Abstract
MicroRNAs (miRNAs) regulate gene expression by repressing translation or directing sequence-specific degradation of complementary mRNA. Here, we report that expression of miR-205 is significantly suppressed in melanoma specimens when compared with nevi and is correlated inversely with melanoma progression. miRNA target databases predicted E2F1 and E2F5 as putative targets. The expression levels of E2F1 and E2F5 were correlated inversely with that of miR-205 in melanoma cell lines. miR-205 significantly suppressed the luciferase activity of reporter plasmids containing the 3′-UTR sequences complementary to either E2F1 or E2F5. Overexpression of miR-205 in melanoma cells reduced E2F1 and E2F5 protein levels. The proliferative capacity of melanoma cells was suppressed by miR-205 and mediated by E2F-regulated AKT phosphorylation. miR-205 overexpression resulted in induction of apoptosis, as evidenced by increased cleaved caspase-3, poly-(ADP-ribose) polymerase, and cytochrome c release. Stable overexpression of miR-205 suppressed melanoma cell proliferation, colony formation, and tumor cell growth in vivo and induced a senescence phenotype accompanied by elevated expression of p16INK4A and other markers for senescence. E2F1 overexpression in miR-205-expressing cells partially reversed the effects on melanoma cell growth and senescence. These results demonstrate a novel role for miR-205 as a tumor suppressor in melanoma.
Keywords: Apoptosis, Cell Cycle, E2F Transcription Factor, MicroRNA, Tumor Suppressor, Melanoma, Senescence
Introduction
MicroRNAs (miRNAs)2 are a class of short noncoding RNAs that regulate gene expression by complementary base pairing with the 3′-UTR of target mRNAs and causing their degradation (1) or by directly mediating mRNA degradation (2). In addition, miRNAs can also inhibit translation in the event of imperfect base pair matching with the target. miRNAs are expressed in a tissue-specific manner and are considered to play important roles in cell proliferation, apoptosis, and differentiation (3, 4).
Deregulation of miRNA expression has been identified in a number of cancers (5, 6), and accumulating data indicate that some miRNAs can function as tumor suppressors or oncogenes and as such are important in cancer development. Specific subsets of miRNAs also have been shown to be dysregulated in various solid tumors (7, 8). The discovery of miRNAs at previously identified chromosomal breakpoints, as well as deletion and amplification sites in certain cancers, implies that they may be involved in disease initiation or progression.
The human genome is predicted to encode as many as 1000 miRNAs (9). Although it is difficult to identify accurately individual miRNA-target interactions, computational predictions of miRNA target genes indicate that as many as one-third of all human protein-encoding genes may be regulated by miRNAs (10). Due to their tremendous regulatory potential and tissue- and disease-specific expression patterns, there is increasing evidence that miRNA expression profiles could be indicative of disease risk or burden. Thus, miRNAs are being assessed as possible biomarkers to aid in the diagnosis and prognosis of different cancers, including melanoma (11, 12). However, to date, very few miRNA expression profiling analyses have been performed on human melanoma samples.
E2F1, the best characterized member of the E2F family, is a master regulator of the G1/S transition phase in the cell cycle that is tightly regulated by the retinoblastoma protein (Rb). E2F1 transactivates a variety of genes involved in chromosomal DNA replication and cell cycle progression (13). Overexpression of E2F1 is an oncogenic event that predisposes cells to transformation (14). E2F1 gene amplification and/or abnormalities in its expression have been reported in various tumor types, including melanoma (15, 16). E2F5, another E2F family member, is an oncogenic cell cycle regulator demonstrated to be amplified or overexpressed in various tumors (17, 18).
In this study, we report that miR-205 is significantly down-regulated in melanoma tumor samples and cell lines. In addition, we examine the consequences of miR-205 overexpression and identify E2F1 and E2F5 as downstream targets of miR-205 action in melanoma.
MATERIALS AND METHODS
Cell Culture, Plasmids, and Transfection
The WM3211, DO4, WM278, and 1205-Lu (kindly provided by Dr. Boris Bastian) and Lox (kindly provided by Dr. Oystein Fodstad) melanoma cell lines were grown in RPMI with 10% fetal bovine serum. C8161.9 cells (obtained from Dr. Danny Welch) were grown in DMEM/F12 with 5% fetal bovine serum (Invitrogen) at 37 °C in an atmosphere containing 5% CO2. Normal human melanocytes were purchased from Lifeline Cell Technology and grown in LL-0027 medium (Lifeline Cell Technology, Walkersville, MD). Plasmids pMax-GFP (Lonza, Walkersville, MD), pMax-E2F1 (Addgene, Cambridge, MA), miRNASelectTM pEP-miR null control vector (pEP Null), and miRNASelectTM pEP-hsa-mir-205 expression vector (pEP miR-205) (Cell Biolabs, Inc., San Diego, CA) were purchased. TaqMan probes for hsa-miR-205 and negative control pre-miR were purchased from Applied Biosystems (Foster City, CA). Transient transfection was carried out by Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Quantitative Real-time PCR
Mature miRNAs and other mRNAs were assayed using the TaqMan MicroRNA Assays and Gene Expression Assays, respectively, in accordance with the manufacturer's instructions (Applied Biosystems). All RT reactions, including no-template controls and RT minus controls, were run in a 7500 Fast Real-time PCR System (Applied Biosystems). RNA concentrations were determined with a NanoDrop (Thermo Scientific, Rockford, IL). Samples were normalized to RNU48 or HPRT (Applied Biosystems), as indicated. Gene expression levels were quantified using 7500 Fast Real-time Sequence detection system Software. Comparative real-time PCR was performed in triplicate, including no-template controls. Relative expression was calculated using the comparative Ct method.
RNA and miRNA Extraction from Tissue Samples and Cell Lines
Samples from patients with primary (n = 20), metastatic melanoma (n = 27), and benign nevi (n = 20) were obtained under a protocol approved by the Institutional Review Board. RNA was extracted by using RNeasy Mini Kit (Qiagen, Valencia, CA) following the manufacturer's protocol. miRNAs were extracted by using the mirVana miRNA extraction kit (Applied Biosystems) from tissues and cell lines following the manufacturer's instructions.
Cell Viability, Colony Formation, and Cell Cycle Analysis
Cells were plated in 96-well plates at a density of 3 × 103 cells per well. Cell viability was assessed at 24, 48, 72, and 96 h post-transfection using Cell Counting Kit-8 (Dojindo, Rockville, MD) following the manufacturer's protocol. For the colony formation assay, 500 cells were plated in a p100 plate and allowed to grow until visible colonies appeared. Colonies were stained with Giemsa and counted. Cell cycle analysis was performed as described previously (19).
Western Blot Analysis
Cell lysates were prepared in PBS containing 1× Halt protease inhibitor mixture and 1× Halt phosphatase inhibitor mixture (Pierce) centrifuged at 3500 rpm for 10 min at 4 °C. Proteins (10–15 ug) from each sample were subjected to SDS-PAGE and transferred onto a nitrocellulose membrane. Target proteins were detected by using specific antibodies against E2F5, PARP, p16INK4a, and GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA), E2F1, AKT, pAKT (Ser-473), pRB (Ser-807/811), MCM3, caspase-9, BAD, pBAD, PCNA, cytochrome c, and cleaved caspase-3 (Cell Signaling Technology, Danvers, MA) and p-caspase-9 (Abcam, Cambridge MA).
Luciferase Assays
The 3′-UTR region of E2F1 and E2F5 containing target site sequences complementary to the seed sequence of miR-205 were cloned downstream of the luciferase gene in the pMIR-REPORT luciferase vector (Ambion, Cambridge, MA), and the resultant vectors were named E2F1–3′-UTR and E2F5–3′-UTR. Mutated 3′-UTR sequences of E2F1 and E2F5 complementary to miR-205 were cloned in the same vector, and the resultant vectors were named E2F1-Mut 3′-UTR and E2F5-Mut 3′-UTR, respectively. For reporter assays, cells were transiently transfected with wild-type or mutant reporter plasmid and miR-205. Firefly luciferase activities were measured by using the Dual-Luciferase Assay (Promega, Madison, WI) 48 h after transfection, and the results were normalized with Renilla luciferase. Each reporter plasmid was transfected at least three times (on different days), and each sample was assayed in triplicate.
Stable Cell Generation and in Vivo Study
C8161.9 cells were transfected with pEP Null and pEP miR-205 vectors (Cell Biolabs, San Diego, CA) and selected with puromycin (1 μg/ml). For in vivo studies as described previously (20), 1 × 106 cells were injected into nude mice subcutaneously, and tumor growth was followed for 28 days. All animal care was in accordance with the institutional guidelines.
SA-β-gal Staining
Staining for senescence-associated β-galactosidase (SA-β-gal) was performed using the Senescence Cell Histochemical Staining Kit from Sigma as per the manufacturer's protocol. In brief, cells were fixed with formalin, washed with PBS, and incubated at 37° C with staining mixture until the cells were stained blue. The percentage of SA-β-gal-positive cells was determined by counting the number of stained cells with bright field illumination.
Statistical Analysis
All quantified data represent an average of at least triplicate samples or as indicated. Error bars represent S.E. Statistical significance was determined by the Student's t test, and two-tailed p values <0.05 were considered significant.
RESULTS
miR-205 Is Down-regulated in Melanoma, and Its Expression Is Inversely Correlated with That of E2F1 and E2F5
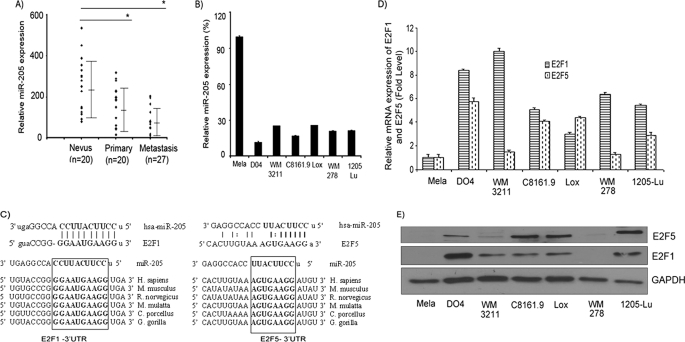
To determine the expression pattern of miRNAs in melanoma, we performed a miRNA microarray on a small number of nevi, primary, and metastatic tumor samples (n = 5 per group) using the Agilent platform. In this analysis, miR-205 emerged as the miRNA with the highest degree of down-regulation in melanoma metastases (data not shown), and its expression showed a significant decline from nevus to primary to metastatic tumor samples (mean values of 8.2, 6.8, and 0.8 for nevus, primary, and metastatic samples, respectively). We aimed to validate the microarray data by miRNA quantitative RT-PCR (miR qRT-PCR) analysis on an independent cohort of nevus and melanoma tissues. miR qRT-PCR of nevus (n = 20), primary (n = 20), and metastatic (n = 27) samples indicated that miR-205 expression is down-regulated significantly in primary and metastatic samples when compared with nevi (Fig. 1A). A similar pattern of reduced expression from primary to metastatic tumor samples was observed. Overall, this analysis demonstrated the utility of miR-205 as a tumor progression marker in melanoma. We then determined the expression levels of miR-205 in a panel of human melanoma cell lines and normal melanocytes. Our results indicate a significant down-regulation in expression of miR-205 in melanoma cells as compared with normal melanocytes (Fig. 1B), thereby suggesting a potential tumor suppressor role for miR-205 in melanoma. To identify the effectors of miR-205, we used different algorithms that predict the mRNA targets of a miRNA: miRanda (21), microRNA target predictions (22), TargetScan (10), and PicTar (23). Among the list of potential targets of miR-205 were the mRNAs encoding E2F1 and E2F5. The seed sequence of miR-205 was complementary to the 3′-UTR of these genes and was conserved highly among six different species (Fig. 1C). To investigate the correlation between expression of miR-205 and that of E2F1 and E2F5, we determined expression of E2F1 and E2F5 at the mRNA and protein levels in the same panel of cell lines. The expression levels of E2F1 and E2F5 mRNA and protein were higher when compared with the normal melanocyte line (Fig. 1, D and E), although the absolute level of expression varied among different melanoma cell lines. These data demonstrated an inverse correlation between the expression of miR-205 and that of E2F1 and E2F5 in melanoma, supporting a potential role for E2F1 and E2F5 as targets of miR-205.
FIGURE 1.
miR-205 expression is down-regulated in melanoma and is inversely correlated with expression of E2F1 and E2F5. A, miRNA quantitative RT-PCR analysis of miR-205 expression level in a cohort of nevus and melanoma tissues. B, miR-205 expression in a panel of human melanoma cells and normal melanocytes (Mela). C, the miR-205 seed sequence is complementary to the 3′-UTR of E2F1 and E2F5 and is conserved in six different species. D and E, E2F1 and E2F5 expression at the mRNA and protein levels in different human melanoma cell lines and normal melanocytes. *, p < 0.05.
3′-UTR of E2F1 and E2F5 Are Direct Targets of miR-205
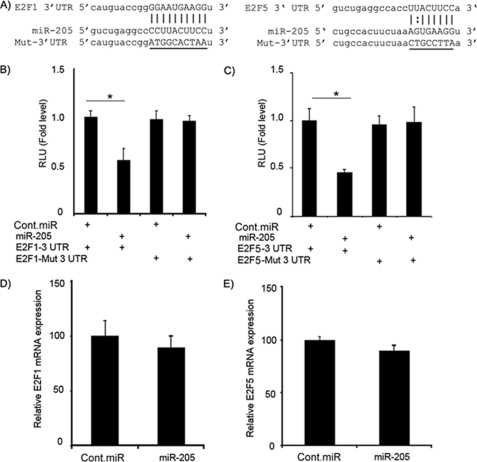
Next, we investigated whether the 3′-UTR of E2F1 and E2F5 are functional targets of miR-205. We cloned the 3′-UTR of E2F1 and E2F5 harboring the complementary sequence to the miR-205 seed sequence in a reporter plasmid vector. In a parallel experiment, the 3′-UTR of E2F1 and E2F5 complementary to the miR-205 seed sequence was mutated and cloned in the same reporter plasmid (Fig. 2A). Transient transfection of human C8161.9 melanoma cells with the E2F1-3′-UTR or E2F5-3′-UTR construct along with miR-205 led to a significant decrease in reporter expression when compared with the control vector (Fig. 2, B and C). The luciferase activity of the reporter vector containing a mutated 3′-UTR of E2F1 or E2F5 was unaffected by a simultaneous transfection with miR-205 (Fig. 2, B and C). These results indicated that the conserved nucleotides in the 3′-UTR of E2F1 and E2F5 were responsible for miR-205 targeting in vitro. Taken together, these results demonstrate E2F1 and E2F5 as targets of miR-205 action in melanoma. Transient transfection of miR-205 had no significant effect on E2F1 and E2F5 mRNA levels (Fig. 2, D and E).
FIGURE 2.
E2F1 and E2F5 3′-UTRs are targets of miR-205. A, the 3′-UTR of E2F1 or E2F5 and mutant 3′-UTR sequences that abolished binding. B and C, luciferase assay showing decreases in reporter activity after co-transfection of either E2F1–3′-UTR or E2F5–3′-UTR with miR-205 in C8161.9 cells. The mutant UTRs of either E2F1 or E2F5 had no effect on reporter activity. D and E, relative mRNA expression levels of E2F1 and E2F5 after transfection with miR-205 or negative control pre-miR. Each experiment was performed in triplicate. *, p < 0.05. Cont.miR, negative control pre-miR.
miR-205 Suppresses E2F1 and E2F5 and Negatively Regulates the AKT Pathway in Melanoma Cell Lines
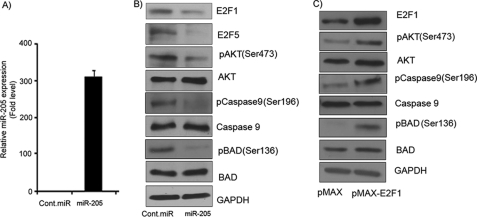
We then sought to determine whether the overexpression of miR-205 in melanoma cell lines can regulate E2F1 and E2F5 protein levels and alter downstream signaling events. C8161.9 cells were transfected with miR-205, resulting in miR-205 overexpression as determined by miR qRT-PCR analysis (Fig. 3A). Western blot analysis confirmed the down-regulation of E2F1 and E2F5 at the protein level following miR-205 overexpression (Fig. 3B). These results support the notion that miR-205 binds to the 3′-UTR of these genes and regulates their expression at the protein level. Expression of E2F3, another putative target of miR-205, was unaffected by miR-205 overexpression (data not shown). As E2F1 is reported to be the most abundant E2F family member in melanoma (24), we further analyzed its role in the response to miR-205 overexpression. E2F1 has been reported to activate the AKT pathway and transduce a proliferative signal (25). To determine whether the AKT pathway is affected by miR-205-mediated suppression of E2F1, C8161.9 cells were transfected with miR-205 or negative control pre-miR. Western blot analysis showed reduced levels of phosphorylated AKT (Ser-473) (Fig. 3B) in cells with suppressed E2F1 expression following miR-205 overexpression. AKT plays an important role in cell proliferation and is known to inhibit apoptosis by phosphorylating and inactivating apoptotic factors such as BAD and caspase-9 (26). As shown in Fig. 3B, BAD phosphorylation at Ser-136 and caspase-9 phosphorylation at Ser-196 are decreased significantly in miR-205-overexpressing cells in which both E2F1 protein levels and AKT phosphorylation are markedly reduced. We next overexpressed E2F1 in melanoma cell lines to determine whether its overexpression restores the down-regulation of phosphorylated AKT. Overexpression of E2F1 rescued miR-205-induced down-regulation of AKT, BAD, and caspase-9 (Fig. 3C). These data indicate that miR-205 targets E2F1, which in turn results in suppression of the AKT pathway and activation of a proapoptotic signaling cascade.
FIGURE 3.
miR-205 suppresses E2F1 and E2F5 expression at the protein level and regulates the AKT pathway. A, relative miR-205 expression level in C8161.9 cells after transfection with miR-205 as determined by miR qRT-PCR. B, Western blot analysis showing decreases in the phosphorylation levels of AKT, BAD, and caspase-9 in C8161.9 cells after miR-205 transfection. C, Western blot analysis showing up-regulation in the phosphorylation levels of AKT, BAD, and caspase-9 after E2F1 overexpression in C8161.9 cells. Cont.miR, negative control pre-miR.
miR-205 Inhibits Melanoma Cell Proliferation and Colony Formation
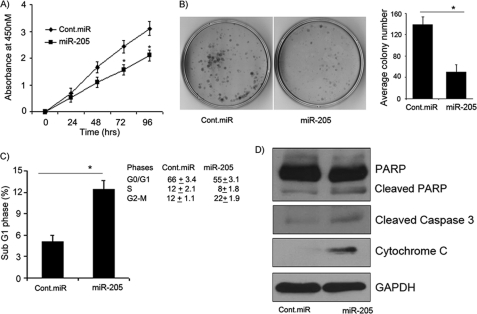
To determine whether activation of the proapoptotic cascade induced by miR-205 overexpression affects cell proliferation, melanoma cells were transfected transiently with miR-205. A significant decrease in cell proliferation was observed over time in C8161.9 cells expressing miR-205 (Fig. 4A) as compared with cells expressing negative control pre-miR. We further examined the effects of miR-205 on melanoma cell viability using a colony formation assay. The miR-205-transfected C8161.9 cells showed low colony formation ability, as both the size and number of foci in miR-205-expressing cells were suppressed when compared with negative control pre-miR expressing cells (Fig. 4B). These results indicate that suppression of the AKT pathway mediated by miR-205 is accompanied by reduced melanoma cell proliferation and survival.
FIGURE 4.
miR-205 inhibits melanoma cell proliferation and colony formation, and induces apoptosis. A, the proliferative ability of C8161.9 cells after miR-205 transfection is reduced significantly as compared with negative control pre-miR (Cont.miR). B, miR-205 overexpression significantly inhibits the colony formation ability of melanoma cells. C, cell cycle analysis showing increase in the sub-G1 phase of C8161.9 cells overexpressing miR-205. D, Western blot analysis showing increases in the cleavage of caspase-3 and PARP and increased release of cytochrome c in miR-205-transfected cells. *, p < 0.05 (experiments depicted in A–C performed in triplicate).
E2F1 Suppression by miR-205 Induces Apoptosis in Melanoma Cell Lines
To determine whether miR-205 overexpression causes apoptosis, melanoma cells were transfected with miR-205 or negative control pre-miR. As shown in Fig. 4C, a significant increase in the sub-G1 phase was observed in C8161.9 cells transfected with miR-205 after 72 h, suggesting activation of apoptosis upon miR-205 overexpression. We also observed cell cycle arrest at the G2-M phase in miR-205-transfected cells (Fig. 4C). These results are supported by our observation of activation of BAD and caspase-9 following down-regulation of E2F1 by miR-205. Activation of BAD has been shown to initiate apoptosis by causing cytochrome c release into the cytoplasm after binding to Bcl-xL and Bcl-2 (27). Accordingly, cytochrome c expression was up-regulated in C8161.9 cells expressing miR-205 when compared with negative control pre-miR-expressing cells, accompanied by significant cleavage of procaspase-3. Finally, there was an increased proportion of cleaved PARP in relation to caspase-3 cleavage, indicating that the observed miR-205-induced apoptosis is also dependent on caspase-3 (Fig. 4D). Taken together, these data strongly suggest that E2F1-mediated suppression of AKT by miR-205 plays an important role in inducing melanoma cell apoptosis.
To confirm the effect of miR-205 on melanoma cell proliferation and apoptosis, miR-205 was transfected into Lox human melanoma cells. As shown in supplemental Fig. 1, A–C, a significant decrease in cell proliferation and colony formation, along with an increase in the apoptotic index, was observed in Lox cells transfected with miR-205. These results confirm the phenotypic effects of miR-205 overexpression in human melanoma cells.
Stable Overexpression of miR-205 Inhibits Cell Survival, Colony Formation, and in Vivo Tumor Cell Growth
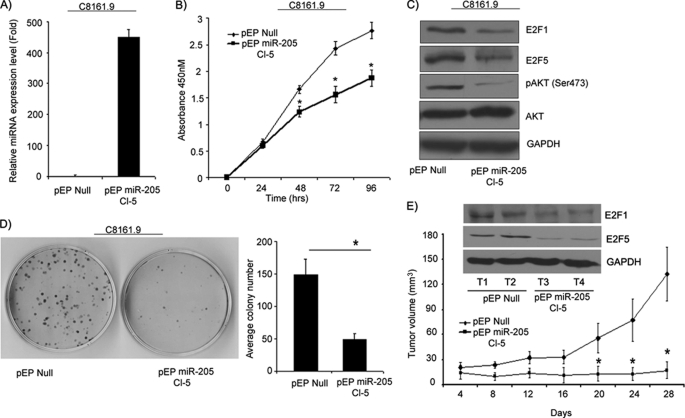
Next, we examined the effects of stable expression of miR-205 in melanoma. C8161.9 cells stably expressing miR-205 were generated, and overexpression of miR-205 was confirmed by miR qRT-PCR analysis (Fig. 5A). C8161.9 cells expressing miR-205 exhibited a significant suppression in cell proliferation as compared with control vector-expressing cells (Fig. 5B). We observed a marked reduction in the expression of E2F1 and in AKT phosphorylation (Fig. 5C), as was observed with transient transfection of miR-205. A significant decrease in the colony formation capability of miR-205-overexpressing cells was observed as compared with control vector-expressing cells (Fig. 5D). Stable overexpression of miR-205 dramatically suppressed tumor growth in vivo upon subcutaneous injection into nude mice when compared with cells expressing control vector (Fig. 5E). E2F1 and E2F5 were suppressed at the protein level in miR-205-overexpressing tumors when compared with control tumors (Fig. 5E).
FIGURE 5.
Stable overexpression of miR-205 inhibits cell proliferation in vitro and in vivo. A, relative miR-205 expression levels in C8161.9 cells stably expressing miR-205 as determined by miR qRT-PCR. B, stable overexpression of miR-205 in melanoma cell lines significantly suppressed cell proliferation. C, Western blot analysis showing suppression in E2F1, E2F5, and AKT phosphorylation levels. D, colony formation ability is significantly reduced by miR-205. E, tumor volume following subcutaneous injection of C8161.9 cells expressing miR-205 was reduced significantly (n = 10 mice per group). Western blot showing expression of E2F1 and E2F5 from tumors from pEP-Null and pEP-miR-205 mice.*, p < 0.05.
miR-205 Overexpression Induces Senescence
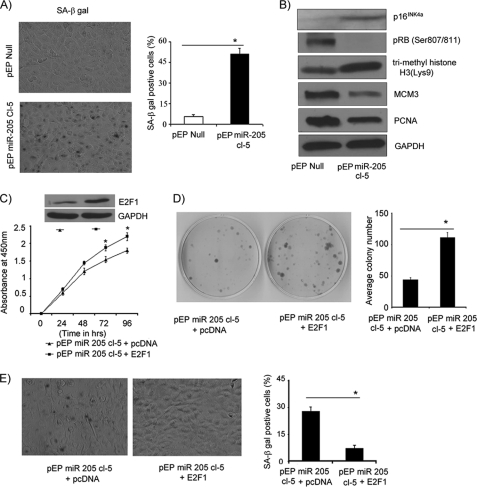
An unexpected phenotypic change, namely cell enlargement and vacuolization, observed in miR-205-expressing C8161.9 cells, prompted us to explore the possibility that overexpression of miR-205 can effectively activate senescence pathways. As shown in Fig. 6, A and B, miR-205-expressing C8161.9 cells exhibited significantly increased staining for SA-β-gal activity when compared with control vector-expressing cells. Thus, 52% of C8161.9 cells stably expressing miR-205 expressed this senescence marker, when compared with 8% of control vector-expressing cells. Overexpression of miR-205 also induced a marked increase in expression of p16INK4a and trimethyl-histone H3 (Lys-9), suggesting activation of both classical senescence as well as alternate pathways that involve heterochromatin-associated histone modification (Fig. 6B). Hypophosphorylation of Rb (Ser-807/811) and down-regulation of key downstream targets of E2F1, MCM3 and PCNA, further reinforces the activation of Rb-mediated senescence (28) upon overexpressing miR-205 in this melanoma cell line (Fig. 6B).
FIGURE 6.
miR-205 overexpression induces senescence that is reversed by E2F1. A, senescence associated-β-galactosidase activity was increased significantly in cells overexpressing miR-205 as compared with cells expressing control vector. The bar graph shows quantification of stained cells. *, p < 0.05. B, Western blot analysis showing up-regulation in p16INK4a, down-regulation in Rb phosphorylation (Ser-807/811), MCM3 and PCNA in miR-205-expressing cells. *, p < 0.05. C, E2F1 overexpression in miR-205-expressing cells significantly increased cell proliferation. Western blot showing expression of E2F1 in miR-205-expressing cells. D, colony formation assay of melanoma cells expressing miR-205 following transfection with plasmid encoding vector only or vector expressing E2F1 cDNA. E, senescence associated-β-galactosidase activity was decreased in miR-205-expressing cells following transfection with vector expressing E2F1 cDNA. *, p < 0.05. (Experiments depicted in A, C, D, and E were performed in triplicate.).
E2F1 Overexpression Rescues Phenotypes Induced by miR-205
Overexpression of E2F1 in C8161.9 cells stably expressing miR-205 resulted in significantly increased cell proliferation and colony formation (Fig. 6, C and D), indicating reversal of the inhibitory effects of miR-205 on melanoma cell growth. E2F1 overexpression also resulted in reversion of the senescence induced by miR-205 in melanoma cells (Fig. 6E).
DISCUSSION
As miRNAs modulate gene expression, it is not surprising that they have been implicated in regulating a wide variety of biological processes. Various studies have characterized the modulation of miRNA expression in cancers and identified a number of miRNAs that are up- or down-regulated in tumors (29, 30). An increasing number of oncogenes or tumor suppressor genes have now been identified as targets of aberrantly expressed miRNAs (31, 32). For example, miR-21 has been shown to be involved in invasion and metastasis in many cancer types by its action on numerous genes involved in extracellular matrix modification (33). In addition, the miR-200 family, along with miR-205, has been shown to regulate epithelial to mesenchymal transition, a key process for initiating metastasis (34).
Despite these advances, little is known about the repertoire and function of miRNAs in melanoma, and few targets of miRNAs in melanoma have been identified. A handful of studies have assessed the levels of various miRNAs through microarray expression profiling (30, 35) or have studied the mechanisms of action of selected miRNAs in this tumor type (36). For example, miR-221 and miR-222 have been implicated in melanoma progression through the regulation of p27 expression (37). Aberrant expression of miR-182 promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor (36). Finally, miR-532–3p has been shown to regulate RUNX3 in cutaneous melanoma (38). In the present study, we examined the expression pattern and functional significance of miR-205 in melanoma progression. Expression of miR-205 in cancer is controversial because it has been found to be either up-regulated (39) or down-regulated (40) in tumors. However, we found miR-205 to be significantly down-regulated in metastatic melanomas when compared with primary tumors or nevi. The reduced levels of miR-205 in metastatic samples in an independent tissue set confirmed its role as a tumor progression marker in melanoma. The down-regulation of miR-205 expression was also shown in melanoma cell lines when compared with normal melanocytes. This is consistent with a recent microarray analysis of melanoma cell lines and tissues that also reported down-regulation of miR-205 in melanoma (35). The significant suppression of miR-205 expression in metastatic tumors and melanoma cell lines suggests a tumor suppressor role in melanoma. Intriguingly, the gene encoding miR-205 is located on 1q32, a locus that is reported to be lost in melanoma and that harbors the known melanoma tumor suppressor genes KISS1 (41) and mda-7 (42), thus providing a possible mechanism for down-regulation of miR-205 expression observed here.
To determine targets of miR-205 action, in silico algorithms were utilized to identify E2F1 and E2F5. The E2F family of transcription factors plays an important role in the regulation of cellular proliferation and cell cycle progression (43). Our results indicate an inverse correlation between expression of miR-205 and that of E2F1 and E2F5 in a panel of melanoma cell lines. We demonstrated that miR-205 directly targets the 3′-UTR of E2F1 and E2F5, as its overexpression was associated with suppression of luciferase activity. In addition, a significant down-regulation of E2F1 and E2F5 protein (but not RNA) levels was observed after miR-205 overexpression, indicating the post-transcriptional regulation of E2F1 and E2F5 via targeting of their 3′-UTRs.
Due to the demonstrated importance of E2F1 in melanoma (24), we further characterized its role in response to miR-205. E2F1 has long been considered an oncoprotein, and shown to promote breast cancer cell and hepatocarcinoma cell proliferation (44). However, E2F1 also has been reported to either induce or inhibit apoptosis (45). E2F1-directed prosurvival signals are mediated by the AKT pathway, which plays an important role in regulating cell growth, survival, and inhibition of apoptosis (46). Recently, E2F1-epidermal growth factor receptor interaction was reported to play a significant role in melanoma progression (47). We observed that miR-205 overexpression reduced AKT phosphorylation, which was restored by E2F1 overexpression, confirming that the miR-205-mediated down-regulation of AKT phosphorylation was due to E2F1. miR-205 overexpression suppressed the phosphorylation of caspase-9 and BAD, thus initiating a caspase cascade to induce apoptosis. Our results indicate that miR-205 inhibited cell proliferation and induced apoptosis in melanoma cells, an observation that was confirmed in two different human melanoma cell lines. miR-205-mediated induction of apoptosis was found to be associated concomitantly with the cleavage of caspase-3 and PARP and the release of cytochrome c. These antiproliferative effects of miR-205, mediated, at least in part, by suppression of E2F1 and AKT phosphorylation, were confirmed following stable overexpression of miR-205 in C8161.9 cells. In addition, in vivo studies demonstrated a striking reduction in subcutaneous tumor cell growth in mice injected with C8161.9 melanoma cells overexpressing miR-205.
Furthermore, stable overexpression of miR-205 induced senescence in melanoma cells. C8161.9 cells overexpressing miR-205 had increased SA-β-gal activity, as well as cell enlargement and vacuolization, which represent conventional markers for senescence (48). Trimethyl-histone H3 (Lys-9), another marker of senescence at the chromatin level, was up-regulated following miR-205 overexpression. The p16INK4a-Rb tumor suppressor pathway plays an important role in the initiation and maintenance of senescence (49). Up-regulation of p16INK4a activates Rb by suppressing its phosphorylation through the inhibition of CDK4/6 kinase activities (50). Rb activation induces senescence and suppresses the expression of E2F1 gene targets such as MCM3 and PCNA in senescent cells (28). Our results suggest that miR-205-induced senescence is mediated, at least in part, by this pathway, as evidenced by the up-regulation of p16INK4a and the suppression of Rb phosphorylation. In addition, both MCM3 and PCNA were down-regulated in miR-205-overexpressing melanoma cells. Our study, for the first time, implicates miR-205 in the senescence of human melanoma cells.
Importantly, the significant effects induced by miR-205 overexpression on melanoma cell growth and senescence were reversed partially following restoration of E2F1 expression. These mechanistic analyses indicate that miR-205 mediates its effects on melanoma cell proliferation and senescence via regulation of E2F1 expression.
Finally, these results help explain previously described differential patterns of Rb and E2F1 activity in melanoma progression. Rb has been reported to be constitutively inactive in metastatic melanoma cells (24), whereas E2F1 and its gene targets are overexpressed in metastases when compared with primary melanomas (51). Our data is consistent with these findings and suggests that the down-regulation of miR-205 in metastatic melanomas may be responsible for the previously observed activation of E2F1 and inactivation of Rb.
In conclusion, our study demonstrates a potent and important tumor suppressor role for miR-205 in melanoma.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health, United States Public Health Service Grants CA114337 and CA122947 (to M. K.-S.). This work was also supported by the Herschel and Diana Zackheim Endowment Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- miRNA
- microRNA
- qRT-PCR
- quantitative RT-PCR
- PCNA
- proliferating cell nuclear antigen
- SA-β-gal
- senescence-associated β-galactosidase
- PARP
- poly-(ADP-ribose) polymerase
- Rb
- retinoblastoma protein
- AKT
- v-akt murine thymoma viral oncogene homolog
- HPRT
- hypoxanthine phosphoribosyltransferase 1
- MCM3
- minichromosome maintenance complex component 3.
REFERENCES
- 1. Ambros V., Chen X. (2007) Development 134, 1635–1641 [DOI] [PubMed] [Google Scholar]
- 2. He L., Hannon G. J. (2004) Nat. Rev. Genet. 5, 522–531 [DOI] [PubMed] [Google Scholar]
- 3. Sempere L. F., Freemantle S., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V. (2004) Genome Biol. 5, R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 5. Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Porkka K. P., Pfeiffer M. J., Waltering K. K., Vessella R. L., Tammela T. L., Visakorpi T. (2007) Cancer Res. 67, 6130–6135 [DOI] [PubMed] [Google Scholar]
- 7. Michael M. Z., O' Connor S. M., van Holst, Pellekaan N. G., Young G. P., James R. J. (2003) Mol. Cancer Res. 1, 882–891 [PubMed] [Google Scholar]
- 8. Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., Yi M., Stephens R. M., Okamoto A., Yokota J., Tanaka T., Calin G. A., Liu C. G., Croce C. M., Harris C. C. (2006) Cancer Cell 9, 189–198 [DOI] [PubMed] [Google Scholar]
- 9. Berezikov E., Guryev V., van de Belt J., Wienholds E., Plasterk R. H., Cuppen E. (2005) Cell 120, 21–24 [DOI] [PubMed] [Google Scholar]
- 10. Lewis B. P., Burge C. B., Bartel D. P. (2005) Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 11. Yi R., Fuchs E. (2010) Cell Death Differ 17, 229–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bartels C. L., Tsongalis G. J. (2009) Clin. Chem. 55, 623–631 [DOI] [PubMed] [Google Scholar]
- 13. DeGregori J. (2002) Biochim. Biophys. Acta 1602, 131–150 [DOI] [PubMed] [Google Scholar]
- 14. Pierce A. M., Schneider-Broussard R., Gimenez-Conti I. B., Russell J. L., Conti C. J., Johnson D. G. (1999) Mol. Cell. Biol. 19, 6408–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Saito M., Helin K., Valentine M. B., Griffith B. B., Willman C. L., Harlow E., Look A. T. (1995) Genomics 25, 130–138 [DOI] [PubMed] [Google Scholar]
- 16. Nelson M. A., Reynolds S. H., Rao U. N., Goulet A. C., Feng Y., Beas A., Honchak B., Averill J., Lowry D. T., Senft J. R., Jefferson A. M., Johnson R. C., Sargent L. M. (2006) Cancer Biol. Ther. 5, 407–412 [DOI] [PubMed] [Google Scholar]
- 17. Kothandaraman N., Bajic V. B., Brendan P. N., Huak C. Y., Keow P. B., Razvi K., Salto-Tellez M., Choolani M. (2010) BMC Cancer 10, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Umemura S., Shirane M., Takekoshi S., Kusakabe T., Itoh J., Egashira N., Tokuda Y., Mori K., Osamura Y. R. (2009) Br. J. Cancer 100, 764–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dar A. A., Zaika A., Piazuelo M. B., Correa P., Koyama T., Belkhiri A., Washington K., Castells A., Pera M., El-Rifai W. (2008) Cancer 112, 1688–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dar A. A., Majid S., Nosrati M., de Semir D., Federman S., Kashani-Sabet M. (2010) J. Invest. Dermatol. 130, 2071–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. John B., Enright A. J., Aravin A., Tuschl T., Sander C., Marks D. S. (2004) PLoS Biol. 2, e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Betel D., Wilson M., Gabow A., Marks D. S., Sander C. (2008) Nucleic Acids Res. 36, D149–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. (2005) Nat. Genet. 37, 495–500 [DOI] [PubMed] [Google Scholar]
- 24. Halaban R., Cheng E., Smicun Y., Germino J. (2000) J. Exp. Med. 191, 1005–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ladu S., Calvisi D. F., Conner E. A., Farina M., Factor V. M., Thorgeirsson S. S. (2008) Gastroenterology 135, 1322–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Datta S. R., Dudek H., Tao X., Masters S., Fu H., Gotoh Y., Greenberg M. E. (1997) Cell 91, 231–241 [DOI] [PubMed] [Google Scholar]
- 27. Guo X., Chen K. H., Guo Y., Liao H., Tang J., Xiao R. P. (2007) Circ. Res. 101, 1113–1122 [DOI] [PubMed] [Google Scholar]
- 28. Narita M., Nũnez S., Heard E., Narita M., Lin A. W., Hearn S. A., Spector D. L., Hannon G. J., Lowe S. W. (2003) Cell 113, 703–716 [DOI] [PubMed] [Google Scholar]
- 29. Jiang J., Lee E. J., Gusev Y., Schmittgen T. D. (2005) Nucleic Acids Res. 33, 5394–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gaur A., Jewell D. A., Liang Y., Ridzon D., Moore J. H., Chen C., Ambros V. R., Israel M. A. (2007) Cancer Res. 67, 2456–2468 [DOI] [PubMed] [Google Scholar]
- 31. Harfe B. D. (2005) Curr. Opin. Genet. Dev. 15, 410–415 [DOI] [PubMed] [Google Scholar]
- 32. Tong A. W., Nemunaitis J. (2008) Cancer Gene Ther. 15, 341–355 [DOI] [PubMed] [Google Scholar]
- 33. Nicoloso M. S., Spizzo R., Shimizu M., Rossi S., Calin G. A. (2009) Nat. Rev. Cancer 9, 293–302 [DOI] [PubMed] [Google Scholar]
- 34. Gregory P. A., Bracken C. P., Bert A. G., Goodall G. J. (2008) Cell Cycle 7, 3112–3118 [DOI] [PubMed] [Google Scholar]
- 35. Philippidou D., Schmitt M., Moser D., Margue C., Nazarov P. V., Muller A., Vallar L., Nashan D., Behrmann I., Kreis S. (2010) Cancer Res. 70, 4163–4173 [DOI] [PubMed] [Google Scholar]
- 36. Segura M. F., Hanniford D., Menendez S., Reavie L., Zou X., Alvarez-Diaz S., Zakrzewski J., Blochin E., Rose A., Bogunovic D., Polsky D., Wei J., Lee P., Belitskaya-Levy I., Bhardwaj N., Osman I., Hernando E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1814–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Felicetti F., Errico M. C., Bottero L., Segnalini P., Stoppacciaro A., Biffoni M., Felli N., Mattia G., Petrini M., Colombo M. P., Peschle C., Carè A. (2008) Cancer Res. 68, 2745–2754 [DOI] [PubMed] [Google Scholar]
- 38. Kitago M., Martinez S. R., Nakamura T., Sim M. S., Hoon D. S. (2009) Clin. Cancer Res. 15, 2988–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gottardo F., Liu C. G., Ferracin M., Calin G. A., Fassan M., Bassi P., Sevignani C., Byrne D., Negrini M., Pagano F., Gomella L. G., Croce C. M., Baffa R. (2007) Urol. Oncol. 25, 387–392 [DOI] [PubMed] [Google Scholar]
- 40. Sempere L. F., Christensen M., Silahtaroglu A., Bak M., Heath C. V., Schwartz G., Wells W., Kauppinen S., Cole C. N. (2007) Cancer Res. 67, 11612–11620 [DOI] [PubMed] [Google Scholar]
- 41. Lee J. H., Miele M. E., Hicks D. J., Phillips K. K., Trent J. M., Weissman B. E., Welch D. R. (1996) J. Natl. Cancer Inst. 88, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 42. Huang E. Y., Madireddi M. T., Gopalkrishnan R. V., Leszczyniecka M., Su Z., Lebedeva I. V., Kang D., Jiang H., Lin J. J., Alexandre D., Chen Y., Vozhilla N., Mei M. X., Christiansen K. A., Sivo F., Goldstein N. I., Mhashilkar A. B., Chada S., Huberman E., Pestka S., Fisher P. B. (2001) Oncogene 20, 7051–7063 [DOI] [PubMed] [Google Scholar]
- 43. Attwooll C., Lazzerini Denchi E., Helin K. (2004) EMBO J. 23, 4709–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Louie M. C., Zou J. X., Rabinovich A., Chen H. W. (2004) Mol. Cell. Biol. 24, 5157–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jiang Y., Saavedra H. I., Holloway M. P., Leone G., Altura R. A. (2004) J. Biol. Chem. 279, 40511–40520 [DOI] [PubMed] [Google Scholar]
- 46. Thomas G. V., Horvath S., Smith B. L., Crosby K., Lebel L. A., Schrage M., Said J., De Kernion J., Reiter R. E., Sawyers C. L. (2004) Clin. Cancer Res. 10, 8351–8356 [DOI] [PubMed] [Google Scholar]
- 47. Alla V., Engelmann D., Niemetz A., Pahnke J., Schmidt A., Kunz M., Emmrich S., Steder M., Koczan D., Pützer B. M. (2010) J. Natl. Cancer Inst. 102, 127–133 [DOI] [PubMed] [Google Scholar]
- 48. Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A. J., Barradas M., Benguría A., Zaballos A., Flores J. M., Barbacid M., Beach D., Serrano M. (2005) Nature 436, 642. [DOI] [PubMed] [Google Scholar]
- 49. Haferkamp S., Becker T. M., Scurr L. L., Kefford R. F., Rizos H. (2008) Aging Cell 7, 733–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Serrano M., Hannon G. J., Beach D. (1993) Nature 366, 704–707 [DOI] [PubMed] [Google Scholar]
- 51. Tuve S., Wagner S. N., Schittek B., Pützer B. M. (2004) Int. J. Cancer 108, 162–166 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.