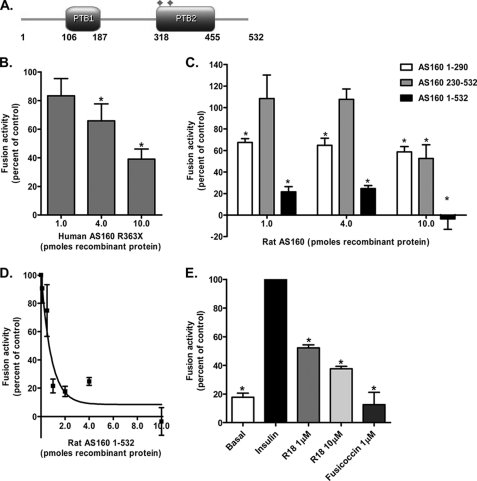
FIGURE 1.
Inhibition of in vitro fusion of GLUT4 vesicles with plasma membranes by N-terminal AS160 constructs and 14-3-3-quenching inhibitors. A, rat N-terminal AS160 PTB domain construct. Shown is the predicted domain organization of rat AS160 transcript variant 4 (LOC686547). The protein sequence was subjected to a domain search using the Pfam tool (42). The diamonds indicates predicted Akt phosphorylation sites. Ser-326 and Ser-350 in the rat sequence correspond to Ser-318 and Ser-341 in the human sequence. B, effect of the human truncated form of AS160 R363X on insulin-stimulated fusion activity. Immunoisolated GLUT4 vesicles from basal cells were pretreated with increasing amounts of FLAG-AS160 R363X for 1 h at 4 °C before measuring the fusion activity in the presence of plasma membranes and cytosol from insulin-stimulated cells. Results are means ± S.E. from four independent experiments. *, p < 0.05 (comparison with the insulin control). C, inhibition of fusion activity by rat N-terminal AS160 constructs. GLUT4 vesicle fusion activity was determined after pretreating immunoisolated GLUT4 vesicles from basal adipocytes with increasing amounts of recombinant His-tagged N-terminal AS160(1–290), AS160(230–532), or AS160(1–532) for 1 h at 4 °C. Fusion activity was determined in the presence of plasma membranes and cytosol from insulin-treated adipocytes. Results are means ± S.E. from four to eight independent experiments. *, p < 0.05 (comparison with the insulin control). D, dose response for the inhibition of fusion activity by His-AS160(1–532). Immunoisolated GLUT4 vesicles isolated from basal cells were pretreated with increasing amounts of His-AS160(1–532) for 1 h at 4 °C before measuring the fusion activity in the presence of plasma membranes and cytosol from insulin-stimulated cells. Results are means ± S.E. from four to five independent experiments. E, effect of 14-3-3-quenching inhibitors R18 and fusicoccin on the in vitro fusion activity. Insulin-stimulated cytosol was pretreated with the indicated concentrations of R18 or fusicoccin for 1 h at 4 °C. Fusion activity was determined in the presence of plasma membranes and cytosol from insulin-treated adipocytes. Result are means ± S.E. from five independent experiments. *, p < 0.05 (comparison with the insulin control).
We tested the human protein construct AS160 R363X, comprising mainly the first PTB domain, as a potential inhibitor of GLUT4 vesicle fusion. Addition of increasing protein concentrations to the fusion assay led to a progressive inhibition of fusion, reaching a maximum ≈50% inhibition at the highest protein concentration tested (Fig. 1B). Protein constructs from rat AS160 incorporating the separate first (AS160(1–290)) and second (AS160(230–532)) PTB domains inhibited vesicle fusion by ∼50% at the maximum protein level tested (Fig. 1C). A protein construct incorporating both tandem PTB domains (AS160(1–532)) inhibited fusion very potently, and the highest protein concentration tested inhibited fusion completely (Fig. 1D).
14-3-3 proteins can potentially interact with phosphorylated AS160 (31), and we have therefore examined whether quenching of 14-3-3 interaction can influence insulin-stimulated GLUT4 vesicle fusion with the plasma membrane target. The R18 peptide, which is a pseudo-substrate for 14-3-3, led to inhibition of in vitro fusion of basal GLUT4 vesicles with the insulin-stimulated plasma membrane (Fig. 1E). A similarly potent inhibition was observed upon treatment of the fusion components with fusicoccin, a known 14-3-3-binding reagent (Fig. 1E). This compound is particularly potent in inhibition of the interaction of 14-3-3 with the plant H+-ATPases (32).
Insulin Stimulates Specific Interactions between 14-3-3γ and AS160 in Rat Adipocytes
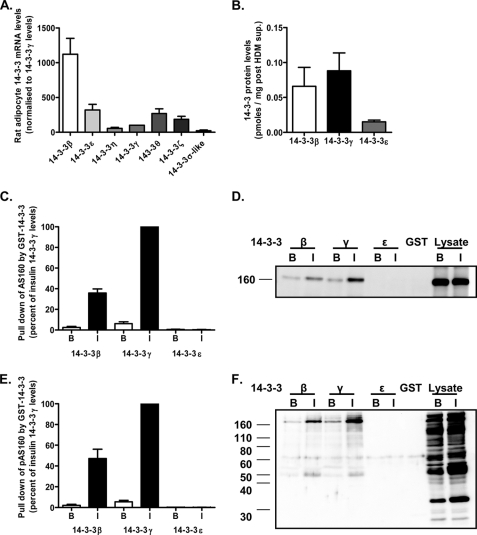
To investigate the possible 14-3-3 isoform involved in insulin action on GLUT4 traffic and GLUT4 vesicle activation, we first determined the expression of the message levels of 14-3-3 isoforms present in rat adipose cells. 14-3-3β was found to be the most abundant isoform, with much higher message levels compared with 14-3-3 isoforms ϵ, η, γ, θ, ξ, and σ (Fig. 2A). However, the high level of 14-3-3β message was not reflected at the protein level, and quantitative blotting against recombinant protein standards of 14-3-3β, -γ, and -ϵ revealed high protein levels of the γ isoform (Fig. 2B). We next compared the extent to which these three isoforms, as GST constructs, could bind to and precipitate AS160 from rat adipocyte cell lysates. Both 14-3-3β and 14-3-3γ precipitated AS160, but the γ isoform was much more effective and precipitated higher amounts (Fig. 2, C and D). The levels of precipitation by 14-3-3γ were 15–17-fold greater from insulin-treated lysates compared with basal lysates, reflecting the higher level of phosphorylation and therefore more available sites for interaction with 14-3-3. In addition, the γ isoform relatively selectively pulled down phosphorylated AS160 from adipocyte lysates as determined using an antibody that recognizes a wide range of phosphorylated Akt substrates (anti-PAS antibody) (Fig. 2, E and F). The extent of precipitation of AS160 as determined by anti-PAS antibody blotting was also 15–17-fold greater in the lysates from insulin-treated cells compared with basal cells.
FIGURE 2.
Analysis of the abundance of 14-3-3 isoforms in rat adipocytes and their interactions with AS160. A, quantitative real-time PCR analysis of the relative abundance of mRNA for the different known 14-3-3 isoforms in rat adipocytes. B, quantification of the 14-3-3β, -γ, and -ϵ proteins in rat adipocytes as detected by immunoblotting and quantified by comparison with a recombinant protein standard curve as illustrated in Fig. 4. C, comparison of the relative affinity of recombinant 14-3-3 isoforms β, γ, and ϵ for AS160. Equal amounts of GST-14-3-3β, -γ, or -ϵ immobilized on glutathione beads were incubated with total cell lysate prepared from basal (B) or insulin (I)-stimulated rat adipocytes. Relative amounts of AS160 pulled down by the recombinant proteins were detected by immunoblotting with anti-AS160 antibody. Results are means ± S.E. from three independent experiments and with a representative immunoblot (D). E, comparison of the interaction of recombinant 14-3-3 isoforms β, γ, and ϵ with phosphorylated AS160. Equal amounts of GST-14-3-3β, -γ, or -ϵ immobilized on glutathione beads were incubated with cell lysates prepared from basal or insulin-stimulated rat adipocytes. Relative amounts of phosphorylated AS160 pulled down by the recombinant proteins were detected by immunoblotting with anti-PAS antibody. Results are means ± S.E. from three independent experiments with a representative immunoblot (F).
14-3-3γ Is Enriched on GLUT4 Vesicles
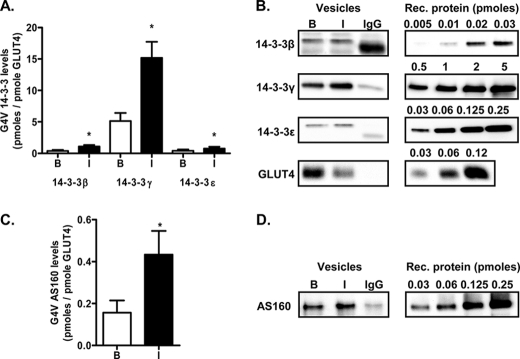
Although the ratios of 14-3-3β to 14-3-3γ message and protein in rat adipocytes were 10 and 0.7, respectively, the γ isoform was found to be highly enriched on GLUT4 vesicles. Quantification of the protein levels against recombinant protein standards revealed that the levels of the γ isoform were 10–15-fold higher than those of the β isoform. Furthermore, the levels of 14-3-3γ were 3-fold higher on GLUT4 vesicles from insulin-treated cells compared with basal cells (Fig. 3, A and B). The levels of AS160 were also higher on GLUT4 vesicles from insulin-treated cells compared with basal cells (Fig. 3, C and D). 14-3-3γ binding to GLUT4 vesicles cannot be solely dependent on the concomitant binding of AS160, as the levels of 14-3-3γ associated with the vesicle were much higher than those of AS160.
FIGURE 3.
Quantification of the amounts of 14-3-3 and AS160 present on GLUT4 vesicles. A, quantification of the amounts of 14-3-3 isoforms β, γ, and ϵ detected on GLUT4 vesicles (G4V) isolated from basal (B) or insulin (I)-stimulated adipocytes. Results are means ± S.E. from three independent experiments. *, p < 0.05 (comparison of basal versus insulin-stimulated adipocytes for each isoform). B, representative immunoblots for the 14-3-3 isoforms and comparison with recombinant (Rec.) protein standard curves. C, quantification of the amount of AS160 detected on GLUT4 vesicles isolated from basal or insulin-stimulated adipocytes. Results are means ± S.E. from three independent experiments. *, p < 0.05 (comparison of basal versus insulin-stimulated adipocytes). D, representative immunoblots for AS160 and comparison with a recombinant protein standard curve.
Our data on AS160 association with GLUT4 vesicles are consistent with a recent study by Stöckli et al. (13), who showed that it is unnecessary for AS160 to dissociate from GLUT4 vesicles. However, working with immunoisolated GLUT4 vesicles from an isolated low density microsome fraction of 3T3-L1 adipocytes, Larance et al. (11) reported an insulin-dependent AS160 dissociation from GLUT4 vesicles. By contrast, we found that, in rat adipocytes, there was an insulin-dependent increase in the level of AS160 associated with GLUT4 vesicles. Differences in methodology for isolation of GLUT4 vesicles may account for this discrepancy. Larance et al. (11) showed that much of the low density microsome fraction of 3T3-L1 cells is a proteinaceous mixture of proteins that are only loosely associated with intracellular membranes and that this material is not recovered following membrane vesicle resuspension. We prepared our vesicles from rat adipocytes using a mild isolation technique that does not involve sedimentation and resuspension of membranes from a low density microsome fraction. Our vesicles are specifically associated with large amounts of 14-3-3γ that we know is lost from the vesicles upon centrifugation or high salt washes.4 It seems likely therefore that we do not observe loss of AS160 from vesicles on insulin action as the 14-3-3 associated AS160 remains loosely associated with a vesicle scaffolding and chaperoning protein network.
AS160 PTB Domains Block 14-3-3γ Recruitment to GLUT4 Vesicles
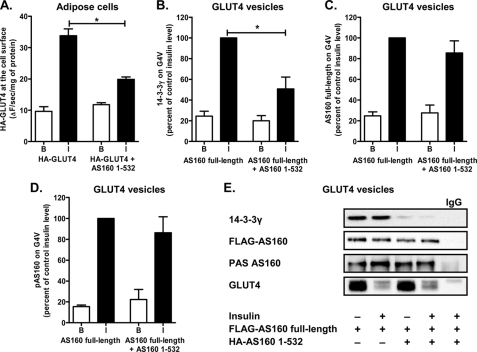
The tandem PTB domain construct potently inhibited insulin-stimulated (but not basal) GLUT4 translocation to the cell surface in the intact adipose cell (Fig. 4A). GLUT4 vesicle loading with AS160 and its association with 14-3-3γ may be important in facilitating the fusion competency of these vesicles and consequently inhibitory effects on cell-surface GLUT4. The vesicles may also be a site of action for the inhibitory effects of AS160 PTB domains. We therefore carried out a series of experiments in which rat adipocytes were cotransfected with a FLAG-tagged version of full-length AS160 together with the HA-tagged AS160 tandem PTB domains (AS160(1–532)). This transfection led to the association of large amounts of expressed FLAG-tagged AS160 on GLUT4 vesicles to an extent that exceeded the levels of endogenous AS160 and was comparable with the level of endogenous 14-3-3. We subsequently isolated the GLUT4 vesicles to determine the extent to which the binding of 14-3-3γ, the binding of FLAG-AS160, and FLAG-AS160 phosphorylation were perturbed by the tandem PTB domain construct. The association of 14-3-3γ with insulin-activated GLUT4 vesicles was significantly inhibited by the expression of the PTB domain construct (Fig. 4B). By contrast, the cellular expression of the PTB domain construct did not alter the level of FLAG-tagged AS160 and did not change the level of FLAG-AS160 phosphorylation as detected with anti-PAS antibody (Fig. 4, C and D). The levels of association of AS160 and 14-3-3γ with GLUT4 vesicles were normalized for expression of GLUT4. The transfection experiments involved a 12-h incubation of the adipocytes, and the recovery of GLUT4 vesicles was found to be abnormally low from cells that were treated with insulin after transfection. This can be seen by comparing Fig. 4E (from transfected adipocytes) with Fig. 3 (from non-transfected adipocytes). The reason for this change is unknown, but it leads to a greater reliance on normalization to the recovered GLUT4 levels for estimating the levels of insulin-dependent changes in vesicle-associated proteins.
FIGURE 4.
AS160(1–532) interacts with full-length AS160 and reduces the relative amount of 14-3-3γ bound to AS160 and associated with GLUT4 vesicles. A, primary rat adipocytes were cotransfected by electroporation with HA-tagged GLUT4 and FLAG-tagged AS160(1–532). After 5 h of expression, cells were maintained in the basal state (B) or stimulated with 60 nm insulin (I). HA-GLUT4 present at the cell surface was detected after incubating the intact cells with anti-HA antibody for 1 h and detection using a β-galactosidase-coupled secondary antibody generating a fluorescent product. Results are means ± S.E. from four replicates from a single experiment representative of two independent experiments. *, p < 0.05 (comparison of HA-GLUT4 alone versus HA-GLUT4 plus FLAG-AS160(1–532)). B–D, primary rat adipocytes were cotransfected by electroporation with FLAG-tagged full-length AS160 and HA-tagged AS160(1–532). After 12 h of expression, cells were left unstimulated or stimulated with 60 nm insulin and then homogenized. GLUT4 vesicles (G4V) were immunoisolated, and the relative amounts of 14-3-3γ (B), AS160 (C), and phosphorylated AS160 (pAS160; D) were detected by immunoblotting with anti-14-3-3γ, anti-AS160, and anti-PAS antibodies, respectively. Results are means ± S.E. from three independent experiments. E, representative immunoblots for data quantified in B–D. *, p < 0.05 (comparison of FLAG-AS160 alone versus FLAG-AS160 plus HA-AS160(1–532)).
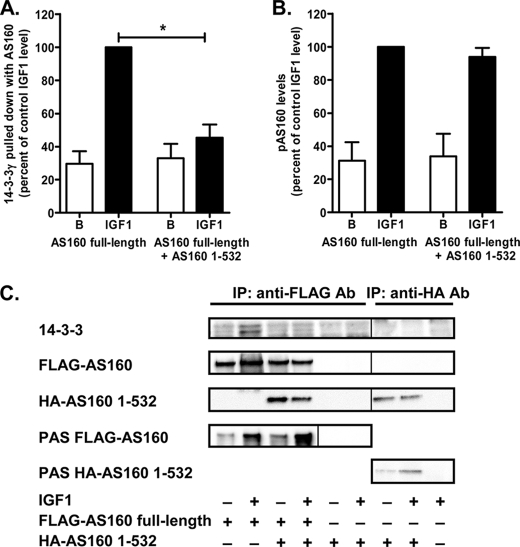
As the main response to the PTB domain expression was a reduced binding of 14-3-3γ, we further investigated the direct protein interaction between AS160, the tandem PTB domain construct, and 14-3-3 isoforms. To do this, we chose the HEK293 cell expression system, which is known to be useful in studying such interactions (10). We were able to show that heterodimer formation occurred between full-length AS160 and the tandem PTB domains. Precipitation of expressed FLAG-tagged AS160 with anti-FLAG antibody revealed that HA-tagged PTB domains (AS160(1–532)) associated with the full-length protein (Fig. 5C). Consistent with the data obtained with rat adipocyte GLUT4 vesicles, the heterodimer formation did not alter IGF1-stimulated phosphorylation of FLAG-AS160 (Fig. 5, B and C) but significantly reduced the combination of the heterodimer with endogenous 14-3-3 that was present in the HEK293 cells and recognized by anti-pan-14-3-3 antibody (Fig. 5A). Most 14-3-3 isoforms are present in HEK293 cells, but 14-3-3ϵ is the most abundant (33). This reduction in 14-3-3 association was related to heterodimer formation, as the PTB domain construct alone (which was phosphorylated) did not lead to association with 14-3-3 (as detected in immunoprecipitates using anti-HA antibody) (Fig. 5C).
FIGURE 5.
AS160(1–532) interacts with full-length AS160, forms heterodimers, and inhibits 14-3-3 binding to AS160 in HEK293 cells. HEK293 cells were cotransfected with FLAG-tagged full-length AS160 and HA-AS160(1–532). After 18 h of expression, cells were serum-starved, left unstimulated (basal (B)) or stimulated with 50 ng/ml IGF1, and then harvested and lysed in detergent. FLAG-AS160 or HA-AS160(1–532) was immunoprecipitated (IP) with anti-FLAG or anti-HA tag antibody, respectively. The immunoprecipitated proteins were subjected to SDS-PAGE and immunoblotted with anti-FLAG, anti-HA, anti-pan-14-3-3, or anti-PAS antibody. The relative levels of 14-3-3 (A) and FLAG-tagged phosphorylated AS160 (pAS160; B) pulled down were quantified and normalized to the level of expression of FLAG-tagged full-length AS160. Results are means ± S.E. from three independent experiments. *, p < 0.05 (comparison of FLAG-AS160 alone versus FLAG-AS160 plus HA-AS160(1–532)). C, representative immunoblots.
DISCUSSION
Recent studies have revealed that the Rab GAP proteins TBC1D1 and AS160 (TBC1D4) have important roles in the metabolic pathways relevant to obesity and type 2 diabetes. A mutant form of TBC1D1 occurs in a rare form of human obesity (34, 35). Furthermore, truncation and loss of TBC1D1 in a mouse strain have revealed that the presence of TBC1D1 confers leanness when crossed with an obese and insulin-resistant mouse strain (36). In this mouse model, loss of TBC1D1 reverses whole body insulin resistance and obesity in an artificial situation in which mice have high access to nutrients. Interestingly, differences in TBC1D1 gene sequence have been genetically linked to differences in growth and metabolic activity occurring within chicken lines bred for high meat content or egg-laying activity (37). The mouse model in which a non-phosphorylatable mutant of AS160 (AS160 T649A) has been expressed leads to a glucose-intolerant phenotype with reduced insulin sensitivity and altered GLUT4 traffic (9). Human insulin resistance has also been found to be associated with AS160 mutations. Families with AS160 variants including N1206S, N655Y, and N785K have been identified and may contribute to varying degrees of insulin resistance. However, complete co-segregation with the phenotype was difficult to achieve in studies on these families (38). In addition, a human truncated variant of AS160 leads to acanthosis nigricans and postprandial hyperinsulinemia (10). The effects of this truncation are associated with impaired GLUT4 translocation in insulin target cells. In 3T3-L1 cells, expression of this truncated mutant leads to elevation of basal and reduction of insulin-stimulated GLUT4 traffic (10). We have confirmed here that the AS160 tandem PTB domain construct inhibits insulin-stimulated GLUT4 translocation to the cell surface in rat adipose cells.
We found inhibition of fusion by the PTB domains of AS160 and by the 14-3-3-quenching inhibitors R18 and fusicoccin, thus identifying the fusion reaction as an important site for action of these proteins, their mutants, and truncated forms. Previous studies have suggested a prefusion role for AS160 in which the delivery of GLUT4 vesicles to the vicinity of the plasma membrane prior to initiation of fusion is one of the means by which translocation is activated by insulin action (3, 16, 19). In total internal reflection fluorescence studies that allow a distinction between vesicle docking and fusion, it has been suggested that a block in docking of GLUT4 vesicles by the AS160–4P construct indirectly affects downstream fusion (17). Insulin action on fusion machinery beyond and downstream of AS160 has been proposed (18, 19), possibly involving the motor protein Myo1c (39, 40). The inhibition of GLUT4 vesicle docking and fusion with the plasma membrane by the PTB domain constructs of AS160 revealed here provides direct evidence that AS160 itself is required for in vitro docking and fusion, where release from intracellular compartments is not occurring.
Our studies on the mechanism by which the PTB domain constructs and 14-3-3-quenching compounds inhibit the fusion reaction have led us to the discovery that 14-3-3γ is highly enriched on GLUT4 vesicles and has marked selectivity for interaction with phospho-AS160. The combination of AS160 with 14-3-3γ appears to be critical, and the PTB domains of AS160 can act as inhibitors of fusion by inhibiting this interaction. This inhibitory effect of the PTB domains on 14-3-3γ binding occurs on GLUT4 vesicles but can also be reproduced in studies on direct protein-protein interaction.
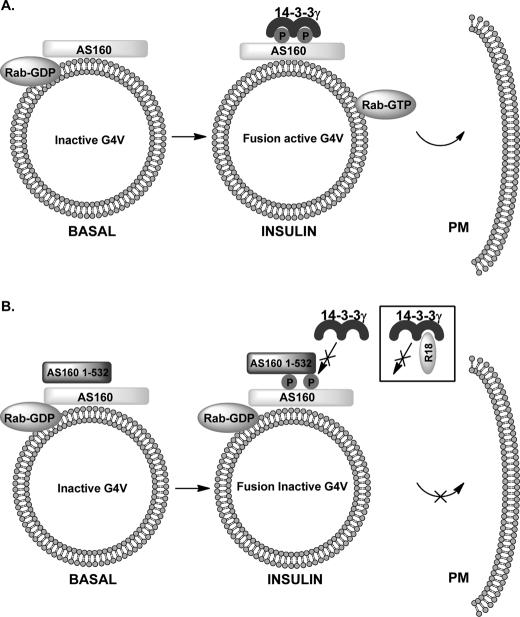
These data led us to propose a model for regulation of GLUT4 vesicle fusion in which insulin-stimulated 14-3-3γ interaction, but not AS160 phosphorylation alone, generates a fusion-activating GAP-inactive form of AS160 (Fig. 6A). We suggest that the PTB domain constructs form heterodimers with endogenous AS160 and that this interaction prevents the binding of 14-3-3 to phosphorylated AS160 (Fig. 6B). Therefore, AS160 remains in its GAP-active form, the associated Rab remains in the GDP form, and fusion activity is therefore low. The basis for the R18 inhibition of fusion is likely to be due to quenching of all available 14-3-3, so, again, although AS160 is phosphorylated, the GAP is active, the associated Rab remains in its GDP form, and fusion activity is therefore low. Our data appear to be inconsistent with a receptor-blocking mode of action of the fusion inhibitory PTB domains. If interaction with a receptor, such as IRAP (Insulin-regulated aminopeptidase) (11, 41), were blocked by the PTB domain constructs, then AS160 would be unable to be recruited to the vesicles, and the vesicle Rab would be expected to be in the fusion-facilitating and fusion-active form. Fusion would be expected to be high, but this has not been observed. In contrast to a study on GLUT4 vesicles in 3T3-L1 cells (11), we found that, in rat adipose cells, AS160 is continuously associated with GLUT4 vesicles and that insulin action does not lead to its dissociation. However, our data support the hypothesis that 14-3-3 interaction with AS160 is largely responsible for its functional inactivation as proposed by Ramm et al. (12).
FIGURE 6.
Model for inhibitory effects of AS160 N-terminal PTB domain constructs and 14-3-3-quenching reagents on GLUT4 vesicle fusion. A, insulin-stimulated generation of fusion-active GLUT4 vesicles (G4V). Insulin action leads to AS160 phosphorylation and 14-3-3γ binding and consequently generation of a GLUT4 vesicle Rab in an active GTP-loaded form. B, proposed mechanism for fusion inhibition by the N-terminal tandem PTB domain construct AS160(1–532). It is proposed that heterodimer formation with full-length AS160 prevents 14-3-3γ binding. The R18 peptide prevents 14-3-3γ binding through direct quenching reaction. PM, plasma membrane.
In conclusion, this investigation of the inhibitory effects of N-terminal PTB domain constructs of AS160, including a human truncated variant, has revealed that the inhibitory PTB domains stop the loading of vesicles with 14-3-3γ but do not stop AS160 phosphorylation. This suggests that phosphorylation alone, without 14-3-3γ combination, does not generate GLUT4 vesicles that can fuse with the insulin-activated plasma membrane in adipocytes. The emerging roles of the Rab GAPs TBC1D1 and AS160 (TBC1D4) in the control of whole body and muscle glucose metabolism in human subjects with obesity and type 2 diabetes indicate that further studies on the roles of both of these proteins in the process of GLUT4 vesicle fusion in muscle are warranted.
Acknowledgments
We thank Gustav Lienhard for FLAG-tagged human AS160, David Savage for FLAG-tagged human AS160 R363X, Samuel Cushman for the HA-tagged GLUT4 construct, and Carol MacKintosh for the HEK293 cells.
This work was supported by the Wellcome Trust and Diabetes UK.
F. Koumanov and G. D. Holman, unpublished data.
- GAP
- GTPase-activating protein
- PTB
- phosphotyrosine-binding
- PAS
- phospho-Akt substrate.
REFERENCES
- 1. Sano H., Kane S., Sano E., Mîinea C. P., Asara J. M., Lane W. S., Garner C. W., Lienhard G. E. (2003) J. Biol. Chem. 278, 14599–14602 [DOI] [PubMed] [Google Scholar]
- 2. Mîinea C. P., Sano H., Kane S., Sano E., Fukuda M., Peränen J., Lane W. S., Lienhard G. E. (2005) Biochem. J. 391, 87–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eguez L., Lee A., Chavez J. A., Miinea C. P., Kane S., Lienhard G. E., McGraw T. E. (2005) Cell Metab. 2, 263–272 [DOI] [PubMed] [Google Scholar]
- 4. Sakamoto K., Holman G. D. (2008) Am. J. Physiol. Endocrinol. Metab. 295, E29–E37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bruss M. D., Arias E. B., Lienhard G. E., Cartee G. D. (2005) Diabetes 54, 41–50 [DOI] [PubMed] [Google Scholar]
- 6. Karlsson H. K., Zierath J. R., Kane S., Krook A., Lienhard G. E., Wallberg-Henriksson H. (2005) Diabetes 54, 1692–1697 [DOI] [PubMed] [Google Scholar]
- 7. Sano H., Roach W. G., Peck G. R., Fukuda M., Lienhard G. E. (2008) Biochem. J. 411, 89–95 [DOI] [PubMed] [Google Scholar]
- 8. Sano H., Eguez L., Teruel M. N., Fukuda M., Chuang T. D., Chavez J. A., Lienhard G. E., McGraw T. E. (2007) Cell Metab. 5, 293–303 [DOI] [PubMed] [Google Scholar]
- 9. Chen S., Wasserman D. H., MacKintosh C., Sakamoto K. (2011) Cell Metab. 13, 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dash S., Sano H., Rochford J. J., Semple R. K., Yeo G., Hyden C. S., Soos M. A., Clark J., Rodin A., Langenberg C., Druet C., Fawcett K. A., Tung Y. C., Wareham N. J., Barroso I., Lienhard G. E., O'Rahilly S., Savage D. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9350–9355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Larance M., Ramm G., Stöckli J., van Dam E. M., Winata S., Wasinger V., Simpson F., Graham M., Junutula J. R., Guilhaus M., James D. E. (2005) J. Biol. Chem. 280, 37803–37813 [DOI] [PubMed] [Google Scholar]
- 12. Ramm G., Larance M., Guilhaus M., James D. E. (2006) J. Biol. Chem. 281, 29174–29180 [DOI] [PubMed] [Google Scholar]
- 13. Stöckli J., Davey J. R., Hohnen-Behrens C., Xu A., James D. E., Ramm G. (2008) Mol. Endocrinol. 22, 2703–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geraghty K. M., Chen S., Harthill J. E., Ibrahim A. F., Toth R., Morrice N. A., Vandermoere F., Moorhead G. B., Hardie D. G., MacKintosh C. (2007) Biochem. J. 407, 231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen S., Murphy J., Toth R., Campbell D. G., Morrice N. A., MacKintosh C. (2008) Biochem. J. 409, 449–459 [DOI] [PubMed] [Google Scholar]
- 16. Zeigerer A., McBrayer M. K., McGraw T. E. (2004) Mol. Biol. Cell 15, 4406–4415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang L., Fan J., Bai L., Wang Y., Chen Y., Yang L., Chen L., Xu T. (2008) J. Biol. Chem. 283, 8508–8516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bai L., Wang Y., Fan J., Chen Y., Ji W., Qu A., Xu P., James D. E., Xu T. (2007) Cell Metab. 5, 47–57 [DOI] [PubMed] [Google Scholar]
- 19. Gonzalez E., McGraw T. E. (2006) Mol. Biol. Cell 17, 4484–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lizunov V. A., Matsumoto H., Zimmerberg J., Cushman S. W., Frolov V. A. (2005) J. Cell Biol. 169, 481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang S., Lifshitz L. M., Jones C., Bellve K. D., Standley C., Fonseca S., Corvera S., Fogarty K. E., Czech M. P. (2007) Mol. Cell. Biol. 27, 3456–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stenkula K. G., Lizunov V. A., Cushman S. W., Zimmerberg J. (2010) Cell Metab. 12, 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koumanov F., Jin B., Yang J., Holman G. D. (2005) Cell Metab. 2, 179–189 [DOI] [PubMed] [Google Scholar]
- 24. Quon M. J., Guerre-Millo M., Zarnowski M. J., Butte A. J., Em M., Cushman S. W., Taylor S. I. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5587–5591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kane S., Sano H., Liu S. C., Asara J. M., Lane W. S., Garner C. C., Lienhard G. E. (2002) J. Biol. Chem. 277, 22115–22118 [DOI] [PubMed] [Google Scholar]
- 26. Rozen S., Skaletsky H. (2000) Methods Mol. Biol. 132, 365–386 [DOI] [PubMed] [Google Scholar]
- 27. Simpson I. A., Yver D. R., Hissin P. J., Wardzala L. J., Karnieli E., Salans L. B., Cushman S. W. (1983) Biochim. Biophys. Acta 763, 393–407 [DOI] [PubMed] [Google Scholar]
- 28. Satoh S., Nishimura H., Clark A. E., Kozka I. J., Vannucci S. J., Simpson I. A., Quon M. J., Cushman S. W., Holman G. D. (1993) J. Biol. Chem. 268, 17820–17829 [PubMed] [Google Scholar]
- 29. Al-Hasani H., Hinck C. S., Cushman S. W. (1998) J. Biol. Chem. 273, 17504–17510 [DOI] [PubMed] [Google Scholar]
- 30. Somwar R., Niu W., Kim D. Y., Sweeney G., Randhawa V. K., Huang C., Ramlal T., Klip A. (2001) J. Biol. Chem. 276, 46079–46087 [DOI] [PubMed] [Google Scholar]
- 31. Pozuelo Rubio M., Geraghty K. M., Wong B. H., Wood N. T., Campbell D. G., Morrice N., MacKintosh C. (2004) Biochem. J. 379, 395–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moorhead G., Douglas P., Morrice N., Scarabel M., Aitken A., MacKintosh C. (1996) Curr. Biol. 6, 1104–1113 [DOI] [PubMed] [Google Scholar]
- 33. Liang S., Xu Y., Shen G., Liu Q., Zhao X., Xu Z., Xie X., Gong F., Li R., Wei Y. (2009) Electrophoresis 30, 4152–4162 [DOI] [PubMed] [Google Scholar]
- 34. Stone S., Abkevich V., Russell D. L., Riley R., Timms K., Tran T., Trem D., Frank D., Jammulapati S., Neff C. D., Iliev D., Gress R., He G., Frech G. C., Adams T. D., Skolnick M. H., Lanchbury J. S., Gutin A., Hunt S. C., Shattuck D. (2006) Hum. Mol. Genet. 15, 2709–2720 [DOI] [PubMed] [Google Scholar]
- 35. Meyre D., Farge M., Lecoeur C., Proenca C., Durand E., Allegaert F., Tichet J., Marre M., Balkau B., Weill J., Delplanque J., Froguel P. (2008) Hum. Mol. Genet. 17, 1798–1802 [DOI] [PubMed] [Google Scholar]
- 36. Chadt A., Leicht K., Deshmukh A., Jiang L. Q., Scherneck S., Bernhardt U., Dreja T., Vogel H., Schmolz K., Kluge R., Zierath J. R., Hultschig C., Hoeben R. C., Schürmann A., Joost H. G., Al-Hasani H. (2008) Nat. Genet. 40, 1354–1359 [DOI] [PubMed] [Google Scholar]
- 37. Rubin C. J., Zody M. C., Eriksson J., Meadows J. R., Sherwood E., Webster M. T., Jiang L., Ingman M., Sharpe T., Ka S., Hallböök F., Besnier F., Carlborg O., Bed'hom B., Tixier-Boichard M., Jensen P., Siegel P., Lindblad-Toh K., Andersson L. (2010) Nature 464, 587–591 [DOI] [PubMed] [Google Scholar]
- 38. Dash S., Langenberg C., Fawcett K. A., Semple R. K., Romeo S., Sharp S., Sano H., Lienhard G. E., Rochford J. J., Howlett T., Massoud A. F., Hindmarsh P., Howell S. J., Wilkinson R. J., Lyssenko V., Groop L., Baroni M. G., Barroso I., Wareham N. J., O'Rahilly S., Savage D. B. (2010) Diabetologia 53, 1239–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bose A., Robida S., Furcinitti P. S., Chawla A., Fogarty K., Corvera S., Czech M. P. (2004) Mol. Cell. Biol. 24, 5447–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yip M. F., Ramm G., Larance M., Hoehn K. L., Wagner M. C., Guilhaus M., James D. E. (2008) Cell Metab. 8, 384–398 [DOI] [PubMed] [Google Scholar]
- 41. Peck G. R., Ye S., Pham V., Fernando R. N., Macaulay S. L., Chai S. Y., Albiston A. L. (2006) Mol. Endocrinol. 20, 2576–2583 [DOI] [PubMed] [Google Scholar]
- 42. Finn R. D., Tate J., Mistry J., Coggill P. C., Sammut S. J., Hotz H. R., Ceric G., Forslund K., Eddy S. R., Sonnhammer E. L., Bateman A. (2008) Nucleic Acids Res. 36, D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]