Abstract
Alginate is a major cell wall polymer of brown algae. The precursor for the polymer is GDP-mannuronic acid, which is believed to be derived from a four-electron oxidation of GDP-mannose through the enzyme GDP-mannose dehydrogenase (GMD). So far no eukaryotic GMD has been biochemically characterized. We have identified a candidate gene in the Ectocarpus siliculosus genome and expressed it as a recombinant protein in Escherichia coli. The GMD from Ectocarpus differs strongly from related enzymes in bacteria and is as distant to the bacterial proteins as it is to the group of UDP-glucose dehydrogenases. It lacks the C-terminal ∼120 amino acid domain present in bacterial GMDs, which is believed to be involved in catalysis. The GMD from brown algae is highly active at alkaline pH and contains a catalytic Cys residue, sensitive to heavy metals. The product GDP-mannuronic acid was analyzed by HPLC and mass spectroscopy. The Km for GDP-mannose was 95 μm, and 86 μm for NAD+. No substrate other than GDP-mannose was oxidized by the enzyme. In gel filtration experiments the enzyme behaved as a dimer. The Ectocarpus GMD is stimulated by salts even at low molar concentrations as a possible adaptation to marine life. It is rapidly inactivated at temperatures above 30 °C.
Keywords: Carbohydrate Biosynthesis, Cell Wall, Dehydrogenase, Enzyme Catalysis, Metabolism, NAD, Nucleoside Nucleotide Metabolism, Plant, Alginate, Nucleotide Sugar Conversion
Introduction
Ectocarpus siliculosus is a marine photoautotrophic brown alga (Phaeophyceae) which, together with oomycetes and diatoms, belongs to the phylum Stramenopiles. This phylum originated approximately 1 billion years ago as a result of a secondary endosymbiotic event, in which a unicellular red alga was captured by an ancestral protist (1).
The cell walls of brown algae differ from those of land plants (for a recent review, see Ref. 2). Along with a minor amount of cellulose, the major cell wall polysaccharide of brown algae is alginate, which accounts for up to 45% of the dry weight (3). The function of alginate in the cell walls of brown algae is similar to the function of cellulose in the cell walls of land plants. Alginate is an unbranched polysaccharide initially synthesized as a β-1,4-d-mannuronic acid chain (M-alginate). The polymer is later modified by C-5 epimerases, which convert single residues or larger blocks of the polymer from d-mannuronic acid into l-guluronic acid (G-alginate). The M- and G-blocks in alginate vary during algae development. G-block-rich alginate will form a high strength gel in the presence of divalent cations (e.g. Ca2+). M-blocks are more flexible and are preferentially found in blades exposed to wave action (3). The genes for C-5 epimerase converting M- into G-alginate have been identified in the brown alga Laminaria digitata (4). L. digitata possesses six different genes for such epimerases, which all show homology to characterized alginate epimerases (AlgG) from Pseudomonas aeruginosa. Apart from these epimerases, no other enzyme of the alginate pathway has been characterized at the molecular level (2). Bacterial alginate has similar properties to brown algal alginate but is often additionally modified by O-acetyl groups at C2 or C3 (5).
The commercial interest in alginate is manifold. Alginates are widely used to form gels e.g. to immobilize enzymes, to thicken food, or to cover organs during transplantation as a barrier between the transplant and the host immune system (6).
The precursor of alginate for alginate-producing bacteria such as P. aeruginosa (7) and the brown algae Fucus gardneri (8) is GDP-mannuronic acid (GDP-ManA).2 So far, no eukaryotic enzyme for the production of GDP-ManA has been identified and characterized biochemically. GDP-ManA is derived by a four-electron oxidation of GDP-mannose (GDP-Man). The enzyme GDP-mannose dehydrogenase (GMD; EC 1.1.1.132) catalyzes the reaction GDP-Man + 2NAD+ → GDP-ManA + 2 NADH and was first purified and characterized from Pseudomonas (9). The enzyme belongs to the superfamily of UDP-glucose/GDP-mannose dehydrogenases which are ubiquitously present in all organisms. In bacteria this superfamily includes enzymes that oxidize different sugars such as UDP-glucose, GDP-mannose, UDP-N-acetylglucosamine, UDP-N-acetyl-d-mannosaminuronic acid, and UDP-galactose. All known eukaryotic members of the group convert only UDP-glucose. Therefore the recently published genome of the brown algae E. siliculosus (10) provides an excellent tool to search for and characterize a first eukaryotic GMD.
We are studying UDP-glucose dehydrogenases from plants, which are involved in providing UDP-sugars for pectic polymers, arabinans, and xyloglucans. All of these enzymes are highly substrate-specific and convert only UDP-glucose into UDP-glucuronic acid. The genome of E. siliculosus contains a gene with high homology to UDP-glucose dehydrogenases, suggesting to us that the alginate precursor GDP-ManA is likely generated by a specific GMD enzyme and not by UDP-glucose dehydrogenase with broad substrate specificity. Here, we characterize a unique recombinant GMD enzyme from Ectocarpus at the molecular level and compare its structure and properties with bacterial enzymes.
EXPERIMENTAL PROCEDURES
Chemicals
LB medium, IPTG, and glycine were from Duchefa, Tris buffer, all nucleotide sugars, nucleotides, and inhibitors were from Sigma. Phosphate buffers salts and NAD+ were from Carl Roth (Karlsruhe, Germany). All chemicals were analytical grade or of higher purity.
Cloning of the GMD Gene
To identify candidate genes for GDP-Man dehydrogenase we searched the E. siliculosus genome sequence and the GenBank EST library using the soybean UDP-glucose dehydrogenase gene as a query. The blast search revealed four significant hits (see Fig. 1), a close homolog of the UDP-glucose dehydrogenase and three distantly related sequences, considered to be candidates for GDP-Man dehydrogenases. One candidate gene for GDP-Man dehydrogenase was amplified by PCR from E. siliculosus cDNA. RNA was reverse-transcribed using the RevertAid enzyme (Fermentas, Thermo Scientific). The following primers (forward, atggatccatgccaggaaaggagaacg; reverse, tgaagcttccagcgagctcaacgtgt) were used to amplify the GMD gene with Phusion proofreading polymerase. The PCR program was as follows: 98 °C 30s; 25× (98 °C 5s; 56 °C 15s; 72 °C 20s); 72 °C 2 min. The PCR product was cleaved with BamHI and HindIII and cloned into the Escherichia coli expression vector pMBP-Parallel1 (11), which was modified to allow fusions with a His6-tagged maltose-binding protein (MBP) to be generated. The fusion proteins encoded by this construct possessed a TEV protease cleavage site between the MBP and the EsGMD1 protein. The sequence of the fusion construct was verified by DNA sequencing (Eurofins).
FIGURE 1.
Unrooted phylogenetic tree of selected nucleotide sugar dehydrogenases and structural motifs. A, amino acid sequences were aligned with ClustalX, and a bootstrapped neighbor joining tree was used to calculate the distance between the sequences. UGDs: soybean, U53418; Arabidopsis, At3g29360; Ectocarpus, Esi0101_0043; human, NP_003350; Drosophila, AAC97125; Physcomitrella patens, XP_001761641; Chlorella variabilis, EFN57705; Volvox carteri, XP_002945908; Ostreococcus tauri, XP_003082645; Branchiostoma floridae, XP_002606188. Eukaryotic GMDs: Ectocarpus GMD1, Esi0164_0053; Ectocarpus GMD2, Esi0051_0113; Ectocarpus GMD3, Esi0051_0092; Sargassum binderi, ESTs DV668856 + DV669914; Fucus serratus, ESTs GH701299 + GH69914; Laminaria digitata, ESTs CN466747 + CN468196 + CN466724; Fucus vesiculosus, GH704760. Prokaryotic GMDs: P. aeruginosa, P11759; Pseudomonas syringae, P59793; Azotobacter, P51585; Mycobacterium smegmatis, YP_890183; Vibrio sp., ABP49558; Marinobacter algicola, ZP_01893737; Sphaerobacter thermophilus, YP_003319392; Streptomyces violaceusniger, ZP_07604805. B, from the sequences shown in the tree in A the NAD+ binding site close to the N terminus and the region around the catalytic Cys residue are shown. Note that the two sequence motifs are highly conserved in all three groups of proteins but still show group specific differences. C, two representative sequences from each subgroup were compared for sequence identity and sequence similarity. The identity among different subgroups is low ∼20%. D, ligand-binding amino acid residues from different UGDs and GMDs are compared.
Expression in E. coli and Protein Purification
The expression vector was transformed into T7-Express E. coli cells (NEB Biolabs). For the production of recombinant proteins, 400 ml of E. coli culture in a 2-liter flask was grown to an A600 nmof ∼0.4 at 37 °C, cooled to 25 °C, and induced with 0.5 mm IPTG for 16 h. Cells were then collected by centrifugation and stored frozen. The protein was purified with a Protrino 1000 column (Machery-Nagel) following the manufacturer's protocol, except that is was necessary to supplement all buffers with 200 μm NAD+ to maintain the enzyme in an active form. All purification procedures were carried out on ice. The eluate of the Protrino column was applied to a small gel filtration column (PD10; GE Healthcare) to bring the enzyme into a suitable storage buffer (20 mm Tris-Cl, pH 8, 50 mm KCl, 200 μm NAD+, 1 mm DTT, 1 mm EDTA, 20% glycerol). The enzyme was snap frozen in aliquots in liquid nitrogen and stored at −80 °C. Aliquots retain their activity over several months when stored at −80 °C.
Enzyme Assay
The routine enzyme assay is based on the increase of NADH measured as an increase in A340 nm. This can be performed easily in microcuvettes as well as in microtiter plates. The standard assay buffer consisted of 50 mm Tris-glycine buffer, pH 8.75, 1 mm NAD+, 0.5 mm GDP-Man, to which the EsGMD1 enzyme (typically 20–30 μg) was added. We compared the kinetic parameters of the MBP-EsGMD1 fusion protein with the TEV-cleaved EsGMD1 alone but did not find any differences in the catalytic properties. Therefore, the MBP fusion protein was used routinely in the experiments.
Cleavage at the TEV Site
One mg of EsGMD1 was incubated in 50 mm NaCl, 1 mm EDTA, 2 mm DTT, 50 mm Tris-Cl, pH 7.5, with 75 μg recombinant TEV protease (a kind gift from Prof. Hans Brandstetter, Department of Structural Biology, University of Salzburg) at 10 °C for 5 h. The activity of the GMD remains constant over this period. The TEV protease releases the pure EsGMD1 in an active form. The protein was applied to a Superose 12 column (GE Healthcare) to determine the size of the protein by gel permeation chromatography. A 50 mm Tris-Cl, pH 8, buffer with 150 mm NaCl was used at a flow rate of 0.4 ml min−1 to separate proteins by size. Fractions of 0.33 ml were analyzed for enzymatic activity. Standard proteins of known molecular mass were run under identical conditions.
HPLC Analysis of Products of the Enzyme Assay
The standard EsGMD1 enzyme assay was stopped by adding 1 volume of sodium phosphate buffer (100 mm, pH 3). The assay was cleared by a 5-min centrifugation and applied to a Partisil 10 SAX column (3 × 125 mm). Buffer A was 10 mm sodium phosphate, pH 3. Buffer B was 750 mm sodium phosphate, pH 3.7. The following method was used for separation: flow rate 0.75 ml min−1; t0 min 3% B; t25 min 40% B; t33 min 75% B; t35 min 75% B; t36 min 3% B. A UV spectrum (240–300 nm) was recorded with a photodiode array (Dionex Ultimate 3000) and analyzed with the Chromeleon software.
Direct Infusion Experiments and MS Parameters
To prepare metabolites for ESI-Orbitrap-MS measurements a standard assay was incubated for 2 h to allow the formation of sufficient GDP-ManA. The assay (1 ml) was applied to a 100-mg EnviCarb solid phase extraction column (Supelco) that had been activated before use by a short wash with 80% acetonitrile/0.1% TFA and equilibrated with 3 ml of water. The column was washed with 2 ml of H2O to wash out buffer salts. The bound nucleotide sugars were eluted with 2 ml of 50% acetonitrile and concentrated 5-fold in an Eppendorf vacuum centrifugator.
For direct infusion experiments, an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific) equipped with an Ion Max ESI source was fine-tuned in negative ionization mode, using 0.5 μg/ml GDP-Man at m/z = 604.07 as the tuning mass. Finally, the solid-phase extraction column purified assay was diluted 1/1 with acetonitrile containing 0.1% formic acid as additive. Acetonitrile (Optigrade for LC/MS) was purchased from Promochem (Wesel, Germany) and formic acid (98–100%, puriss.) was from Riedel-de Haën (Hannover, Germany). The solution was directly infused using a syringe pump with a flow rate of 5.0 μl · min−1. The mass spectrometric parameters were as follows: mass analyzer, FTMS; resolution, 30,000; scan rate, normal; maximal injection time, 10 ms; ion polarity, negative. The optimized ESI source parameters were: source voltage, 2.5 kV; capillary voltage, −35 V; capillary temperature, 280 °C; sheath gas flow, 15 arb. units; aux gas flow, 5 arb. units; tube lens, −89.32 V. The molecular formulas and mass deviations were calculated by using XCalibur software version 2.0.7 (Thermo Fisher Scientific).
RESULTS
Phylogenetic Analysis of Ectocarpus GMD
The recent sequencing of the alginate producing brown alga E. siliculosus (10) allowed us to search for nucleotide sugar dehydrogenases, which provide the alginate precursor GDP-ManA. So far in eukaryotes, only one type of nucleotide sugar dehydrogenase, UDP-glucose dehydrogenase (UGD), has been identified and characterized biochemically. UGDs are ubiquitously present in animals, fungi, and plants. Indeed, E. siliculosus contains a single gene, which clusters with UGD sequences from plants and animals (Fig. 1A) but in addition has three further genes that are clearly distinct from the UGD and are thus candidate genes for a GMD. A multiple alignment of bacterial GMDs, the E. siliculosus candidate GMD, related candidate genes from brown algae assembled from the GenBank EST database and a selection of UGDs is shown as a tree in Fig. 1A and as full alignment in supplemental Fig. S1. As expected, all UGDs cluster in one branch of the unrooted tree, as do the bacterial GMDs. However, the GMDs from E. siliculosus and four other brown algae fall into a distinct third branch almost as distant from the bacterial GMDs as from the UGDs. Because the three E. siliculosus GMD sequences are highly similar to each other, we decided to focus on one of the genes EsGMD1, which is encoded by Esi0164_0053 and for which there are several EST sequences (supplemental Fig. S2).
Structural Motif Conservation
The full alignment of all sequences from the tree in Fig. 1A is shown in supplemental Fig. S1. The protein sequences of the GMD from P. aeruginosa and EsGMD1 from E. siliculosus are broadly collinear with the exception of a 19-amino acid gap in the EsGMD1 sequence. However, EsGMD1 is considerably shorter than the bacterial GMDs and lacks the C-terminal extensions of these proteins. All three E. siliculosus GMDs as well as the GMDs from Laminaria, Fucus, and Sargassum, lack the C-terminal extension typically found in all bacterial GMDs (supplemental Fig. S1). The sequence of EsGMD1 was corroborated by finding eight overlapping EST clones spanning the full-length coding sequence of the gene (supplemental Fig. S2). Therefore, we consider the EsGMD1 sequence as a full-length open reading frame.
Two signatures of nucleotide sugar dehydrogenases, the NAD+ binding site at the N terminus and the catalytic center around a highly conserved cysteine residue are shown in Fig. 1B for all the proteins of the phylogenetic tree from Fig. 1A. The three clusters, eukaryotic UGDs, eukaryotic GMDs, and prokaryotic GMDs each have distinct sequence motifs around the conserved NAD+ binding site as well around the catalytic site, which can be considered as typical for each of the subgroups (Fig. 1B). The overall sequence identity/similarity between two bacterial GMDs, two brown algae GMDs, and two plant UGDs is shown in Fig. 1C. The identity between bacterial and brown algae GMDs is ∼17% (∼26% similarity).
The GMD protein from P. aeruginosa (12) and the UGD protein from S. pyogenes (13) were recently crystallized. The structure prediction tool Phyre indicates a similar overall structure of the Ectocarpus GMD compared with the crystallized UGD and GMD proteins. Thus substrate interacting amino acid residues were identified. By combining the information from the publications by Snook et al. (12) and Campbell et al. (13) along with our sequence alignment we were able to predict putative substrate interacting residues of the E. siliculosus GMD (Fig. 1D). Substrate-interacting amino acid residues of the bacterial GMD (Protein Data Bank ID code 1MUU) are shown in supplemental Fig. S3. Most of the substrate binding residues are identical in UGD and GMD sequences, indicating a common evolutionary origin. Notably, the Arg244 from S. pyogenes UGD which makes hydrogen bonds to the OH groups at C2 and C3 from the glucose of UDP-glucose is replaced by a Glu residue in EsGMD1. This could be a structural requirement for binding GDP-Man, which differs in the steric orientation of the C2-OH group from mannose relative to glucose.
Expression and Purification of Recombinant GMD
To characterize and confirm the GMD function of EsGMD1, we expressed the enzyme as a recombinant fusion protein with the MBP. Attempts to obtain EsGMD1 with a His tag alone were unsuccessful and resulted in inclusion body formation under a variety of conditions (5–500 μm IPTG as inductor; 37 °C −15 °C for induction; different E. coli strains). The engineered fusion protein contains a TEV protease cleaving site in front of the GMD protein which allows it to be cleaved from the MBP fusion partner (Fig. 2). On a 10% SDS-PAGE the purified EsGMD1 ran as a 34-kDa protein consistent with the predicted theoretical mass of 34.528 kDa.
FIGURE 2.
A, SDS-PAGE of recombinant MBP-GMD from Ectocarpus. First lane, recombinant EsGMD1 as MBP fusion protein. Second lane, EsGMD1 after cleavage and removal of the MBP (marked with an asterisk). Third lane, molecular mass markers. Fourth lane, recombinant TEV protease used to cleave off the MBP. B, size exclusion chromatography of EsGMD1 (without MBP). 200-μl samples were separated on a Superose 12 column, and fractions of 0.33 ml were collected and analyzed for activity (black bars). The column was calibrated with standard proteins of known molecular mass (A, cytochrome c; B, β-lactoglobulin; C, BSA; D, catalase). The EsGMD1 protein (34 kDa) elutes at approximately 68 kDa, suggesting that it runs as a dimer.
The Pseudomonas GMD is believed to be active as a hexamer (9, 14). We therefore separated EsGMD1 protein on a Superose 12 FPLC size exclusion column. GMD activity plotted against the molecular mass is shown in Fig. 2B, revealing a maximum between 65 and 70 kDa. This fits very well with the hypothesis that EsGMD1 forms a dimeric complex.
Enzyme Assay and Product Confirmation
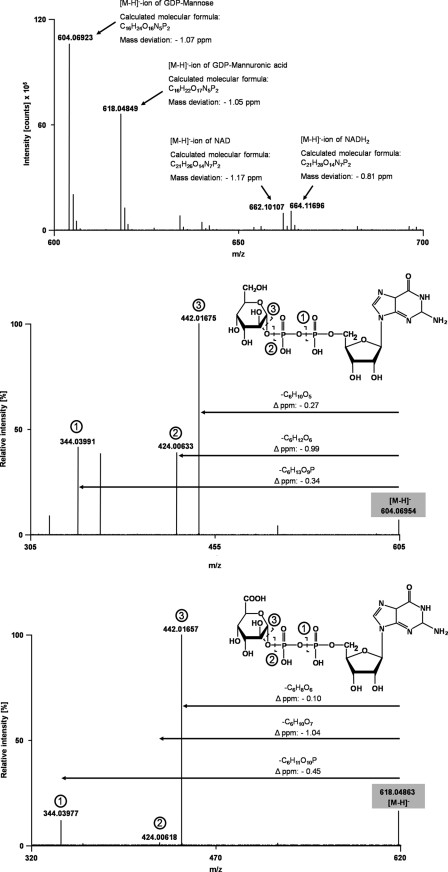
The enzymatic activity of EsGMD1 was confirmed by HPLC and ESI-MS analysis. A series of HPLC chromatograms is shown in Fig. 3. The commercial substrate GDP-Man (4) was separated in the uppermost trace. The inset shows a typical guanosine UV spectrum. The second trace shows a control assay without the recombinant enzyme added. Compound 1 is NAD+, 2 is an unknown impurity in the NAD+. The two bottom chromatograms show two time points of a typical GMD assay after 5 and 60 min, respectively. Compound 3 is NADH, a product of the reaction. The novel compound 5 is GDP-ManA and has the same UV spectrum as GDP-Man. A comparison of the peak areas of the substrate GDP-Man (4) and the product GDP-ManA (5) for several assays shows a constant peak area sum of both metabolites indicating the direct conversion of 4 into 5. To verify the identity of the GMD-ManA we purified the nucleotide sugars from an enzyme assay by solid phase extraction on an EnviCarb column. The salt-free extract was then analyzed by ESI-Orbitrap-MS (Fig. 4). The signals of GDP-Man, GDP-ManA, NAD+, and NADH were readily identified with high abundance and mass accuracies in the sub-ppm range. GDP-ManA shows the predicted mass increase of 14 mass units resulting from the oxidation of the alcoholic C6 group into a carboxylic acid group. CID fragmentation of GDP-Man and GDP-ManA results in the loss of Man/ManA either with or without the oxygen atom at C1 of the sugar (Fig. 4, middle and bottom).
FIGURE 3.
HPLC chromatography of enzyme assays. The top trace shows the separation of the substrate GDP-Man and a UV spectrum as the inset. The second trace shows an enzyme assay, from which the EsGMD1 enzyme was omitted. The two bottom traces show a full enzyme assay after 5 and 60 min, respectively. The UV spectrum of the product GDP-ManA (5) is shown in the lower inset (1). NAD+; 2, unknown impurity from NAD+; 3, NADH; 4, GDP-Man; 5, GDP-ManA.
FIGURE 4.
Top, high resolution full-scan mass spectrum of the solid-phase extraction column-purified assay mixture in a mass range of m/z = 600–700, applying negative ionization mode. Middle, high resolution centroid CID fragmentation spectrum of GDP-Man in a mass range of m/z = 305–605, applying a relative CID fragmentation energy of 25%. The precursor mass of the GDP-Man is highlighted in gray. The dashed lines in the structural formula indicate heterolytic cleavage sites due to CID fragmentation. Bottom, high resolution centroid CID fragmentation spectrum of GDP-ManA in a mass range of m/z = 320–620, applying a relative CID fragmentation energy of 19%. The precursor mass of the GDP-ManA is highlighted in gray. The dashed lines in the structural formula indicate heterolytic cleavage sites due to CID fragmentation. Molecular formulas of neutral losses and corresponding mass deviations were calculated using the XCalibur software version 2.0.7.
Biochemical Characterization of the Enzyme
We next proceeded with a detailed biochemical characterization of EsGMD1. The enzyme has an alkaline pH optimum around 8.75–9 and is inactive below pH 7 (supplemental Fig. S4). EsGMD1 is quite sensitive to certain buffers, such as diethanolamine, which totally inhibits all enzyme activity. All further assays were performed in Tris-glycine buffer, pH 8.75.
EsGMD1 is temperature-sensitive. The activity steadily increases from 0 °C to 30 °C but strongly declines above 30 °C. An incubation period of 15 min at 45 °C fully inactivates EsGMD1.
The kinetic parameters of EsGMD1 follow typical Michealis-Menten kinetics. Using the minimum square deviation method to fit the hyperbolic data, we obtained a Km of 95 μm for the substrate GDP-Man and a Km of 86 μm for the cofactor NAD+ (supplemental Fig. S5). The kcat value of the purified recombinant GMD is 0.21 · s−1. The specific activity of recombinant EsGMD1 is 3.3 nanokatals mg−1 protein. E. siliculosus is a marine alga living in a salty environment. This prompted us to analyze the effect of mono- and divalent salts on GMD activity. All salts led to an increase in enzymatic activity. The optimum is approximately 500 mm for Na2SO4 and NaCl and slightly lower for KCl. Even the presence of 1.5 m salts increases the GMD activity compared with the 50 mm buffer alone (supplemental Fig. S6).
The substrate specificity of EsGMD1 was tested with several other nucleotide sugars, but none except GDP-Man is converted by the enzyme (Table 1). The data suggest that EsGMD1 is highly specific for the production of the alginate precursor GDP-ManA. The cofactor NAD+ cannot be replaced by NADP+, addition of the latter instead of NAD+ resulted in zero activity.
TABLE 1.
Substrate specificity of Ectocarpus EsGMD1
| Substrates | Enzyme activity |
|---|---|
| % of control | |
| GDP-mannose (Control) | 100 ± 0.7 |
| GDP-glucose | 0 |
| UDP-glucose | 0 |
| UDP-N-acetylglucoseamine | 0 |
| UDP-galactose | 0 |
Some potential inhibitors of GMD were tested, and the data are summarized in Table 2. NADH is a weak inhibitor of the reaction. The decrease in enzyme activity observed after longer incubation times cannot be fully explained by the accumulation of NADH. Presumably, the product GDP-ManA is another inhibitor of the reaction. We did not determine the Ki for GDP-ManA because to our knowledge this compound is not commercially available. The catalytic cysteine is sensitive to heavy metals. In particular, copper ions and the organic mercury compound p-chloromercuribenzoate totally inhibit GMD at micromolar concentrations. In contrast, EDTA prevents binding of heavy metals to the Cys residue and therefore slightly increases the enzymatic activity by chelating residual heavy metals present in the buffers etc.
TABLE 2.
Inhibitors of EsGMD1
| Treatment | Enzyme activity |
|---|---|
| % of control | |
| Control | 100 ± 0.7 |
| Nucleotides | |
| ATP (1 mm) | 97 ± 0.8 |
| UTP (1 mm) | 96 ± 0.5 |
| UDP (1 mm) | 102 ± 0.6 |
| UMP (1 mm) | 101 ± 0.8 |
| Chelator, metals, and modifying reagents | |
| EDTA (1 mm) | 105 ± 0.9 |
| CoCl2 (1 mm) | 44.8 ± 0.6 |
| CuSO4 (1 mm) | 0 ± 0.01 |
| FeCl3 (1 mm) | 49 ± 0.5 |
| FeSO4 (1 mm) | 77.4 ± 0.5 |
| pCMB (1 μm) | 99 ± 0.7 |
| pCMB (5 μm) | 87.9 ± 0.6 |
| pCMB (10 μm) | 20.6 ± 0.2 |
| pCMB (25 μm) | 0.8 ± 0.04 |
DISCUSSION
Properties of EsGMD1
Here we characterize the first GMD from a eukaryotic organism, the brown alga E. siliculosus. Previously, only the GMD from the prokaryote P. aeruginosa has been partially analyzed biochemically (9). Both enzymes catalyze the same reaction, the conversion of GDP-Man into GDP-ManA, but they differ widely in their biochemical properties. The Km of EsGMD1 is roughly 5-fold higher than the reported Km for GDP-Man of the bacterial enzyme. This difference can possibly be explained by the different roles of alginate in bacteria and algae. E. siliculosus does not need GDP-Man only for alginate biosynthesis, but also for the synthesis of fucan polymers destined for the cell wall. In flowering plants, GDP-Man is also an important precursor for ascorbic acid biosynthesis (15). This use of GDP-Man in several pathways in brown algae, which are all of major quantitative importance, may explain the higher Km of the EsGMD1 to maintain the supply of GDP-Man for the other pathways. The Vmax of EsGMD1 is ∼3-fold lower than that reported for the Pseudomonas enzyme (9).
EsGMD1 is sensitive to higher temperatures and was rapidly inactivated at 35 °C, whereas the bacterial enzyme has a temperature optimum at 50 °C. This suggests there might be unique structural features of the bacterial enzymes that impart greater enzyme stability.
EsGMD1 is a member of a small group of NAD+-dependent four-electron transfer dehydrogenases all of which contain a conserved catalytic Cys residue. This residue is believed to be involved in the formation of a thiohemiacetal intermediate with the substrate GDP-Man after the first two-electron oxidation step, which has also been observed in the sister enzyme UGD (13, 16). The alkaline pH optimum for EsGMD1 is approximately pH 8.5- and thus near to the pKa-value of the Cys residue. Similar pH optima were observed for UGDs from plants (17, 18). For the Pseudomonas GMD Roychoudhury et al. (9) report a significantly lower and very sharp pH optimum of pH 7.7 which seems to be an exception for this group of enzymes.
Structural Differences between Pro- and Eukaryotic GMDs
The most striking difference between pro- and eukaryotic GMDs is the structure of the enzyme. The bacterial enzyme has been crystallized, and from this dataset several binding sites for the substrates NAD+ and GDP-Man were predicted (12). The structure of Pseudomonas GMD is composed of two large but mirrored domains that are connected by a long α-helix. The N-terminal domain (residues 1–154) shows a typical Rossmann fold and contains a complete dinucleotide binding motif with six-stranded parallel β-sheets and five α-helices. The swapped domain at the C terminus (residue 315–436), however, represents an incomplete Rossmann fold, in which the dinucleotide-binding motif is missing the third β-strand and the final α-helix (12). This second domain is completely absent in the GMDs from E. siliculosus, suggesting that the N-terminal Rossmann fold from the brown algal enzyme is sufficient to bind its substrates. Some speculative models for substrate binding have been proposed for the bacterial enzyme, in which the N-terminal Rossmann fold of enzyme A completes the substrate binding sites with the C-terminal incomplete Rossmann fold from enzyme B. This model was supported by the finding that GMD genes from bacteria are all highly similar in their domain organization. Because EsGMD1 lacks the C-terminal Rossmann fold, a different mechanism of substrate binding is likely, or the N-terminal domains from two GMD-molecules completes one binding site. The latter hypothesis seems possible as EsGMD1 is active as a dimer. In this case the structure of the GMD must be very flexible, as we did not detect differences in biochemical properties between the pure EsGMD1 enzyme and the fusion protein with MBP. Discerning between these possibilities will likely require structural elucidation of the EsGMD1 enzyme.
Evolution of Ectocarpus GMD
The brown algae are phylogenetically related to another group of algae, the diatoms. Two diatom genomes have been published recently, but database homology searches produced no convincing evidence of the presence of GMD genes in diatoms. Furthermore, none of the currently more than 280 other eukaryotic genomes in GenBank, including further genomes of the Stramenopiles lineage, contains a GMD-like gene.
In contrast, several different bacteria have genes for GMD, which are highly similar to that characterized from P. aeruginosa. This could be a hint for a single evolutionary event in the Stramenopiles lineage, in which a bacterial GMD was taken up by early brown algae, modified drastically, and afterward maintained in different brown algae. All characteristic structural and sequence features of EsGMD1 are highly conserved between different order of brown algae like the Ectocarpels, Fucales, and Laminariales. Ectocarpus can be infected by a large genome virus EsV-1 (335-kbp genome) which has distant copy of a UGD or GMD gene in its genome (19). Such a virus could have caused a lateral gene transfer in the past, in which genes for an alginate pathway were transferred into a brown alga. A convergent evolution of the GMD in brown algae is very unlikely, as for example most of the substrate binding amino acids and the residues of the catalytic center are identical or very similar in bacterial and brown algae GMDs.
Supplementary Material
Acknowledgments
We thank Doris Wittmann and Delphine Scornet for technical help and Hans Brandstetter for valuable discussions about the structural analysis of GMD proteins and for providing the His-tagged MBP expression vector.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- GDP-ManA
- GDP-d-mannuronic acid
- CID
- collision-induced dissociation
- ESI
- electrospray ionization
- EST
- expressed sequence tag
- GDP-Man
- GDP-d-mannose
- GMD
- GDP-mannose dehydrogenase
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- MBP
- maltose-binding protein
- TEV
- tobacco etch virus
- UGD
- UDP-glucose dehydrogenase.
REFERENCES
- 1. Reyes-Prieto A., Weber A. P., Bhattacharya D. (2007) Annu. Rev. Genet. 41, 147–168 [DOI] [PubMed] [Google Scholar]
- 2. Michel G., Tonon T., Scornet D., Cock J. M., Kloareg B. (2010) New Phytol. 188, 82–97 [DOI] [PubMed] [Google Scholar]
- 3. Kloareg B., Quatrano R. S. (1988) Oceanogr. Marine Biol. 26, 259–315 [Google Scholar]
- 4. Nyvall P., Corre E., Boisset C., Barbeyron T., Rousvoal S., Scornet D., Kloareg B., Boyen C. (2003) Plant Physiol. 133, 726–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Skjåk-Braek G., Grasdalen H., Larsen B. (1986) Carbohydr. Res. 154, 239–250 [DOI] [PubMed] [Google Scholar]
- 6. Draget K. I., Smidsrod O., Skjak-Braek G. (2002) in Biopolymers Polysaccharides II (Vandamme E. J., De Baets S., Steinbüchel A. eds) pp. 215–244, Wiley-VCH, Weinheim [Google Scholar]
- 7. Deretic V., Gill J. F., Chakrabarty A. M. (1987) J. Bacteriol. 169, 351–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin T. Y., Hassid W. Z. (1966) J. Biol. Chem. 241, 5284–5297 [PubMed] [Google Scholar]
- 9. Roychoudhury S., May T. B., Gill J. F., Singh S. K., Feingold D. S., Chakrabarty A. M. (1989) J. Biol. Chem. 264, 9380–9385 [PubMed] [Google Scholar]
- 10. Cock J. M., Sterck L., Rouzé P., Scornet D., Allen A. E., Amoutzias G., Anthouard V., Artiguenave F., Aury J. M., Badger J. H., Beszteri B., Billiau K., Bonnet E., Bothwell J. H., Bowler C., Boyen C., Brownlee C., Carrano C. J., Charrier B., Cho G. Y., Coelho S. M., Collén J., Corre E., Da Silva C., Delage L., Delaroque N., Dittami S. M., Doulbeau S., Elias M., Farnham G., Gachon C. M., Gschloessl B., Heesch S., Jabbari K., Jubin C., Kawai H., Kimura K., Kloareg B., Küpper F. C., Lang D., Le Bail A., Leblanc C., Lerouge P., Lohr M., Lopez P. J., Martens C., Maumus F., Michel G., Miranda-Saavedra D., Morales J., Moreau H., Motomura T., Nagasato C., Napoli C. A., Nelson D. R., Nyvall-Collén P., Peters A. F., Pommier C., Potin P., Poulain J., Quesneville H., Read B., Rensing S. A., Ritter A., Rousvoal S., Samanta M., Samson G., Schroeder D. C., Ségurens B., Strittmatter M., Tonon T., Tregear J. W., Valentin K., von Dassow P., Yamagishi T., Van de Peer Y., Wincker P. (2010) Nature 465, 617–621 [DOI] [PubMed] [Google Scholar]
- 11. Sheffield P., Garrard S., Derewenda Z. (1999) Protein Expr. Purif. 15, 34–39 [DOI] [PubMed] [Google Scholar]
- 12. Snook C. F., Tipton P. A., Beamer L. J. (2003) Biochemistry 42, 4658–4668 [DOI] [PubMed] [Google Scholar]
- 13. Campbell R. E., Mosimann S. C., van De Rijn I., Tanner M. E., Strynadka N. C. (2000) Biochemistry 39, 7012–7023 [PubMed] [Google Scholar]
- 14. Naught L. E., Gilbert S., Imhoff R., Snook C., Beamer L., Tipton P. (2002) Biochemistry 41, 9637–9645 [DOI] [PubMed] [Google Scholar]
- 15. Conklin P. L., Norris S. R., Wheeler G. L., Williams E. H., Smirnoff N., Last R. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4198–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ge X., Penney L. C., van de Rijn I., Tanner M. E. (2004) Eur. J. Biochem. 271, 14–22 [DOI] [PubMed] [Google Scholar]
- 17. Klinghammer M., Tenhaken R. (2007) J. Exp. Bot. 58, 3609–3621 [DOI] [PubMed] [Google Scholar]
- 18. Hinterberg B., Klos C., Tenhaken R. (2002) Plant Physiol. Biochem. 40, 1011–1017 [Google Scholar]
- 19. Delaroque N., Müller D. G., Bothe G., Pohl T., Knippers R., Boland W. (2001) Virology 287, 112–132 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.