Abstract
Death receptors (DRs) induce apoptosis but also stimulate proinflammatory “non-apoptotic” signaling (e.g. NF-κB and mitogen-activated protein kinase (MAPK) activation) and inhibit distinct steps of DR-activated maturation of procaspase-8. To examine whether isoforms of cellular FLIP (cFLIP) or its cleavage products differentially regulate DR signaling, we established HaCaT cells expressing cFLIPS, cFLIPL, or mutants of cFLIPL (cFLIPD376N and cFLIPp43). cFLIP variants blocked TRAIL- and CD95L-induced apoptosis, but the cleavage pattern of caspase-8 in the death inducing signaling complex was different: cFLIPL induced processing of caspase-8 to the p43/41 fragments irrespective of cFLIP cleavage. cFLIPS or cFLIPp43 blocked procaspase-8 cleavage. Analyzing non-apoptotic signaling pathways, we found that TRAIL and CD95L activate JNK and p38 within 15 min. cFLIP variants and different caspase inhibitors blocked late death ligand-induced JNK or p38 MAPK activation suggesting that these responses are secondary to cell death. cFLIP isoforms/mutants also blocked death ligand-mediated gene induction of CXCL-8 (IL-8). Knockdown of caspase-8 fully suppressed apoptotic and non-apoptotic signaling. Knockdown of cFLIP isoforms in primary human keratinocytes enhanced CD95L- and TRAIL-induced NF-κB activation, and JNK and p38 activation, underscoring the regulatory role of cFLIP for these DR-mediated signals. Whereas the presence of caspase-8 is critical for apoptotic and non-apoptotic signaling, cFLIP isoforms are potent inhibitors of TRAIL- and CD95L-induced apoptosis, NF-κB activation, and the late JNK and p38 MAPK activation. cFLIP-mediated inhibition of CD95 and TRAIL DR could be of crucial importance during keratinocyte skin carcinogenesis and for the activation of innate and/or adaptive immune responses triggered by DR activation in the skin.
Keywords: Apoptosis, Cell Surface Receptor, Death Domain, Membrane Proteins, Protease Inhibitor, Tumor Necrosis Factor (TNF), CD95L, TRAIL, cFLIP, Gene Induction
Introduction
Death receptors (DR)3 activate the “extrinsic” apoptotic signaling pathway and are crucial regulators of cellular homeostasis during embryogenesis, autoimmunity, and cancer development (1). Initiation of cell death by DRs such as CD95, TRAIL-R1, and TRAIL-R2 requires recruitment of the adapter protein FADD to the death-inducing signaling complex (DISC). FADD in turn mediates recruitment and activation of initiator caspases such as caspase-8 or caspase-10 in the DISC (for review, see Refs. 2 and 3). Due to the potential deleterious effect of deregulated apoptosis and the ubiquitous expression of DRs these processes have to be tightly regulated. Anti-apoptotic molecules able to block initiator caspases are crucial regulators and are widely expressed in mammals (4, 5). In particular, different isoforms of the cellular FLICE-inhibitory protein (cFLIP) have been described and studied as inhibitors of DISC-associated caspase-8 activation (for review, see Refs. 6–8). cFLIP isoforms inhibit the most membrane-proximal steps of DR-mediated apoptosis at the DISC. Two isoforms of cFLIP are commonly detected in human cells: a long form (cFLIPL) and a short form (cFLIPS). cFLIPL, a 55-kDa protein, contains two death effector domains (DEDs) and an inactive caspase-like domain, which resembles caspase-8 in structure. In contrast, cFLIPS, a 26-kDa protein, consists of two DEDs and has several viral homologues (9). Both isoforms of cFLIP are recruited to the DISC, form heterodimers with procaspase-8 and prevent its activation. Complicating the signaling capabilities of cFLIP, the caspase-8/cFLIP heterodimer has enzymatic activity (10). Each cFLIP isoform has a specific mechanism of action: whereas cFLIPS blocks caspase-8 cleavage by preventing the initial cleavage step of procaspase-8, cFLIPL permits this initial processing step but inhibits further maturation resulting in the generation of an intermediary p43 fragment (11, 12). Thus, cFLIPL does not interfere with the enzymatic activity within the DISC, but rather interferes with the release of active caspase-8 from the DISC (12) suggesting a more complex picture for the physiological role of cFLIP proteins beyond apoptosis protection.
In addition to apoptosis, activation of the transcription factor NF-κB, the PKB/Akt pathway, and mitogen-activated protein kinases (MAPK) such as c-Jun N-terminal kinase (JNK), ERK, and p38 can also be induced by DRs (e.g. hereafter referred to as “non-apoptotic” DR signals; for review, see Ref. 2). Thus it appears plausible that cFLIP proteins are involved in the regulation of these DR-associated non-apoptotic pathways. Emerging evidence suggests that these DR-induced non-apoptotic signals initiate a number of different cellular responses rather than simply modulating apoptosis sensitivity. For example, activation of MAPK as well as NF-κB signaling pathways by DRs in the skin have been demonstrated (13, 14). These DR functions may have important and distinct stage-specific consequences during skin pathophysiology such as tumorigenesis or inflammatory diseases (15).
In this report, we have investigated HaCaT keratinocytes expressing either different isoforms of cFLIP or cFLIP mutants that are resistant to caspase-8 cleavage or mimic the DISC-associated caspase-cleaved p43 fragment of cFLIPL. Whereas all cFLIP variants blocked TRAIL- and CD95L-induced apoptosis, a distinct DISC-associated cleavage pattern of caspase-8 was detected. Only cells expressing full-length cFLIPL (irrespective of cFLIP cleavage) induced proteolysis of caspase-8 to its p43/41 fragments. In contrast, cFLIPS or cFLIPp43 blocked procaspase-8 cleavage. All cFLIP variants blocked death ligand-induced NF-κB activation and the induction of the NFκB target gene IL-8, and caspase-8 was critical and necessary for gene induction. In line with these findings, down-regulation of endogenous cFLIP expression in primary human keratinocytes (PK) enabled death ligand-induced gene expression independent of cell death induction. Taken together our data demonstrate that cleavage of cFLIPL or caspase-8 in the DISC is neither a prerequisite for DR-induced NF-κB signaling nor necessary for the inhibitory function of cFLIP on DR-induced activation of NF-κB or MAPK.
EXPERIMENTAL PROCEDURES
Materials
The following antibodies (Abs) were used: for Caspase-8 (C15), cFLIP (NF-6; Alexis, San Diego, CA), and FADD (Transduction Laboratories, San Diego, CA), and IκBα (C-21), p38 (N-20), JNK-1 (C-17) from Santa Cruz Biotechnology Inc., Santa Cruz, CA. CPP32 (kindly provided by H. Mehmet, Merck Frost, Quebec, Canada), RIP (BD Biosciences), phospho (p)-JNK, JNK, phospho-p38, p-HSP-27, and p-IκBα were purchased from Cell Signaling (Frankfurt am Main, Germany), β-tubulin Abs were from Sigma. Horseradish peroxidase (HRP)-conjugated donkey anti-rabbit and goat anti-mouse IgG Abs and HRP-conjugated goat anti-mouse IgG1 and IgG2b, IgG2a, IgG1κ Abs were obtained from Southern Biotechnology (Birmingham, AL). TRAIL-R1 (HS 101), TRAIL-R2 (HS 201), TRAIL-R3 (HS 301), and TRAIL-R4 (HS 402) monoclonal Abs for FACScan analysis of surface receptor expression were used as described (16) and are available from Alexis. Recombinant TRAIL and TNF were produced as reported (17). For expression of Fc-CD95L, CD95-Fc, and TRAIL-R2-Fc we used published constructs (kindly provided by P. Schneider, Epalinges, Switzerland) (18). One unit of Fc-CD95L was determined at a 1:500 dilution of the stock Fc-CD95L supernatant, and one unit/ml of Fc-CD95L supernatant was sufficient to kill 50% (LD50) of the A375 melanoma cells, as recently described (12). TNC-TRAIL was produced and purified as described elsewhere (19). The caspase inhibitor Z-Val-Ala-Asp-fluoromethyl ketone (Z-VAD-fmk) was purchased from Bachem (Heidelberg, Germany), and quinolyl-valyl-O-methylaspartyl-[2,6-difluoro-phenoxy]-methyl ketone (QVD) was purchased from Enzyme Systems Products, Inc., Livermore, CA. siRNA duplexes for Caspase-8 (5′-AAGAGTCTGTGCCCAAATCAA-3′) an a control siRNA was purchased from Qiagen (Hilden, Germany). JNK inhibitor SP600125 were purchased from Merck.
Cell Culture
The spontaneously transformed keratinocyte line HaCaT and the derived metastatic clone A5RT3 (20) were kindly provided by Dr. P. Boukamp (DKFZ, Heidelberg) and cultured as described (21). PK were cultured as previously reported (22). Cells were maintained in PK medium from Cellntec Advanced Cell Systems (Bern, Switzerland) in a 5% CO2 humidified atmosphere at 37 °C. All experiments were performed between passages 3 and 8 after primary cell culture. The human melanoma cell line IGR was generously provided by Randy H. Byers (Department of Dermatology, Boston University School of Medicine) and cultured as previously described (12).
Vectors
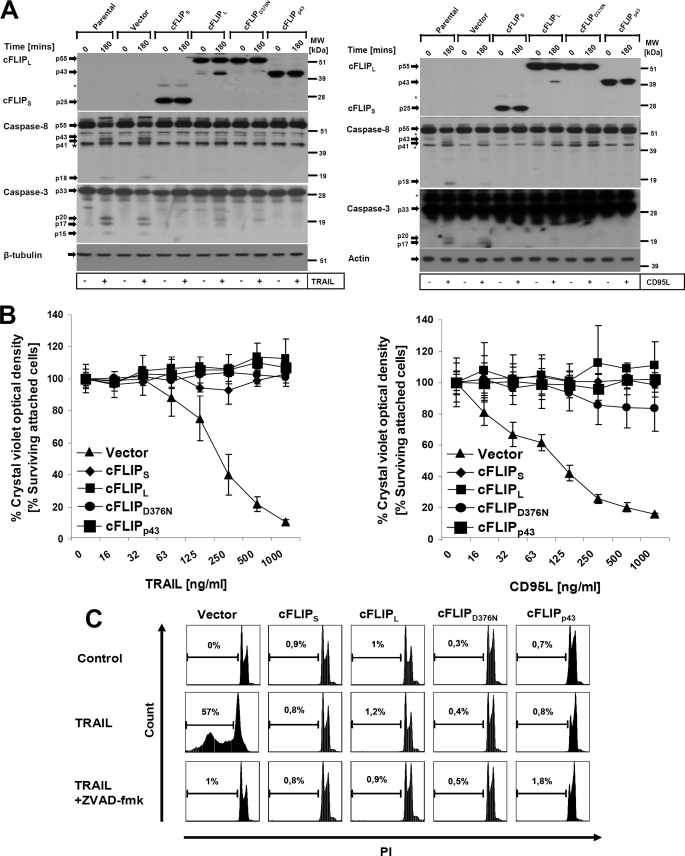
Retroviral vector pEGZ containing cDNA inserts of cFLIPS and cFLIPL were generated as described previously (12). cDNAs of cFLIPD376N and cFLIPp43 were amplified with Pfu Taq polymerase (Promega GmbH, Mannheim, Germany) by polymerase chain reaction (PCR) and subcloned into pJET.blunt vector (Fermentas, St. Leon-Rot, Germany). Following sequence verification, cDNAs were subsequently subcloned into the pEGZ retroviral vector into BglII and XbaI restriction sites. The authenticity of the inserts was confirmed by restriction digestion analysis of the final constructs (data not shown) as well as by protein expression analysis following stable expression (Fig. 1).
FIGURE 1.
cFLIPL, cFLIPS, cFLIPp43, and cFLIPD376N are equally effective for the protection of keratinocytes to DL-mediated apoptosis. A, HaCaT keratinocytes were retrovirally transduced with different forms of cFLIP or the respective mutants as described under “Experimental Procedures.” Cells were left untreated or incubated with TRAIL (0.5 μg/ml, left panel) or CD95L (0.5 μg/ml, right panel) for 3 h. Subsequently, 3 μg of total cellular protein lysates were analyzed by Western blotting for cFLIP, caspase-8, and caspase-3. β-Tubulin served as a control for comparable loading of protein. The asterisk indicates a nonspecific band. B, cells were seeded in 96-well plates in triplicates and stimulated with increasing concentrations of recombinant TRAIL (left panel) or CD95L (right panel) for 16–24 h. Cellular viability was assessed using crystal violet assay as described under “Experimental Procedures.” The percentage of living cells was normalized to mock-stimulated cells (∼100%). Shown is mean ± S.E. of a total of three independent experiments. C, infected keratinocyte cell lines were either left untreated or stimulated with the indicated amounts of TRAIL (0.5 μg/ml) for 6 h, harvested, and examined for hypodiploidy by FACScan analysis. PI, propidium iodide.
Retroviral Infection and Generation of HaCaT Cell Lines Stably Expressing cFLIP Variants
Retroviral infection of HaCaT cells was essentially performed as described earlier (16, 23). Briefly, the amphotrophic producer cell line was transfected with 10 μg of the retroviral vectors by calcium phosphate precipitation. Transfected ϕNX cells (always >95% GFP-positive) were incubated for 24 h. Subsequently medium was exchanged with fresh HaCaT medium (DMEM containing 10% FCS) before harvesting the retroviral supernatants 16–24 h later. Following filtration (45 μm filter, Schleicher & Schuell), culture supernatant was added to HaCaT cells seeded in 6-well plates 24 h earlier in the presence of 5 μg/ml of Polybrene. HaCaT cells were centrifuged for 3 h at 21 °C and the viral particle containing the supernatant was subsequently replaced by fresh medium. After 10–14 days of zeocin selection of bulk infected cultures, FACS analysis for GFP expression (always >90%, data not shown) and Western blot analysis (Fig. 1A) were performed on polyclonal cells to confirm ectopic expression of cFLIP.
Stable siRNA Expression
We used retrovirus-mediated gene transfer for stable expression of short hairpin RNA containing either cFLIP targeting sequence or a hyper random sequence as recently utilized (12, 22). Infected cells were selected with puromycin (1 μg/ml; Sigma) for 3–7 days to obtain bulk infected cultures for further analysis. Aliquots of these cells were used for cytotoxicity assays, biochemical characterization, and IL-8 expression analysis between passages 2 and 5 following selection.
FACScan Analysis
For surface staining of TRAIL receptors (TRAIL-R1 to TRAIL-R4), 2 × 105 cells were incubated with monoclonal Abs against TRAIL-R1 to TRAIL-R4, or isotype-matched control IgG for 30 min followed by incubation with biotinylated goat anti-mouse secondary Abs and Cy5-phycoerythrin-labeled streptavidin (Caltag, Burlingame, CA) as described (23), and 104 cells were analyzed by FACScan (BD Biosciences).
Apoptosis and Cytotoxicity Assays
Crystal violet staining of attached, living cells was performed 16–24 h after stimulation with different concentrations of TRAIL (16–1000 ng/ml) or CD95L (16–1000 ng/ml) in 96-well plates (24). Subdiploid DNA content was analyzed as described by Nicoletti et al. (25). Briefly, cells from a 35-mm dish were subsequently stimulated with TRAIL or CD95L for 6 h. Cells were then harvested, washed with cold PBS, and resuspended in buffer N (sodium citrate, 0.1% (w/v), Triton X-100, 0.1% (v/v), propidium iodide, 50 μg/ml). Cells were incubated in the dark at 4 °C for 48 h and then diploidy was measured by FACScan analysis.
Western Blot Analysis
For the preparation of protein lysates, keratinocytes were washed twice with ice-cold PBS and lysed in a buffer containing 20 mm Tris (pH 7.4), 137 mm NaCl, 10% (v/v) glycerol, 1% (v/v) Triton X-100, 2 mm EDTA, 50 mm sodium β-glycerophosphate, 20 mm sodium pyrophosphate, 1 mm Pefabloc, 5 mg/ml of aprotinine, 5 mg/ml of leupeptin, 5 mm benzamidine, and 1 mm sodium orthovanadate on ice for 1 h. Cellular debris was removed by centrifugation at 15,000 × g for 15 min. The supernatant was collected, and protein content was measured by Bio-Rad protein assay. Equal amounts of cellular protein were separated by SDS-PAGE on 4–12% gradient gels (Invitrogen) followed by transfer to nitrocellulose or PVDF membranes. Blocking of membranes and incubation with primary and appropriate secondary Abs were essentially performed as described (26). Bands were visualized with ECL (Amersham Biosciences).
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed using nuclear extracts of HaCaT keratinocytes exactly as described previously (16).
RNase Protection Assay Analysis
Total RNAs were extracted with the peqGOLD RNAPure reagent (PeqLab Biotechnology GmbH, Erlangen, Germany) and analyzed using the hAPO-3b Multi-Probe template kit (BD Biosciences). Probe synthesis, hybridization, and RNase treatment of the RiboQant Multi-probe RNase protection assay system (BD Biosciences) were essentially performed according to the manufacturer's protocol. Transcripts were resolved by denaturing polyacrylamide gel electrophoresis (5% acrylamide) and analyzed using PhosphorImager with ImageQuant software (GE Healthcare).
Real Time Quantitative PCR (RT qPCR)
Total RNA extraction was performed using RNeasy Kit (Qiagen). cDNA was synthesized in 20 μl using a mixture of oligo(dT) primers and random nonamers in a ratio of 1:10 and SuperScript II Reverse Transcriptase (Invitrogen). Primers were designed using Primer3 software. RT qPCR analyses for the genes encoding interleukin-8 (IL-8), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and β-actin (ACTB) were performed in a final volume of 25 μl using IQTM SYBR® Green (Bio-Rad) in an iCycler (Bio-Rad). The following primers were used: IL-8 forward, 5′-CACCCCAAATTTATCAAAGA-3′; reverse, 5′-ACTGGCATCTTCACTGATTC-3′; GAPDH forward, 5′-CCTGGTATGACAACGAATTT-3′; reverse, 5′- GTGAGGGTCTCTCTCTTCCT-3′; ACTB forward, 5′-AGAAAATCTGGCACCACACC-3′; reverse, 5′-GGGGTGTTGAAGGTCTCAAA-3′. Genes of interest and reference gene products were amplified individually, under equal cycling conditions. HotStarTaq DNA Polymerase was launched by an initial 15 min at 95 °C followed by 42 cycles of one step (denaturation) at 94 °C for 15 s, one step (annealing) at 55 °C for 30 s, and one step (extension) at 72 °C for 30 s. The specific amplification of a single product of the expected size was confirmed by melting curve analysis. Consecutive dilutions of cDNA (1, 1:5, and 1:25) were amplified for the construction of a standard curve (plotted as a logarithmic function of the cDNA dilution factor) and used for the calculation of the RT-PCR efficiency using iCycler software. The relative quantification for IL-8 was calculated after dividing the standard curve value of IL-8 by that of the reference gene (GAPDH and ACTB) for each individual sample. Two reference genes were used for data normalization to account for possible variations as a result of DL treatments. The effects on IL-8 expression were calculated by analyzing mean values obtained from three independent experiments. In two independent experiments, RNA was reverse transcribed three times and the cDNA from the three independent reverse transcriptions were assayed for RT qPCR in triplicates. The mean values obtained by the above explained procedure were compared for all different experimental conditions.
Luciferase Assays
Cells were seeded in 96-well plates and transfected using Lipofectamine (Invitrogen) at 50% confluence with either 3× κB firefly (NF-κB promoter elements) or 5× TK (AP-1 promoter elements) luciferase constructs. As internal control, we used a construct expressing Renilla luciferase from a ubiquitin promoter as a control for transfection efficiency as described (27). 20–24 h after transfection, cells were stimulated with DLs for 6 h and subsequently assayed for luciferase activity according to the manufacturer's recommendations (Promega GmbH, Mannheim, Germany).
DISC Analysis
For the precipitation of the TRAIL or CD95L DISC, 5 × 106 HaCaT keratinocytes were used for each condition. Cells were washed once with DMEM at 37 °C and subsequently incubated at 37 °C in the presence of either 2.5 μg/ml of FLAG-TRAIL pre-complexed with 5 μg/ml of anti-FLAG M2 (Sigma) for 30 min or 1 μg/ml of CD95L-Fc for 30 min. Receptor complex formation was stopped by washing the monolayer four times with ice-cold PBS. Cells were lysed on ice by addition of 2 ml of lysis buffer (30 mm Tris-HCl, pH 7.5, at 21 °C, 120 mm NaCl, 50 mm sodium β-glycerophosphate, 20 mm sodium pyrophosphate, 1 mm sodium orthovanadate, 10% glycerol, 1% Triton X-100, Complete® protease inhibitor mixture (Roche Molecular Diagnostics). Lysates were centrifuged twice at 20,000 × g for 10 min, followed by another centrifugation of 30 min to remove cellular debris. A minor fraction of the resulting clear lysates was used to control for the input of the respective proteins. Receptor complexes were precipitated from the lysates by coincubation with 40 μl of protein G beads (Roche Applied Science) for 24 h on an end-over-end shaker at 4 °C. For the precipitation of the non-stimulated receptors, 50 ng of either FLAG-TRAIL precomplexed with anti-FLAG M2 antibody or 20 ng/ml of Fc-CD95L were added to the lysates prepared from non-stimulated cells to control for protein association with non-stimulated receptor(s). Ligand affinity precipitates were performed as described (12, 28) and analyzed by Western blot analysis.
Determination of IL-8 Secretion
IL-8 secretion was analyzed by enzyme-linked immunosorbent assay (BD Biosciences) according to the manufacturer's recommendations.
Caspase-8 Knockdown in HaCaT Keratinocytes
Cells were seeded to have a confluency of 40–50% on the day of transfections. Transfected control and Caspase-8 siRNA duplexes (5′-AAGAGTCTGTGCCCAAATCAA-3′) were transfected at 20 μm final concentration with Lipofectamine (Invitrogen). The transfected cells were left for 24 h at 37 °C and cell culture medium was replaced with fresh DMEM. Cells were cultured for an additional 24 h at 37 °C and subsequently stimulated with death ligands as indicated in the figure legends.
In Vitro Kinase Assay
The activity of JNK was analyzed in an in vitro kinase assay using 0.5 mg of protein. Total cellular lysates were immunoprecipitated with 1 μg of anti-JNK-1 (C-17; Santa Cruz Biotechnology) antibody/sample by permanent shaking for 3 h at 4 °C. Immune complexes were recovered using 50 μl of a 50% protein A-Sepharose bead suspension (Amersham Biosciences) and incubated for 90 min at 4 °C in a permanent upside down shaker. Subsequently, immunoprecipitates were washed six times in lysis buffer and incubated with 1 μg of GST-c-Jun(1–135) in 25 μl of kinase buffer (20 mm HEPES, pH 7.6, 2 mm EGTA, 20 mm MgCl2, 1 mm sodium orthovanadate, 1 mm DTT, 0.1% Triton X-100, 0.25 mm [32P]ATP) for 3 min at 30 °C as previously described (29). For further analysis, lysates were resuspended in loading buffer, separated by SDS-PAGE, and transferred to PVDF membranes. The gels were dried and exposed to Amersham Biosciences TM film at −70 °C using an intensifying screen. Expression and purification of GST-c-Jun (amino acids 1–135) was performed as indicated previously (29). In brief, the pGEX-GST-c-Jun plasmid (kindly provided by Stefan Ludwig, University of Münster) was transformed in Escherichia coli (strain BL21). Cells were induced with isopropyl 1-thio-β-d-galactopyranoside and lysed (50 mm Tris, pH 8.0, 120 mm NaCl, 0.5% Nonidet P-40, 1× protease inhibitors, 1 mm 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride, 1 mm sodium vanadate, 1 mm DTT) on ice by sonication. Bacterial lysates were harvested and GST-tagged proteins were purified with GST columns according to the manufacturer's instructions (Amersham Biosciences). GST fusion proteins were eluted (50 mm Tris, pH 8.0, 10 mm reduced glutathione), and recombinant GST-c-Jun protein was aliquoted and stored at −20 °C.
RESULTS
Different cFLIP Isoforms Protect Keratinocytes from DL-mediated Apoptosis
The role of cFLIP in DR-mediated signaling pathways has been studied extensively (7, 11, 30, 31). It was suggested that cFLIP may activate proinflammatory signaling pathways in a DISC-independent manner (32). To investigate the different signaling capabilities of cFLIP variants without the limitations of transient expression, cFLIPL, cFLIPS, a cleavage-site mutated cFLIPL (cFLIPD376N), and the caspase-8 cleaved fragment of cFLIPL (cFLIPp43) were studied in more detail using retroviral stable overexpression of these gene products. Expression of the different cFLIP isoforms or mutants was confirmed and proved to be comparable in the different polyclonal cell lines (Fig. 1A). When cells were treated with TRAIL or CD95L, we observed robust activation of caspase-8 and caspase-3 within 3 h in control-infected cells. Interestingly, we detected in some experiments (Fig. 1A) a stimulation-dependent caspase-8 reactive band of higher molecular weight, likely explained by stimulation-dependent post-translational modification of caspase-8 such as polyubiquitination (33). All cFLIP isoform/mutants fully blocked caspase-8 cleavage (Fig. 1A) and equally protected HaCaT cells from DL-induced cell death (Fig. 1, B and C), in line with several other reports (11, 13, 34), although the cell line expressing uncleavable cFLIPL (cFLIPD376N) showed a minor sensitivity to CD95L at high concentrations. cFLIPL containing a cleavage site mutation (cFLIPD376N) did not undergo DL-induced cleavage (Fig. 1A, left and right panel), in line with a previous report (11). Our results suggest that cFLIP is able to protect against DL-mediated apoptosis independent of cFLIPL cleavage.
cFLIP Proteins Inhibit CD95L or TRAIL-induced Non-apoptotic Signaling
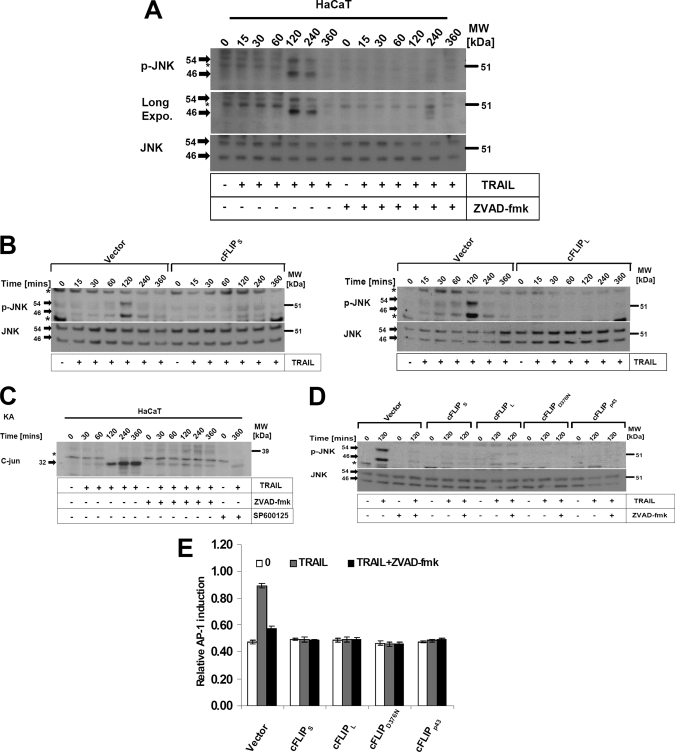
A number of reports have shown that CD95L or TRAIL may activate non-apoptotic signaling pathways such as the MAPK JNK and its downstream target transcription factor AP-1. AP-1 eventually triggers pro-inflammatory or proliferative responses (35). We therefore next asked if cFLIP variants differentially regulated the activation of JNK in our experimental system. JNK was variably activated within 15 min of treatment with DLs with a further robust increase up to 2 h after stimulation in HaCaT (Fig. 2, A–C). TRAIL-mediated induction was largely repressed by overexpression of cFLIPS (Fig. 2B, left panel), cFLIPL (Fig. 2B, right panel), or Z-VAD-fmk (Fig. 2A). To test whether JNK activation is simply a consequence of caspase activation, we used the pan-caspase inhibitor Z-VAD-fmk known to completely protect keratinocytes from TRAIL-mediated apoptosis (16). Remarkably, TRAIL-mediated late JNK activation was completely caspase-dependent (Fig. 2, A and D, and supplemental Fig. S2B), indicative of the activation of JNK downstream of caspases (36). Moreover, caspase activation and JNK activation had a similar dose dependence of caspase activation (as determined by cleavage of caspase-8) and JNK activation (supplemental Fig. S5). Of note, we detected a weak TRAIL-induced JNK activation within 30 min in some experiments, which was not fully inhibited by Z-VAD-fmk. In particular this finding was evident in kinase assays (Fig. 2C). In contrast the later induction of JNK from 60 to 240 min after stimulation was completely and reproducibly blocked by Z-VAD-fmk (Fig. 2, A and C). Different isoforms of cFLIP or the respective mutants were equally effective to block the late TRAIL-induced JNK activation as determined by Western blotting (Fig. 2D) or kinase assays (data not shown). In line with these results, the JNK downstream target AP-1 was induced within 6 h in control cells, whereas all isoforms or mutants of cFLIP fully blocked AP-1 induction (Fig. 2E). Collectively, these data indicate that all cFLIP variants inhibit DL-induced activation of JNK and its downstream target AP-1.
FIGURE 2.
DL-mediated JNK phosphorylation and its downstream target transcriptional factor AP-1 are dependent upon active caspases and blocked by different isoforms/mutants of cFLIP. A, HaCaT keratinocytes were preincubated with the caspase inhibitor (Z-VAD-fmk) for 1 h and stimulated with TRAIL for the indicated time points. Cellular lysates were analyzed for JNK phosphorylation. The asterisk indicates a nonspecific band. B, control infected keratinocytes and keratinocytes expressing cFLIPL or cFLIPS were stimulated with TRAIL (1 μg/ml) for the indicated time points. Cellular lysates were subsequently analyzed for the phosphorylation of JNK using phospho-specific antibodies. Membranes were stripped and total levels of JNK were determined. The asterisk indicates a nonspecific band. C, HaCaT keratinocytes were stimulated with TRAIL (0.5 μg/ml) for the indicated time points and analyzed for JNK phosphorylation by in vitro kinase assay as described under “Experimental Procedures.” The asterisk indicates a nonspecific band. D, HaCaT keratinocytes expressing the different cFLIP isoforms as outlined in the legend to Fig. 1 were preincubated for 1 h in the presence or absence of Z-VAD-fmk (20 μm), and subsequently stimulated for 2 h with TRAIL (1 μg/ml). Total cellular lysates were characterized for JNK phosphorylation as indicated for A. The asterisk indicates a nonspecific band. E, cFLIP overexpressing HaCaT keratinocytes were seeded in a 96-well plate and transfected with AP-1 firefly luciferase reporter constructs along with the internal control reporter construct as described under “Experimental Procedures.” Following transfection, cells were stimulated with TRAIL (0.5 μg/ml) for 6 h and assayed for firefly luciferase activity. Shown is mean ± S.E. of a total of three independent experiments.
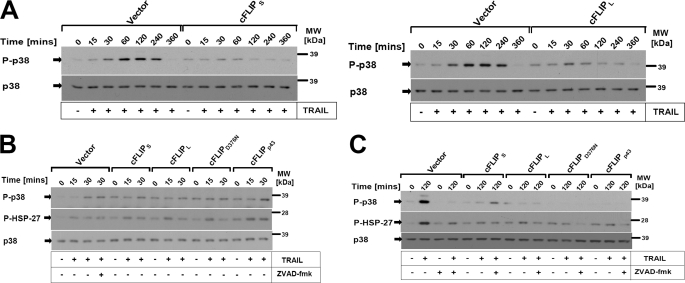
We next investigated if cFLIP modulates MAPK p38, which is relevant for the transcriptional as well as post-transcriptional regulation of gene expression (37). TRAIL rapidly activated p38 within 15 min with a further increase up to 4 h after stimulation. HaCaT expressing different cFLIP isoforms or control cells treated with Z-VAD-fmk showed complete inhibition of the late induction of DL-mediated p38 activation at 2–4 h (Fig. 3 and supplemental Fig. S2B). In contrast, early activation of MAPK p38 was not inhibited by Z-VAD-fmk or cFLIP isoforms (Fig. 3B). Collectively these data indicated that different cFLIP isoforms containing either DED and a caspase-like domain or cFLIP proteins containing solely tandem DEDs are able to block DR-mediated late p38 activation in a comparable fashion.
FIGURE 3.
DLs rapidly activate MAPK p38 and its target gene HSP-27, which is blocked by the caspase inhibitor or cFLIP isoforms/mutants. A, control infected keratinocytes and keratinocytes expressing cFLIPL or cFLIPS were stimulated with TRAIL (0.5 μg/ml) for the indicated time points. Cellular lysates were subsequently analyzed for the phosphorylation of MAPK p38 using phospho-specific antibodies. Membranes were stripped and total levels of p38 were determined to confirm even loading of proteins. B, control cells or cells expressing the different cFLIP isoforms or mutants, respectively, were preincubated for 1 h in the presence or absence of Z-VAD-fmk (20 μm), and subsequently stimulated for 15 and 30 min (B) or 2 h (C), respectively, with TRAIL (0.5 μg/ml). Total cellular lysates were characterized for p38 phosphorylation and HSP-27 phosphorylation. One of three independent experiments is shown.
A recent report has suggested that Z-VAD-fmk induces autocrine production of TNF that may in turn lead to gene induction (38). To investigate this possibility in our experimental setting, we compared Z-VAD-fmk with QVD. Both inhibitors failed to block the TRAIL-induced IκBα phosphorylation that was independent of TNF signaling, but fully inhibited by TRAIL-R2-Fc (supplemental Fig. S2A). TRAIL-induced JNK and p38 phosphorylation was effectively suppressed by Z-VAD-fmk or QVD (supplemental Fig. S2B). When we analyzed TRAIL-induced MAPK and NF-κB activation in the presence of exogenously added TNF-R2-Fc or TRAIL-R2-Fc in HaCaT keratinocytes, only TRAIL-R2-Fc, but not TNF-R2-Fc fully blocked TRAIL-induced MAPK or IκBα phosphorylation, whereas TNF-R2-Fc was ineffective (supplemental Fig. S2, A and C). These data exclude that TRAIL-induced proinflammatory signals are mediated by a Z-VAD-fmk-induced autocrine loop of TNF.
cFLIP Isoforms Are Equally Effective in the Inhibition of CD95L- and TRAIL-induced NF-κB Activation
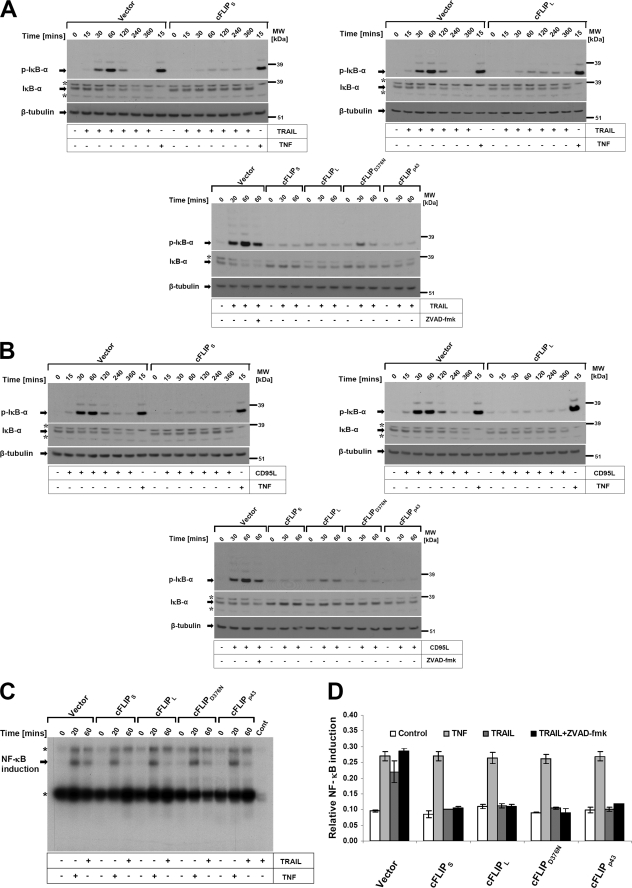
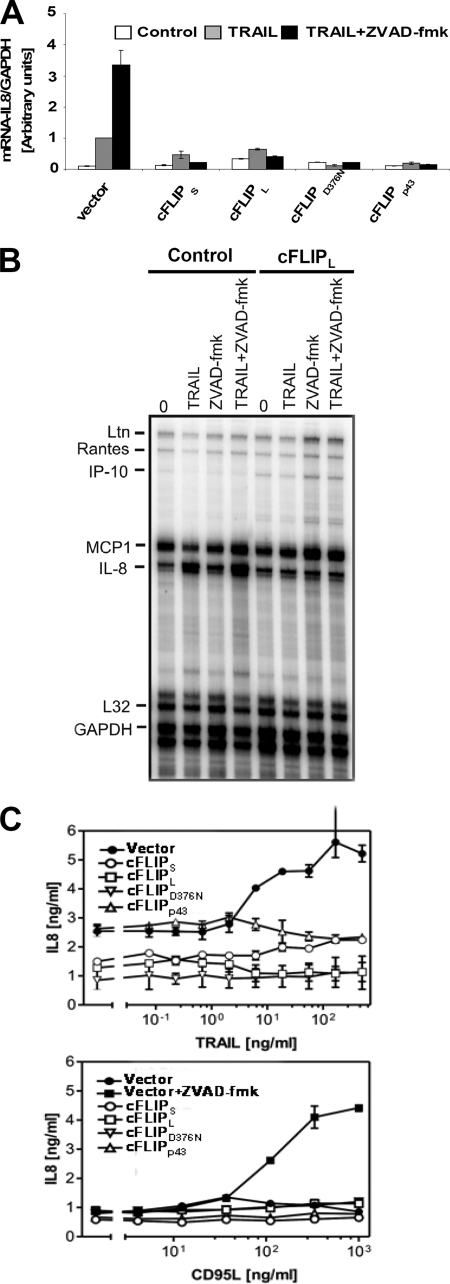
TNF, TRAIL, or CD95L induce a large array of proinflammatory responses (e.g. NF-κB activation) and subsequent target gene expression, especially when apoptotic caspase activation is blocked (14, 16, 39). In contrast to the inhibitory effects of Z-VAD-fmk on TRAIL- and CD95L-induced late activation of JNK and p38, NF-κB activation was not blocked by this compound or an alternative pan-caspase inhibitor QVD (supplemental Fig. S2, A and B) (40). However cFLIPL and cFLIPS potently interfered with TRAIL- or CD95L-induced NF-κB activation, indicative of a DISC-associated regulation of this signaling pathway (13, 34). We next asked if isoforms or distinct cleavage fragments of cFLIP may differentially regulate NF-κB activation and its target gene IL-8 induction upon CD95 and TRAIL-R activation. In these experiments, we used TNF as a control. TRAIL or CD95L rapidly activated phosphorylation of IκBα within 15–30 min in control transduced cells. Detectable phosphorylation and ubiquitination of IκBα persisted up to 2 h after stimulation, paralleled by the loss of total IκBα (Fig. 4A). NF-κB activation was significantly impaired in all cells expressing cFLIP variants or mutants in response to TRAIL or CD95L, but not TNF. Thus, cleavage of cFLIPL does not compromise its inhibitory effect on CD95L- and TRAIL-induced NF-κB activation (Fig. 4B). In line with these results, we also observed suppression of NF-κB DNA binding (Fig. 4C) and NF-κB promoter activity (Fig. 4D) in response to TRAIL and CD95L, but not TNF in HaCaT expressing the different cFLIP variants. To investigate if the inhibitory effect of cFLIPs translated into changes of gene expression, we studied IL-8 expression. mRNA analysis using qPCR and the RNase protection assay demonstrated that all cFLIP proteins completely prevented TRAIL-induced IL-8 mRNA induction (Fig. 5, A and B), or TRAIL/CD95L-induced IL-8 secretion (Fig. 5C). Taken together, our data support a concept that cFLIP is able to block NF-κB activation by CD95 and the TRAIL death receptors but not by the closely related death receptor TNFR1.
FIGURE 4.
DL-mediated phosphorylation and degradation of IκBα are blocked by different cFLIP isoforms and cFLIPL mutants. cFLIPL and cFLIPS expressing keratinocytes as characterized in Fig. 1 were stimulated with TRAIL (0.5 μg/ml) or TNF (0.5 μg/ml) (A) and CD95L (0.5 μg/ml) (B) for the indicated times. Cellular lysates were collected as indicated under “Experimental Procedures” and subsequently investigated for IκBα phosphorylation and IκBα degradation using antibodies specific for phosphorylated IκBα (upper panel) or total IκBα (lower panel). β-Tubulin served as a loading control. One of three independent experiments is shown. The asterisk indicates a nonspecific band. C, in parallel experiments, nuclear extracts were analyzed for κB-specific DNA binding by EMSA. The positions of p65/p50 heterodimers or p50 homodimers are indicated. One of three independent experiments is shown. The asterisk indicates a nonspecific band. D, cFLIP overexpressing HaCaT keratinocytes were seeded in a 96-well plate and transfected with 3× κB firefly (NF-κB promoter elements) along with the internal control vector (Renilla luciferase driven by the ubiquitin promoter) as described under “Experimental Procedures.” 24 h after transfection, cells were stimulated with TRAIL (0.5 μg/ml) or TNF-α (0.5 μg/ml) for 6 h and assayed for NF-κB activation as described under “Experimental Procedures.” Shown is mean ± S.E. of a total of two independent experiments.
FIGURE 5.
DL-mediated IL-8 induction is blocked by cFLIP isoforms as well as cFLIPL mutants. A, TRAIL-induced IL-8 induction is inhibited by cFLIP variants. Following preincubation with either diluent alone or 20 μm Z-VAD-fmk for 1 h, control or cFLIP variants overexpressing HaCaT keratinocytes were treated with 0.5 μg/ml of TRAIL for 3 h and total RNA was isolated and after synthesis of cDNA, qPCR were performed. IL-8 mRNA expression was normalized with the housekeeping gene GAPDH as described under “Experimental Procedures.” Shown are mean ± S.E. of a representative experiment of a total of two independent experiments. B, total RNA of control or cFLIPS cFLIPL-overexpressing keratinocytes stimulated with TRAIL (200 ng/ml) in the presence or absence of Z-VAD-fmk (20 μm) for 6 h was analyzed by RNase protection assay for the indicated genes. The protected fragments for IL-8 mRNA or control mRNA L32 or GAPDH are indicated. C, TRAIL- and CD95L-induced IL-8 secretion is inhibited by different cFLIP isoforms as well as their mutants. Following preincubation with either diluent alone or 20 μm Z-VAD-fmk for 1 h, control keratinocytes or the different cFLIP-overexpressing keratinocytes were treated with TRAIL or CD95L, respectively, for 24 h. Supernatants were assayed for IL-8 protein by ELISA. Shown are mean ± S.E. of a total of three independent experiments.
Caspase-8 Is Critical and Necessary for TRAIL-induced Apoptotic and Non-apoptotic Signaling
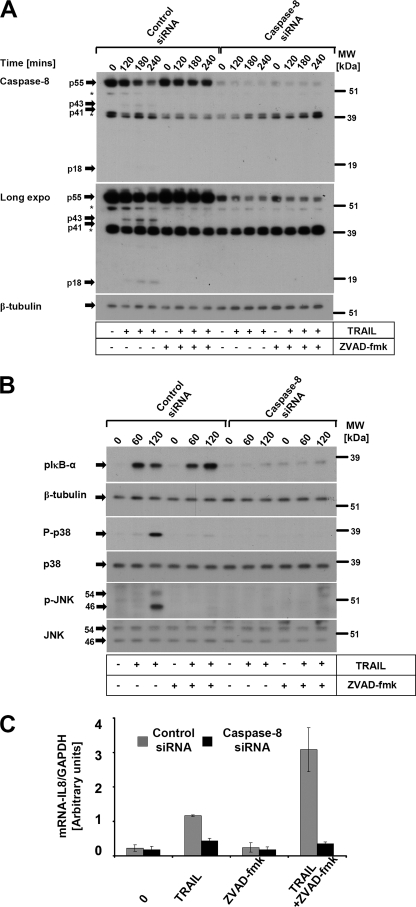
cFLIP is directly recruited to the DISC and the caspase-8/cFLIP heterodimer has enzymatic activity (10). To investigate if caspase-8 is necessary for TRAIL-induced non-apoptotic signaling pathways such as NF-κB, p38, and JNK, we performed knockdown experiments of endogenous caspase-8 using siRNA duplexes. Efficient down-regulation of endogenous caspase-8 (Fig. 6A) completely blocked TRAIL-induced phosphorylation of IκB-α, JNK, and p38 (Fig. 6B). In line with the phosphorylation of Iκβ-α, TRAIL-induced IL-8 mRNA induction was abrogated in caspase-8 knockdown cells (Fig. 6C). These data demonstrate that caspase-8 is critical and necessary for TRAIL-induced apoptosis and gene induction.
FIGURE 6.
Knockdown of caspase-8 fully suppresses apoptotic and non-apoptotic signaling induced by TRAIL death receptors. HaCaT keratinocytes were transfected with control or caspase-8 siRNA duplexes as described under “Experimental Procedures.” Post-transfection, transfectants were stimulated with TRAIL (0.1 μg/ml) for the indicated times. Cellular lysates were analyzed for the efficient knockdown of caspase-8 (A). The asterisk indicates nonspecific bands. B, activation of non-apoptotic proinflammatory signaling pathways (p-IκBα, p-p38, and p-JNK) of caspase-8 knockdown cells or control transfectants. The respective transfected cells were analyzed for phosphorylation of p-IκBα, p-p38, and p-JNK after the indicated times following TRAIL (0.1 μg/ml) stimulation. β-Tubulin, p38, and JNK served as loading controls. C, TRAIL-induced IL-8 requires caspase-8. Following preincubation with either diluent alone or 20 μm Z-VAD-fmk for 1 h, control cells or caspase-8 knockdown cells were treated with TRAIL (0.1 μg/ml) for 3 h. Total RNA was isolated and qPCR analysis was performed. IL-8 mRNA expression was normalized with the housekeeping gene GAPDH as described under “Experimental Procedures.” Shown are mean ± S.D. of a representative experiment of a total of two independent experiments.
cFLIP Isoforms Differentially Affect Post-translational Modification of DISC-associated Proteins
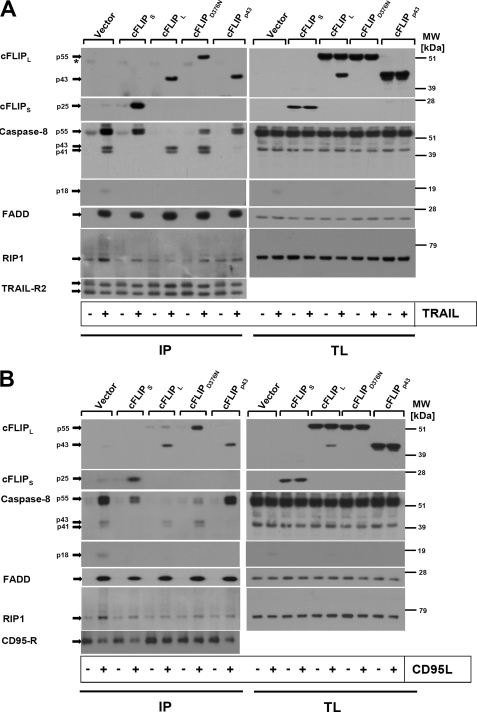
To investigate if cFLIP variants differentially affect TRAIL- and CD95L-induced caspase-8 activation in the DISC, we characterized the DISC recruitment of FADD, caspase-8, receptor interacting protein-1 (RIP-1), and cFLIP in the transduced cells (HaCaT-cFLIPL, HaCaT-cFLIPS, HaCaT-cFLIPp43, or HaCaT-cFLIPD376N) using ligand affinity precipitation (Fig. 7, A and B). FADD recruitment to the TRAIL or CD95L DISC did not significantly differ between parental, control cells, or HaCaT expressing different cFLIP variants (Fig. 7, A and B). In control cells, endogenous cFLIPL was only detected as the p43 cleavage fragment in the DISC, indicative of stoichiometric caspase-mediated cleavage. A marked increase in the association of cFLIP p43 was observed in HaCaT-cFLIPL, in line with our previous report (13). The cleavage site-defective cFLIPL mutant was detected as a p55 protein in the DISC, indicative of the lack of DISC-associated cleavage. Ectopically expressed cFLIPp43 was detected at similar levels in the DISC of HaCaT-cFLIPp43. Recruitment of cFLIPS in HaCaT-cFLIPS was comparably effective as recruitment of the various FLIPL-related FLIP variants. We detected RIP-1 recruitment and its modification in the TRAIL and CD95L DISC of control-infected HaCaT cells, in line with our previous and recent report (13, 28). Notably, we observed a suppression of RIP-1 recruitment in all cells expressing cFLIP variants. The caspase-8 cleavage pattern detected in the DISC was different; in control cells, the proform of caspase-8 (p55/53) and the cleavage product p43/41 were found, whereas the DISC of HaCaT-cFLIPL largely contained p43/41 fragments of caspase-8. Expression of cFLIPS completely inhibited caspase-8 cleavage, but did not interfere with procaspase-8 recruitment to the DISC. These results indicated that cFLIPS blocked procaspase-8 processing in the DISC (Fig. 7, A and B). Similarly, expression of cFLIPp43 yielded an increased level of full-length caspase-8 within the DISC, suggesting that uncleaved cFLIPL is required for procaspase-8 cleavage within the DISC. Last, HaCaT-cFLIPD376N had a higher amount of procaspase-8 p55/53 in the DISC when compared with HaCaT-cFLIPL, suggesting that uncleaved cFLIPL favored procaspase-8 recruitment to the DISC as compared with cleavage-competent cFLIPL. When we examined recruitment of TNF receptor-associated factor 2 (TRAF2), we detected stimulus-dependent recruitment of the molecule in the CD95 DISC of HaCaT-cFLIPL and HaCaT-cFLIPD376N, whereas control cells, HaCaT cFLIPS, and HaCaT-cFLIPp43 lacked specific recruitment of TRAF2. These data indicate that the proform of cFLIPL and/or cleaved caspase-8 favors the direct or indirect recruitment of TRAF2 to the CD95 DISC (supplemental Fig. S1). However, TRAF2 recruitment cannot explain the inhibitory effect of cFLIP on TRAIL- and CD95L-induced NF-κB activation. In summary, our data suggest that different cFLIP isoforms block TRAIL- and CD95L-induced apoptosis of keratinocytes by interference with different steps of caspase-8 processing at the DISC. Full-length cFLIPL is required for efficient procaspase-8 recruitment and the initial cleavage step to caspase-8 p43/41, but these events are not crucial for the inhibitory effect of cFLIP upon TRAIL- and CD95L-induced NF-κB activation.
FIGURE 7.
Different cFLIP isoforms modulate the composition of the TRAIL or CD95 DISC. A, differential composition of the TRAIL DISC (A) or CD95 DISC (B) in keratinocytes expressing different isoforms or mutants of cFLIPL expression. DISC analysis (IP) was performed from a total of 5 × 106 cells. Precipitates of non-stimulated cells served as specificity controls for ligand affinity precipitates and were assayed for comparable immune precipitation of TRAIL-R2 or CD95 as a control. Cells were characterized for cFLIP, caspase-8, FADD, RIP-1 recruitment, and TRAIL-R2 precipitation by Western blotting. Total cellular lysates were analyzed in parallel from all samples. A representative of one of three independent experiments is shown. The asterisk indicates nonspecific bands.
cFLIP Knockdown Enhances TRAIL-induced Apoptosis and Gene Induction in Human PK
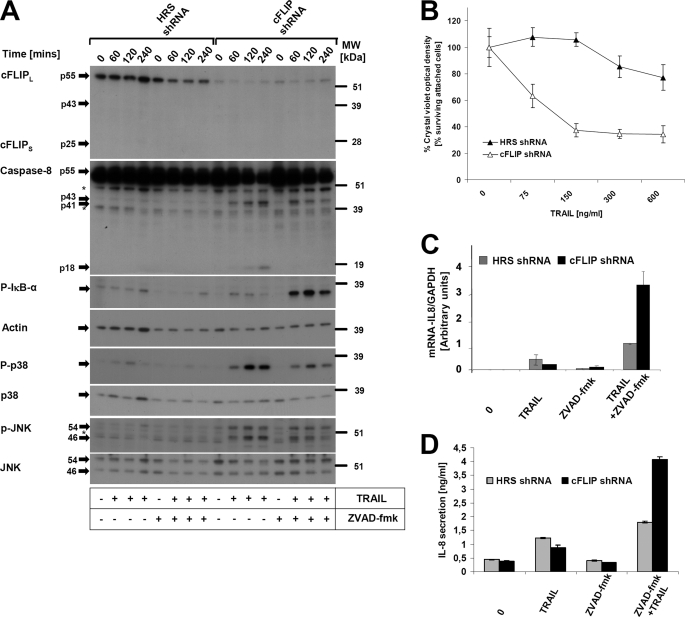
We aimed to study if cFLIP modulates CD95L and TRAIL-induced gene induction and cell death under physiological conditions using PK. Efficient down-regulation of cFLIPL and cFLIPS dramatically sensitized PK to TRAIL-mediated cell death (Fig. 8B) and increased cleavage of initiator caspase-8 (Fig. 8A). Reflecting caspase activity under those conditions, cleavage of the substrate PARP1 (data not shown) was observed. To investigate if loss of cFLIP modulated TRAIL-mediated non-apoptotic signaling pathways (NF-κB, JNK, p38), we analyzed the phosphorylation of IκBα, p38, and JNK in PK with knockdown of cFLIP. TRAIL-induced phosphorylation of IκBα was enhanced in cells with repressed cFLIP levels. This effect was observed in the absence and (more pronounced) in the presence of Z-VAD-fmk (Fig. 8A). Furthermore, we also observed enhanced TRAIL-induced p38 and JNK activation in these cells (Fig. 8A). Interestingly caspase inhibition with Z-VAD-fmk largely repressed TRAIL-induced late JNK and p38 activation (Fig. 8A). Moreover, loss of cFLIP in PK also led to increased expression of IL-8 at mRNA (Fig. 8C) and protein levels (Fig. 8D). Taken together, these results supported our hypothesis that cFLIP has an important physiological role not only for apoptosis protection, but also for inhibition of TRAIL (CD95L)-induced non-apoptotic signaling pathways (NF-κB, p38, and JNK activation) and subsequent target gene induction in primary human keratinocytes. To address if cFLIP modulated TRAIL-mediated NF-κB activation in other cell types, we performed knockdown of cFLIP in a melanoma cell line (IGR; supplemental Fig. S4) and in another cell line of keratinocyte origin (A5RT3; (supplemental Fig. 3). In these cells, efficient cFLIP knockdown enhanced TRAIL-induced apoptosis (supplemental Figs. S3 and S4). However, knockdown of cFLIP only enhanced TRAIL-induced NF-κB activation in A5RT3 cells in the presence of Z-VAD-fmk, whereas knockdown of cFLIP failed to increase NF-κB activation in melanoma cells. These data indicate a cell type-specific pattern and the potential requirement of additional downstream signaling modules necessary for TRAIL (or CD95L)-induced gene induction. Future studies are required to delineate this issue in more detail.
FIGURE 8.
Knockdown of cFLIP protein leads to increased sensitivity to TRAIL-mediated apoptosis in PK and facilitates non-apoptotic signaling pathways (NF-κB, JNK, and p38) in PK. A, PK were infected with cFLIP-specific short hairpin RNA (shRNA). Transduced cells were selected with puromycin (1 μg/ml) for 3 days. Hyper random sequence (HRS)-infected cells (Vector) served as control for cFLIP short hairpin RNA-transduced PK. Cells were subsequently stimulated with recombinant TRAIL (150 ng/ml) for the indicated time points and analyzed for cFLIP and caspase-8 expression and cleavage. Activation of p-IκB-α, p-P38, and p-JNK was monitored by Western blotting with the respective phospho-specific antibodies. Analysis of β-actin, p38, or JNK served as loading control (note relative overloading of control cells). The asterisk indicates nonspecific bands. Molecular weights are indicated on the right side. B, knockdown of cFLIP sensitizes to TRAIL-induced cell death. PK generated as described in A were treated with the indicated concentrations of TRAIL for 24 h. Viability was subsequently examined by a crystal violet assay. Shown are mean ± S.D. of three independent experiments. C, down-regulation of cFLIP facilitates IL-8 induction at the mRNA level. Cells generated as in A were stimulated with TRAIL (150 ng/ml) for 6 h and qPCR was performed. Shown are mean ± S.E. of a total of three independent experiments. D, knockdown of cFLIP leads to increased secretion of IL-8 protein as determined by ELISA. Cells were stimulated with TRAIL (150 ng/ml) for 24 h and supernatants were assayed by ELISA. Shown are mean ± S.E. of a total of three independent experiments.
DISCUSSION
CD95 and TRAIL death receptor-mediated proinflammatory non-apoptotic signaling has been a topic of intense scientific debate over the years. Recombinant forms of TRAIL or TRAIL-R agonists are currently in phase I and II clinical trials for tumor therapy (41) and the potential detrimental effects of TRAIL death receptor stimulation for tumor cells not undergoing apoptosis have raised concerns about the tumor-promoting abilities of DR agonists. Although is well known that cFLIP isoforms confer resistance to apoptosis, their role in non-apoptotic signaling are less clear. Several reports have demonstrated spontaneous NF-κB activation after FLIP expression, in particular viral forms of FLIP. These reports include studies in murine embryonic fibroblasts or RIP1-deficient Jurkat cells (42, 43) and indicated that independent of DISC recruitment, cFLIP isoforms such as vFLIPs may have direct effects on NF-κB activity, in particular in transgenic T cells (44) or murine embryonic fibroblasts (42). Examining the effects of transient cFLIP overexpression in 293T cells, Kataoka et al. (45, 46) concluded that different fragments of cFLIP have distinct signaling capabilities. In these reports, the direct activating effect of cFLIP for NF-κB activity in T cells or Raji cells was demonstrated and the impact of cFLIP for death receptor stimulation-dependent NF-κB activation was implied. Other reports have shown that caspase-8 and RIP1 are critical for death receptor-induced NF-κB activation in Jurkat T cells (34) or HT1080 cells (47). Although we identified cell types that do not increase NF-κB activation following down-regulation of cFLIP (compare supplemental Fig. S4), there is sufficient evidence to claim, together with the data presented in this report, that cFLIP acts as negative regulator of NF-κB activation induced by CD95 or TRAIL death receptors, at least in epithelial cell types.
Our report investigated molecular mechanisms of cFLIP-dependent signaling using stable expression of cFLIP in HaCaT keratinocytes that endogenously contain low levels of cFLIP (24). Comparable amounts of the cFLIP isoforms cFLIPS or cFLIPL, or the cFLIP mutants cFLIPD376N or cFLIPp43 resulted in complete protection against DR-mediated apoptosis at the level of the DISC and emphasize the importance of cFLIP for DR-mediated cell death.
When we examined DISC-associated signaling events, we show that the presence of cFLIPS and cFLIPp43 prevented the cleavage of procaspase-8 and promoted accumulation of full-length procaspase-8 in the DISC. In contrast, cFLIPL and the mutant cFLIPD376N allow the initial cleavage step but block further processing as indicated by the lack of detection of fully active p18 subunit of caspase-8 in total cellular lysates. These results indicate that cFLIP isoforms similarly influence cellular apoptosis signaling, but nonetheless, differentially influence the pattern of DISC-associated proteins, in line with data in other cellular models (11). Clearly, our study shows that in HaCaT cells TRAIL- and CD95L-induced activation of MAPK p38 and JNK are caspase-dependent signaling events based upon experiments with two different pancaspase inhibitors (Z-VAD-fmk and QVD). cFLIPL blocks intracellular, but not DISC-associated caspase activity, which is sufficient to interfere with JNK activation. Interestingly we also observed that a suppression of caspase-8 expression fully blocks DR-induced non-apoptotic signaling pathways. These findings indicate that caspase-8 is necessary and critical for DR-induced gene induction. Also our data suggest that the late TRAIL-induced JNK and p38 activation is activated downstream of both cFLIP and caspase-8, at least in the epithelial cells examined in this report.
The recruitment of additional components of the NF-κB or MAPK signaling cascade to the DISC has been investigated by several groups. Although RIP-1 is uniformly and strongly found in the TNF complex I, diverse results have been published in respect to the recruitment of RIP-1, TRAF2, or other components to the DISC of CD95 or TRAILR1/2 (13, 46). In particular, when caspase activity is blocked, RIP-1 recruitment to the DISC has been robustly detected by several groups (13, 40, 48). We show here that all cFLIP isoforms/mutants suppressed the recruitment of RIP-1 to the DISC. This suggests that cleavage of cFLIP or the initial cleavage of procaspase-8 is irrelevant for the repression of RIP-1 recruitment to the DISC. Moreover cFLIPp43 does not facilitate NF-κB activation by TRAIL or CD95L. Our data contrast a study that used transient transfection assays to suggest that cFLIP isoforms or cFLIP cleavage mutants, in particular cFLIPp43, differentially influence non-apoptotic DR signals (45). While this manuscript was under revision, Oberst et al. (49) suggested that both events, dimerization and cleavage of caspase-8, are required for caspase-8 activity. Clearly, more data are necessary to dissect the role of distinct caspase-8 cleavage events for TRAIL- or CD95-induced gene inductive signaling. Future studies including experiments using ex vivo DISC analysis as recently published will potentially answer this important question in more detail (50). Our data further support that the stoichiometry of caspase-8 and cFLIP, however, is a major determinant of TRAIL- and CD95-induced NF-κB activation (for review, see Ref. 6).
Mechanistically ample evidence exists that RIP-1 is ubiquitinated and interacts with the IKK-γ (NEMO) subunit of the IκB kinase (IKK) complex. The importance of linear ubiquitin chains bound to RIP1 for the activation of the IKK complex has been convincingly demonstrated (51–53). In line with our data that demonstrate that cFLIP interferes with RIP recruitment, knockdown of RIP-1 inhibited TRAIL-induced IκBα degradation in HT1080 cells (54). Our data furthermore show that suppression of RIP-1 recruitment to the CD95/TRAIL DISC by all cFLIP isoforms/mutants leads to suppression of TRAIL-induced MAPK activation. However, RIP-1-deficient murine embryonic fibroblasts are still able to activate NF-κB in response to TNF (55), suggesting that indirectly binding proteins not detected in our DISC analysis may be critical for the RIP1-dependent response, at least for TNF. It is tempting to speculate that a secondary intracellular signaling platforms might regulate the proinflammatory signaling by prototypical death receptors that are distinct from the TNF complex II (56). Current models indicate that proinflammatory signaling in the TNF pathway is regulated upstream and independent of caspase-8/cFLIP, a possible explanation for our findings that TNF-induced NF-κB activation is not blocked by cFLIP isoforms.
There are a number of reports indicating that TRAF2 is recruited to DR complexes and thereby influence NF-κB, JNK, and p38 activation (57). Endogenous TRAF2 was shown to interact with cFLIPp43 and promotes formation of a cFLIPp43·caspase-8·TRAF2 tertiary complex. This complex was suggested to be a prerequisite for NF-κB activation in lymphocytes. Our study now investigated the differential outcomes of DR triggering for NF-κB activation. The CD95 DISC of HaCaT overexpressing cFLIPL or the cFLIPD376N mutant contained TRAF2 in a stimulation-dependent manner (compare supplemental Fig. S1), in line with another report in pancreatic tumor cells (48). However, we noticed that CD95-mediated suppression of NF-κB activation was not altered by the different cFLIP isoforms/mutants regardless of the recruitment of TRAF2. This indicates that TRAF2 recruitment to the CD95 DISC is not critical for DR-induced suppression of NF-κB activation. In line with these results, recent evidence for TNF has demonstrated that NF-κB activation in TRAF2 knock-out murine embryonic fibroblasts is unaltered (58). Thus, TRAF2 may either require other cFLIP-dependent secondary proteins or binding of TRAF2 to the DISC may be indirect. Taken together these observations indicate that cleavage of cFLIPL or caspase-8 in the DISC is neither associated with increased NF-κB signaling nor necessary for the inhibitory function of cFLIP isoforms on DR-induced NF-κB signaling.
Most of the experiments performed in this report have used the caspase inhibitor Z-VAD-fmk. A recent report has suggested that Z-VAD-fmk induces autocrine production of TNF that may in turn lead to gene induction (38). With a number of experimental approaches, this potential explanation for TRAIL-induced gene induction was excluded by comparison of Z-VAD-fmk and QVD. Both inhibitors failed to block TRAIL-induced IκBα phosphorylation although the increased gene induction in the presence of Z-VAD-fmk was less pronounced with QVD (compare supplemental Fig. S2, A and B). These data together with the studies using receptor fusion proteins sufficiently exclude that TRAIL-induced proinflammatory signals are mediated by a Z-VAD-fmk-induced autocrine loop of TNF.
Recent system biology approaches predicted that the stoichiometry of cFLIP and caspase-8 in the DISC is of critical importance for the outcome of receptor triggering (59, 60). Due to our overexpression study we currently cannot exclude that the levels of cFLIP expressed are higher than found under physiological or pathophysiological conditions. However, due to the fact that we can detect DISC-associated endogenous caspase-8 in cFLIPL- or cFLIPS-overexpressing cells, we believe that our experimental system bears physiological relevance. Of note, cFLIP represses the amounts of caspase-8 recruited to the DISC, although we still detect recruitment of endogenous caspase-8, indicative of the physiological relevance of the stoichiometry studied in our transduced HaCaT cells. Nonetheless, we aimed to further extend our studies to the situation of endogenous protein expression and have explored the role of cFLIP for DR signaling in PK that are (a) resistant to TRAIL-induced cell death and (b) express high levels of cFLIP when compared with HaCaT (24). Knockdown of endogenous cFLIP in PK increased the proinflammatory effect of TRAIL and CD95L in PK especially when apoptosis was blocked (Fig. 8). In the skin there is a constant need to repress proinflammatory gene induction and unwanted cell death, at least in the basal cell layer of the epidermis (61). This corresponds well with our data showing that cFLIP isoforms block inflammatory CD95 and TRAIL-R signaling irrespective of cell death induction. In the absence of caspase activity (e.g. inhibition with Z-VAD-fmk) in cells expressing low levels of cFLIP, we detect augmented NF-κB activation and target gene induction. Furthermore, PK with lowered levels of cFLIP showed increased activation of p38 or JNK in a caspase-dependent manner (Fig. 8A). Thus, our experiments in primary keratinocytes demonstrate that cFLIP protects from activation of non-apoptotic signaling pathways (JNK, p38, and NF-κB) at the level of the DISC. Importantly, this is not an epiphenomenon of cell death protection or a phenotype solely seen upon overexpression, arguing for cFLIP as an indispensable protein to protect from death receptor-induced apoptosis and inflammatory responses (Fig. 9). When total cellular caspase activity is inhibited, increased DISC-associated caspase activity in the absence of cFLIP promotes gene induction. We suggest that the gene-inductive signal is otherwise counteracted by cellular caspase activity (Fig. 9). This might be physiologically relevant in cells that do not undergo cell death (by e.g. Bcl-2 family proteins) in the presence of higher levels of intracellular caspase-8 activity. Clearly, alternative hypotheses exist to explain these surprising and discordant results for cFLIP and Z-VAD-fmk and are under current investigation. Nonetheless, the data presented in this report narrow explanations for these discordant phenotypes to differential effects in the DISC.
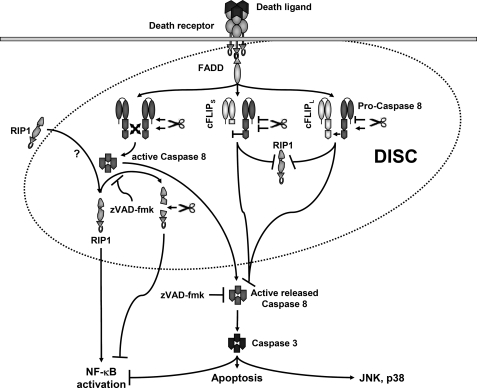
FIGURE 9.
DISC-associated proteins and the impact of inhibitors (cFLIP proteins or pharmacological caspase inhibitors) in DR-mediated signaling pathways in human keratinocytes. Upon stimulation of death receptors, the DISC forms in a stimulation-dependent manner. The adaptor protein FADD recruits caspase-8 and cFLIP isoforms via its DED. Caspase-8 is necessary and critical for apoptotic and the different non-apoptotic signals activated by death receptors CD95 and TRAIL-R. Recruitment of caspase-8 heterodimers lead to DISC-associated active caspase-8 that promotes RIP1 recruitment by thus far unknown mechanisms. Caspase-8 is subsequently released. The release of caspase-8 heterotetramers is required for p38 or JNK activation but inhibits NF-κB activation by caspase-mediated cleavage of NF-κB proteins (64). Active DISC-associated caspase-8 is able to cleave RIP-1 associated with the DISC, thereby counteracting NF-κB activation (65). Z-VAD-fmk increases DR-induced NF-κB activation through inhibition of DISC-associated RIP-1 cleavage by active caspase-8 heterotetramers (13, 40, 66). High cellular levels of cFLIPL or cFLIPS inhibit RIP-1 recruitment to the DISC and the full activation of caspase-8 heterotetramers within the DISC irrespective of partial cleavage of caspase-8 or cFLIPL in the DISC (by cFLIPL-Caspase-8 heterodimers). The caspase activity associated with the heterodimer of cFLIP and caspase-8 remains membrane (DISC)-bound, thereby lacking proapoptotic or gene inductive function. Active caspases downstream of the DISC activate effector caspases, JNK, and p38, whereas NF-κB activation is blocked.
Tumor cells acquire various characteristics to evade the host immune system. Previous reports indicated that, surprisingly, ligation of DRs may result in the activation of chemokine expression (IL-8, MCP-1) via activation of MAPK p38 and JNK by AP-1- and/or NF-κB-dependent transcription. These signals result in granulocyte invasion, angiogenesis, and inflammation at the site of the tumor and may be important for tumor promoting functions such as proliferation, migration, inflammation, or metastasis (62). This mechanism probably explains why DR signals could favor tumor progression under some circumstances when apoptosis induction by DRs is blocked downstream of the DISC, as shown in a pancreatic tumor model (63). It is tempting to speculate that the inhibitory effect of FLIP for TRAIL- and CD95L-induced NF-κB activation acts as a safety mechanism. cFLIP may act by dual action in the DISC: cFLIP reduces apoptosis induction, and also blocks proinflammatory signaling. Such a dual inhibitory role of cFLIP may ultimately be of advantage for tumor cells to evade tumor surveillance. Understanding the detailed mechanism of apoptotic and non-apoptotic responses to DR stimulation is therefore crucial to target tumor cells effectively by DR agonists in the future.
Supplementary Material
Acknowledgments
We are grateful to P. H. Krammer for mAbs to caspase-8 (C-15) and cFLIP (NF-6), H. Walczak and T. Haas for recombinant TRAIL, H. Mehmet for CPP32 antiserum, and P. Schneider for CD95L-Fc, CD95-Fc, and TRAIL-R2-Fc expression plasmids. We thank P. Boukamp, R. Byers, and M. Herlyn for providing cell lines and M. Sprick, T. Haas, and D. Brenner for helpful discussions. We are indebted to B. Baumann and S. Ludwig for methodological advice for kinase assays and constructs.
This work was supported by Deutsche Forschungsgemeinschaft Grants Le 953/5-1 and Le 953/6-1, Deutsche Forschungsgemeinschaft Graduiertenkolleg 1167, Wilhelm-Sander-Stiftung Grant 2008.072.1, and Deutsche Krebshilfe Grant 106849 (to M. L.). S. M. K. and M. F. were supported by the “Center of Excellence Dermatology Mannheim of Baden-Württemberg.”

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- DR
- death receptor
- TRAIL
- tumor necrosis factor-related apoptosis inducing ligand
- AP-1
- activated protein kinase-1
- IL-8
- interleukin-8
- cFLIP
- cellular FLICE-inhibitory protein
- Z
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone
- QVD
- quinolyl-valyl-O-methylaspartyl-[2,6-difluoro-phenoxy]-methyl ketone
- RIP-2
- receptor interacting protein-1
- TRAF
- TNF-receptor associated factor 2
- PK
- primary keratinocytes
- DISC
- death inducing signaling complex
- DED
- death effector domain
- qPCR
- quantitative PCR
- DL
- death ligand.
REFERENCES
- 1. Krammer P. H., Kamiński M., Kiessling M., Gülow K. (2007) Adv. Cancer Res. 97, 111–138 [DOI] [PubMed] [Google Scholar]
- 2. Falschlehner C., Emmerich C. H., Gerlach B., Walczak H. (2007) Int. J. Biochem. Cell Biol. 39, 1462–1475 [DOI] [PubMed] [Google Scholar]
- 3. Park H. H., Lo Y. C., Lin S. C., Wang L., Yang J. K., Wu H. (2007) Annu. Rev. Immunol. 25, 561–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Duckett C. S. (2005) Biochem. J. 385, e1–e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Silke J., Kratina T., Chu D., Ekert P. G., Day C. L., Pakusch M., Huang D. C., Vaux D. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16182–16187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Budd R. C., Yeh W. C., Tschopp J. (2006) Nat. Rev. Immunol. 6, 196–204 [DOI] [PubMed] [Google Scholar]
- 7. Kataoka T. (2005) Crit. Rev. Immunol 25, 31–58 [DOI] [PubMed] [Google Scholar]
- 8. Yu J. W., Shi Y. (2008) Oncogene 27, 6216–6227 [DOI] [PubMed] [Google Scholar]
- 9. Scaffidi C., Schmitz I., Krammer P. H., Peter M. E. (1999) J. Biol. Chem. 274, 1541–1548 [DOI] [PubMed] [Google Scholar]
- 10. Boatright K. M., Deis C., Denault J. B., Sutherlin D. P., Salvesen G. S. (2004) Biochem. J. 382, 651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krueger A., Schmitz I., Baumann S., Krammer P. H., Kirchhoff S. (2001) J. Biol. Chem. 276, 20633–20640 [DOI] [PubMed] [Google Scholar]
- 12. Geserick P., Drewniok C., Hupe M., Haas T. L., Diessenbacher P., Sprick M. R., Schön M. P., Henkler F., Gollnick H., Walczak H., Leverkus M. (2008) Oncogene 27, 3211–3220 [DOI] [PubMed] [Google Scholar]
- 13. Wachter T., Sprick M., Hausmann D., Kerstan A., McPherson K., Stassi G., Bröcker E. B., Walczak H., Leverkus M. (2004) J. Biol. Chem. 279, 52824–52834 [DOI] [PubMed] [Google Scholar]
- 14. Farley S. M., Dotson A. D., Purdy D. E., Sundholm A. J., Schneider P., Magun B. E., Iordanov M. S. (2006) J. Invest. Dermatol. 126, 2438–2451 [DOI] [PubMed] [Google Scholar]
- 15. Leverkus M., Diessenbacher P., Geserick P. (2008) Exp. Dermatol. 17, 614–622 [DOI] [PubMed] [Google Scholar]
- 16. Leverkus M., Sprick M. R., Wachter T., Mengling T., Baumann B., Serfling E., Bröcker E. B., Goebeler M., Neumann M., Walczak H. (2003) Mol. Cell. Biol. 23, 777–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Diessenbacher P., Hupe M., Sprick M. R., Kerstan A., Geserick P., Haas T. L., Wachter T., Neumann M., Walczak H., Silke J., Leverkus M. (2008) J. Invest. Dermatol. 128, 1134–1147 [DOI] [PubMed] [Google Scholar]
- 18. Bossen C., Ingold K., Tardivel A., Bodmer J. L., Gaide O., Hertig S., Ambrose C., Tschopp J., Schneider P. (2006) J. Biol. Chem. 281, 13964–13971 [DOI] [PubMed] [Google Scholar]
- 19. Berg D., Lehne M., Müller N., Siegmund D., Münkel S., Sebald W., Pfizenmaier K., Wajant H. (2007) Cell Death Differ. 14, 2021–2034 [DOI] [PubMed] [Google Scholar]
- 20. Mueller M. M., Peter W., Mappes M., Huelsen A., Steinbauer H., Boukamp P., Vaccariello M., Garlick J., Fusenig N. E. (2001) Am. J. Pathol. 159, 1567–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boukamp P., Petrussevska R. T., Breitkreutz D., Hornung J., Markham A., Fusenig N. E. (1988) J. Cell Biol. 106, 761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schmidt M., Hupe M., Endres N., Raghavan B., Kavuri S., Geserick P., Goebeler M., Leverkus M. (2010) J. Cell Mol. Med. 14, 1760–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leverkus M., Sprick M. R., Wachter T., Denk A., Bröcker E. B., Walczak H., Neumann M. (2003) J. Invest. Dermatol. 121, 149–155 [DOI] [PubMed] [Google Scholar]
- 24. Leverkus M., Neumann M., Mengling T., Rauch C. T., Bröcker E. B., Krammer P. H., Walczak H. (2000) Cancer Res. 60, 553–559 [PubMed] [Google Scholar]
- 25. Nicoletti I., Migliorati G., Pagliacci M. C., Grignani F., Riccardi C. (1991) J. Immunol. Methods 139, 271–279 [DOI] [PubMed] [Google Scholar]
- 26. Leverkus M., Jochim R. C., Schäd S., Bröcker E. B., Andersen J. F., Valenzuela J. G., Trautmann A. (2006) J. Invest. Dermatol. 126, 91–96 [DOI] [PubMed] [Google Scholar]
- 27. Azoitei N., Wirth T., Baumann B. (2005) J. Neurochem. 93, 1487–1501 [DOI] [PubMed] [Google Scholar]
- 28. Geserick P., Hupe M., Moulin M., Wong W. W., Feoktistova M., Kellert B., Gollnick H., Silke J., Leverkus M. (2009) J. Cell Biol. 187, 1037–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schmidt M., Goebeler M., Posern G., Feller S. M., Seitz C. S., Brocker E. B., Rapp U. R., Ludwig S. (2000) J. Biol. Chem. 275, 41011–41017 [DOI] [PubMed] [Google Scholar]
- 30. Chang D. W., Xing Z., Capacio V. L., Peter M. E., Yang X. (2003) EMBO J. 22, 4132–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thome M., Tschopp J. (2001) Nat. Rev. Immunol. 1, 50–58 [DOI] [PubMed] [Google Scholar]
- 32. Golks A., Brenner D., Krammer P. H., Lavrik I. N. (2006) J. Exp. Med. 203, 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jin Z., Li Y., Pitti R., Lawrence D., Pham V. C., Lill J. R., Ashkenazi A. (2009) Cell 137, 721–735 [DOI] [PubMed] [Google Scholar]
- 34. Kreuz S., Siegmund D., Rumpf J. J., Samel D., Leverkus M., Janssen O., Häcker G., Dittrich-Breiholz O., Kracht M., Scheurich P., Wajant H. (2004) J. Cell Biol. 166, 369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peter M. E., Budd R. C., Desbarats J., Hedrick S. M., Hueber A. O., Newell M. K., Owen L. B., Pope R. M., Tschopp J., Wajant H., Wallach D., Wiltrout R. H., Zörnig M., Lynch D. H. (2007) Cell 129, 447–450 [DOI] [PubMed] [Google Scholar]
- 36. Wicovsky A., Müller N., Daryab N., Marienfeld R., Kneitz C., Kavuri S., Leverkus M., Baumann B., Wajant H. (2007) J. Biol. Chem. 282, 2174–2183 [DOI] [PubMed] [Google Scholar]
- 37. Goebeler M., Gillitzer R., Kilian K., Utzel K., Bröcker E. B., Rapp U. R., Ludwig S. (2001) Blood 97, 46–55 [DOI] [PubMed] [Google Scholar]
- 38. Wu Y. T., Tan H. L., Huang Q., Sun X. J., Zhu X., Shen H. M. (2011) Cell Death. Differ. 18, 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Banno T., Gazel A., Blumenberg M. (2005) J. Biol. Chem. 280, 18973–18980 [DOI] [PubMed] [Google Scholar]
- 40. Harper N., Farrow S. N., Kaptein A., Cohen G. M., MacFarlane M. (2001) J. Biol. Chem. 276, 34743–34752 [DOI] [PubMed] [Google Scholar]
- 41. Ashkenazi A. (2002) Nat. Rev. Cancer 2, 420–430 [DOI] [PubMed] [Google Scholar]
- 42. Matta H., Sun Q., Moses G., Chaudhary P. M. (2003) J. Biol. Chem. 278, 52406–52411 [DOI] [PubMed] [Google Scholar]
- 43. Sun Q., Zachariah S., Chaudhary P. M. (2003) J. Biol. Chem. 278, 52437–52445 [DOI] [PubMed] [Google Scholar]
- 44. Dohrman A., Kataoka T., Cuenin S., Russell J. Q., Tschopp J., Budd R. C. (2005) J. Immunol. 174, 5270–5278 [DOI] [PubMed] [Google Scholar]
- 45. Kataoka T., Budd R. C., Holler N., Thome M., Martinon F., Irmler M., Burns K., Hahne M., Kennedy N., Kovacsovics M., Tschopp J. (2000) Curr. Biol. 10, 640–648 [DOI] [PubMed] [Google Scholar]
- 46. Kataoka T., Tschopp J. (2004) Mol. Cell. Biol. 24, 2627–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ehrenschwender M., Siegmund D., Wicovsky A., Kracht M., Dittrich-Breiholz O., Spindler V., Waschke J., Kalthoff H., Trauzold A., Wajant H. (2010) Cell Death. Differ. 17, 1435–1447 [DOI] [PubMed] [Google Scholar]
- 48. Siegmund D., Klose S., Zhou D., Baumann B., Röder C., Kalthoff H., Wajant H., Trauzold A. (2007) Cell Signal. 19, 1172–1184 [DOI] [PubMed] [Google Scholar]
- 49. Oberst A., Pop C., Tremblay A. G., Blais V., Denault J. B., Salvesen G. S., Green D. R. (2010) J. Biol. Chem. 285, 16632–16642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hughes M. A., Harper N., Butterworth M., Cain K., Cohen G. M., MacFarlane M. (2009) Mol. Cell 35, 265–279 [DOI] [PubMed] [Google Scholar]
- 51. Rahighi S., Ikeda F., Kawasaki M., Akutsu M., Suzuki N., Kato R., Kensche T., Uejima T., Bloor S., Komander D., Randow F., Wakatsuki S., Dikic I. (2009) Cell 136, 1098–1109 [DOI] [PubMed] [Google Scholar]
- 52. Tokunaga F., Sakata S., Saeki Y., Satomi Y., Kirisako T., Kamei K., Nakagawa T., Kato M., Murata S., Yamaoka S., Yamamoto M., Akira S., Takao T., Tanaka K., Iwai K. (2009) Nat. Cell Biol. 11, 123–132 [DOI] [PubMed] [Google Scholar]
- 53. Haas T. L., Emmerich C. H., Gerlach B., Schmukle A. C., Cordier S. M., Rieser E., Feltham R., Vince J., Warnken U., Wenger T., Koschny R., Komander D., Silke J., Walczak H. (2009) Mol. Cell 36, 831–844 [DOI] [PubMed] [Google Scholar]
- 54. Varfolomeev E., Maecker H., Sharp D., Lawrence D., Renz M., Vucic D., Ashkenazi A. (2005) J. Biol. Chem. 280, 40599–40608 [DOI] [PubMed] [Google Scholar]
- 55. Wong W. W., Gentle I. E., Nachbur U., Anderton H., Vaux D. L., Silke J. (2010) Cell Death Differ. 17, 482–487 [DOI] [PubMed] [Google Scholar]
- 56. Micheau O., Tschopp J. (2003) Cell 114, 181–190 [DOI] [PubMed] [Google Scholar]
- 57. Devin A., Lin Y., Liu Z. G. (2003) EMBO Rep. 4, 623–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang L., Blackwell K., Thomas G. S., Sun S., Yeh W. C., Habelhah H. (2009) J. Mol. Biol. 389, 495–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Neumann L., Pforr C., Beaudouin J., Pappa A., Fricker N., Krammer P. H., Lavrik I. N., Eils R. (2010) Mol. Syst. Biol. 6, 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fricker N., Beaudouin J., Richter P., Eils R., Krammer P. H., Lavrik I. N. (2010) J. Cell Biol. 190, 377–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kerstan A., Leverkus M., Trautmann A. (2009) Exp. Dermatol. 18, 893–899 [DOI] [PubMed] [Google Scholar]
- 62. Choi C., Xu X., Oh J. W., Lee S. J., Gillespie G. Y., Park H., Jo H., Benveniste E. N. (2001) Cancer Res. 61, 3084–3091 [PubMed] [Google Scholar]
- 63. Trauzold A., Siegmund D., Schniewind B., Sipos B., Egberts J., Zorenkov D., Emme D., Röder C., Kalthoff H., Wajant H. (2006) Oncogene 25, 7434–7439 [DOI] [PubMed] [Google Scholar]
- 64. Kim H. S., Chang I., Kim J. Y., Choi K. H., Lee M. S. (2005) Cancer Res. 65, 6111–6119 [DOI] [PubMed] [Google Scholar]
- 65. Martinon F., Holler N., Richard C., Tschopp J. (2000) FEBS Lett. 468, 134–136 [DOI] [PubMed] [Google Scholar]
- 66. Micheau O., Thome M., Schneider P., Holler N., Tschopp J., Nicholson D. W., Briand C., Grütter M. G. (2002) J. Biol. Chem. 277, 45162–45171 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.