Abstract
Orexin G protein-coupled receptors (OxRs) and their cognate agonists have been implicated in a number of disorders since their recent discovery, ranging from narcolepsy to formation of addictive behavior. Bioluminescence resonance energy transfer assays of agonist-occupied OxRs provided evidence for a strong dose-dependent interaction with both trafficking proteins β-arrestin 1 and 2 that required unusually high agonist concentrations compared with inositol phosphate signaling. This appears to be reflected in functional differences in potency with respect to orexin A (OxA) and OxR2-dependent ERK1/2 phosphorylation after 90 min compared with 2 min, potentially consistent with β-arrestin-mediated versus G protein-mediated signaling, respectively. Furthermore, extended bioluminescence resonance energy transfer kinetic data monitoring OxA-dependent receptor-β-arrestin and β-arrestin-ubiquitin proximity suggested subtype-specific differences in receptor trafficking, with OxR2 activation resulting in more sustained receptor-β-arrestin-ubiquitin complex formation than elicited by OxR1 activation. Enzyme-linked immunosorbent assay (ELISA) data also revealed that OxR1 underwent significantly more rapid recycling compared with OxR2. Finally, we have observed sustained OxA-dependent ERK1/2 phosphorylation in the presence of OxR2 compared with OxR1. Although both OxR subtypes could be classified as class B receptors for β-arrestin usage based on the initial strength of interaction with both β-arrestins, our temporal profiling revealed tangible differences between OxR subtypes. Consequently, OxR1 appears to fit uneasily into the commonly used β-arrestin classification scheme. More importantly, it is hoped that this improved profiling capability, enabling the subtleties of protein complex formation, stability, and duration to be assessed in live cells, will help unlock the therapeutic potential of targeting these receptors.
Keywords: G Protein-coupled Receptors (GPCR), Receptor Desensitization, Receptor Endocytosis, Receptor Recycling, Receptor Regulation
Introduction
The orexin system is intimately involved in regulation and integration of sleep-wake states with metabolic energy levels, as well as the development of addictive behavior (1–3). Over a decade ago, the orexin system was characterized by a single gene encoding precursor peptides orexin A (OxA)4 and orexin B (OxB) in neurons of the lateral hypothalamus (4, 5) and also in peripheral tissues (6). In the central nervous system, neurons send dense projections to nearly all regions of the brain (7), where orexins have specificity for two G protein-coupled receptors (GPCRs), orexin receptors 1 and 2 (OxR1 and OxR2). OxA displays similar potency for both receptors; however, OxB is substantially less potent at OxR1 compared with OxR2, as measured by ability to stimulate Ca2+ release (4).
Agonist-induced GPCR activation promotes phosphorylation of intracellular serine/threonine residues of many GPCRs by G protein-coupled receptor kinases (GRKs) (8). Typically, this event promotes recruitment of the multiadaptor proteins β-arrestin 1 and β-arrestin 2 (also known as arrestin-2 and arrestin-3, respectively) that results in desensitization of G protein-mediated signaling, and it generally provides a scaffold for internalization of the activated GPCR complex into endosomes inside the cell (9).
It has been shown for a number of GPCRs that trafficking of their activated complexes may be β-arrestin-independent (10–16). Indeed, there is notable evidence for some receptors not preferentially internalizing with β-arrestins despite interacting with them (13). However, the majority of GPCRs appear to utilize β-arrestins at some stage in the trafficking process, and it appears that an important factor in determining the ultimate fate of the receptor-β-arrestin complex tends to be the strength of the receptor-β-arrestin interaction.
In general terms, there are believed to be two possible outcomes for the GPCR following internalization. Resensitization may occur by dephosphorylation of the receptor complex allowing recycling back to the cell membrane. Alternatively, the complex may be targeted to late endosomes for proteosomal or lysosomal degradation (17). Additionally, it has been shown that GPCR-β-arrestin complexes can act as scaffolds for alternative downstream kinase signaling pathways, such as Src tyrosine kinases, Akt, and extracellular signal-regulated kinase (ERK) 1/2, independently of G protein coupling of the receptors (18–22). Importantly, this can alter the spatio-temporal nature of receptor-mediated kinase signaling (23).
Further functional outcomes of activated GPCR complexes have been correlated with the stability of ubiquitination of both GPCRs and β-arrestins (24, 25). Ubiquitination involves a multistep process culminating in the covalent attachment of ubiquitin to a protein that is consequently targeted for degradation through either the 26 S proteasome or lysosomes (26, 27). However, it also appears that ubiquitin modification can also compartmentalize protein complexes directly, or through the binding of ubiquitin-binding elements, to host a range of other nonproteolytic processes. These include regulation of endocytosis and cellular signaling (28–30), depending on the conjugation site and multimeric state of ubiquitin (31). Recently, the ubiquitination status of β-arrestin-bound GPCR complexes has been shown to play roles in endosomal targeting, ERK1/2 activation (32), and determining the stability of β-arrestin-receptor complexes (33). In addition, the state of these interactions seems to parallel the duration and magnitude of ERK1/2 phosphorylation through the scaffold activity exhibited by the multiadaptor protein properties of β-arrestin (34, 35).
Here, we show that both human orexin receptor subtypes interact with both β-arrestins 1 and 2 in an agonist dose-dependent manner but with 2 orders of magnitude lower potency than observed for G protein signaling. These potencies are lower than we have observed for other GPCRs interacting with β-arrestins. Furthermore, this difference is reflected in a potency shift between early (2 min) and late (90 min) OxA-induced ERK1/2 phosphorylation that is likely to result from predominantly G protein-mediated versus predominantly β-arrestin-mediated signaling, respectively (34, 35). Furthermore, bioluminescence resonance energy transfer (BRET) data indicate that the nature of the receptor-arrestin-ubiquitin complex differs between OxR subtypes, with OxA-induced BRET kinetics for proximity between receptor and β-arrestin or β-arrestin and ubiquitin correlating with the kinetics of both receptor recycling and ERK1/2 phosphorylation.
EXPERIMENTAL PROCEDURES
Materials
Wild type orexin receptor cDNAs were kindly provided by M. Yanagisawa (Howard Hughes Medical Institute, Dallas, TX); β-arrestin 1 and β-arrestin 2 cDNAs were kindly provided by J. Benovic (Kimmel Cancer Research Institute, Philadelphia), and phosphorylation-independent β-arrestin mutants (R169E and R170E) were generously provided by V. Gurevich (Vanderbilt University Medical Centre, Nashville, TN). cDNA sequences were PCR-amplified and subcloned into pcDNA3.1+ backbone vectors containing enhanced GFP (EGFP), Venus yellow fluorescent protein, or Renilla luciferase (Rluc or Rluc8) cDNA. The stop codon between the sequences was removed to generate constructs capable of being translated into fusion proteins upon transfection, as described previously (36, 37). Venus was kindly provided by Atsushi Miyawaki (RIKEN Brain Science Institute, Wako-city, Japan), and Rluc8 was kindly provided by Andreas Loening and Sanjiv Gambhir (Stanford University, Stanford, CA). cDNA encoding ubiquitin (Addgene plasmid 11928) (38) was similarly subcloned into a Venus (no stop codon)-pcDNA3.1+ vector. Because of the ability of ubiquitin to form polyubiquitin chains, lysine residues at positions 48 and 63 were mutated to alanine. This has been done previously when BRET assays involving ubiquitin have been carried out, to avoid the potential for multiple acceptor moieties causing quenching or interference phenomena (39). Fusion cDNA constructs were verified by ABI Prism BigDye terminator sequencing (Australian Genome Research Facility, Brisbane, Australia) and compared with published sequence data. Ligands used were OxA and OxB (American Peptide Company).
Cell Culture and Transfection
COS-7, HEK293, and HEK293FT cells were maintained at 37 °C in 5% CO2 and complete media (Dulbecco's modified Eagle's medium (DMEM) containing 0.3 mg/ml glutamine, 100 IU/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) supplemented with 5% fetal calf serum (FCS; Invitrogen)). HEK293FT media also contained geneticin (G418; 400 μg/ml; Invitrogen). Transfections were carried out 24 h after cell seeding using GeneJuice (Novagen) according to the manufacturer's instructions. HEK293 stable cell lines were maintained in 400–500 μg/ml G418.
Inositol Phosphate Assays
COS-7 cells were seeded in 100-mm dishes at a density of 700,000 cells/dish, and total inositol phosphate production was measured as described previously (37).
BRET Assays
COS-7 or HEK293FT cells transfected 48 h earlier were harvested and prepared as described previously in white 96-well plates (Nunc) (37). For BRET1 dose-response assays, coelenterazine h substrate was added to a final concentration of 5 μm, and analysis was carried out immediately. Samples were incubated for 5 min in the presence of various concentrations of agonist and then measured for four sequential reads. For extended BRET (eBRET) assays, cells were resuspended in HEPES-buffered (25 mm) phenol-red free DMEM with 5% FCS to maintain viability. EnduRenTM substrate (Promega) was added to each well at a final concentration of 60 μm. Cells were left for 2 h at 37 °C, 5% CO2 in order for the cell-permeable substrate to equilibrate. Samples were sequentially read using either a MithrasTM LB940 luminescence plate reader (Berthold) or VICTOR LightTM 1420 luminescence counter (PerkinElmer Life Sciences) using appropriate filter sets as detailed below. eBRET kinetics were measured for ∼30 min to obtain a basal signal. Cells were then treated with vehicle or ligand and read continuously for several hours. BRET ratios for β-arrestin recruitment to OxRs were calculated by subtracting the ratio of >500 nm emission over the 400–475-nm emission for a cell sample containing only the Rluc construct from the same ratio for a sample containing both the Rluc and EGFP fusion proteins as described previously (37). The ligand-induced BRET signal for β-arrestin-ubiquitin proximity was calculated by subtracting the ratio of 520–540-nm emission over the 400–475-nm emission for a vehicle-treated cell sample containing both Rluc8 and Venus fusion proteins from the same ratio for a second aliquot of the same cells that was treated with ligand as described previously (40). The final pretreatment reading is presented at the zero time point (time of ligand or vehicle addition).
Confocal Microscopy
HEK293 cells were seeded in 6-well plates at a density of 650,000 cells/well, and confocal microscopy procedures were carried out as described previously (37).
Enzyme-linked Immunosorbent Assay (ELISA)
OxR constructs were generated with a hemagglutinin (HA) epitope tag incorporated at the N terminus of OxR1 and OxR2. Stably transfected HEK293 cells expressing either HA-OxR1 or HA-OxR2 were plated into 24-well plates at 200,000 cells/well with complete media. The next day, media were removed, and 0.5 ml of media containing a 1:2500 dilution of anti-HA serum (raised in rabbit and generously provided by S. Schulz, Institute of Pharmacology and Toxicology, University of Jena, Germany) was added to wells. Cells were then incubated with antibody at 4 °C for 2 h. Media were removed and replaced with 0.5 ml of media containing OxA. Cells were then incubated at 37 °C for 1 h. Following treatment, cells were washed with phosphate-buffered saline (PBS). They were then fixed in 0.5 ml of Zamboni's fixative at room temperature for ∼30 min, either immediately (0 min washout) or following incubation for 20, 40, or 60 min in media. Fixative was removed, and the cells were washed twice with PBS before adding 0.5 ml of PBS containing secondary antibody (anti-rabbit IgG, horseradish-peroxidase-linked whole antibody raised in donkey; Amersham Biosciences). Secondary antibody was diluted 1:2000. Samples were incubated at room temperature with gentle rocking for ∼2 h. Finally, cells were washed twice with PBS before 250 μl of horseradish peroxidase substrate 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (Sigma) was added to wells and incubated at room temperature with rocking for ∼15 min. 200-μl aliquots were then transferred to a clear-bottomed 96-well plate, and samples were measured at 405 nm using an Envision 2102 multilabel plate reader (PerkinElmer Life Sciences). All treatments were performed in triplicate, and each assay included a primary antibody control.
Homogeneous Cell-based ERK1/2 Phosphorylation Assay
ERK1/2 phosphorylation was measured using the SureFire ERK1/2 phosphorylation kit (TGR Biosciences) with IgG Protein A AlphaScreen donor/acceptor beads (PerkinElmer Life Sciences) (41). HEK293 cells stably expressing OxR cDNA constructs were seeded in white 96-well plates (Nunc) at a density of 60,000 cells/well in serum-free DMEM. After 16 h, media were replaced, and treatments were carried out 1 h later. Treated cells were assayed according to the manufacturer's instructions, and samples were measured using an Envision 2102 multilabel plate reader (PerkinElmer Life Sciences).
Data Presentation and Statistical Analyses
Data were presented and analyzed using Prism 5.0 software (GraphPad). Sigmoidal dose-response curves were fitted to the data for inositol phosphate production, β-arrestin recruitment to OxRs, and ERK1/2 phosphorylation using nonlinear regression. Statistical comparisons of logEC50 values and recycling time course data were assessed using two-way analysis of variance with Bonferroni post-test analysis or Student's t test where appropriate. Statistical significance for eBRET and ERK1/2 phosphorylation kinetic data was determined using a two-way repeated measure analysis of variance with Bonferroni post-test analysis.
RESULTS
Inositol Phosphate Signaling
Wild type (WT) and EGFP-tagged human OxR constructs for both subtypes were able to induce robust inositol phosphate production in the presence of either orexin ligand. In response to OxA or OxB treatment, the signaling potencies of EGFP-tagged OxR1 and OxR2 constructs were not significantly different from respective WT receptors (Table 1). As expected, a nanomolar effective concentration of agonist was shown to elicit a half-maximal response for OxA at OxR1 and OxR2, as well as OxB at OxR2. Additionally, the potency of OxB was substantially lower at OxR1 compared with that observed with OxA at OxR1 or OxB at OxR2, as has been similarly observed for G protein-mediated Ca2+ signaling (4).
TABLE 1.
EC50 data for inositol phosphate signaling dose-response assays comparing wild type with EGFP-tagged OxR subtypes
Inositol phosphate production was measured in COS-7 whole cell lysates transfected with wild type (WT) or EGFP-tagged OxRs and treated with a range of concentrations of OxA or OxB. Results are presented as mean ± S.E. of three independent experiments. Statistics were carried out using logEC50 values.
| OxR1 EC50 | OxR2 EC50 | |
|---|---|---|
| nm | nm | |
| OxA | ||
| Receptor-WT | 5.2 ± 1.1 | 2.6 ± 0.5 |
| Receptor-EGFP | 9.6 ± 1.8 | 4.3 ± 0.9 |
| OxB | ||
| Receptor-WT | 59.4 ± 14.1a,b | 2.3 ± 1.0 |
| Receptor-EGFP | 92.0 ± 18.3a,b | 2.9 ± 0.7 |
a p < 0.05 compared with respective OxR2.
b p < 0.05 compared with respective receptor treated with OxA.
BRET Dose-Response Analysis of OxR-β-Arrestin Proximity
BRET EC50 values for OxR-β-arrestin proximity were found to be 2 orders of magnitude higher (Table 2) than the EC50 values observed for inositol phosphate production (Table 1). These data were generated using COS-7 cells; however, similar BRET EC50 values were observed using HEK293FT cells (data not shown). The use of phosphorylation-independent β-arrestin mutants had little effect, indicating that receptor phosphorylation was not generally a limiting factor under these conditions. A small but significant difference in potency was observed for OxR1 compared with OxR2 in COS-7 cells treated with OxA (Table 2). This was also observed with the phosphorylation-independent mutants. Again, similar observations were made with HEK293FT cells, although the differences between receptors treated with OxA were not statistically significant in these cells (data not shown).
TABLE 2.
EC50 data for BRET OxR-β-arrestin proximity dose-response assays
COS-7 cells were co-transfected with either OxR1 or OxR2 (EGFP-tagged) and either a wild type (WT) or phosphorylation-independent mutant (R169E or R170E) form of β-arrestin 1 or 2 (Rluc-tagged). Cells were treated with a range of concentrations of OxA or OxB at 37 °C. Results are presented as mean ± S.E. of three independent experiments. Statistics were carried out on logEC50 values.
| OxR1 EC50 | OxR2 EC50 | |
|---|---|---|
| nm | nm | |
| OxA | ||
| β-Arrestin 1-WT | 777.0 ± 54.6a | 375.9 ± 121.5 |
| β-Arrestin 2-WT | 719.4 ± 102.2a | 214.7 ± 75.7 |
| β-Arrestin 1-R169E | 412.1 ± 44.1a | 138.7 ± 22.7b |
| β-Arrestin 2-R170E | 400.8 ± 46.5a | 161.5 ± 18.2 |
| OxB | ||
| β-Arrestin 1-WT | Too high to determine | 334.3 ± 112.6 |
| β-Arrestin 2-WT | Too high to determine | 383.6 ± 203.3 |
| β-Arrestin 1-R169E | 3766.8 ± 546.7a | 228.3 ± 65.5 |
| β-Arrestin 2-R170E | 3788.8 ± 366.4a | 278.2 ± 148.1 |
a p < 0.05 compared with OxR2.
b p < 0.05 compared with the respective β-arrestin WT. Note that as EC50 values for OxB-induced OxR1 proximity with β-arrestin 1 or 2 WT were too high to determine, statistical comparisons involving these values could not be carried out.
BRET EC50 values for OxR2 proximity to mutant β-arrestins in the presence of OxB were significantly less than those observed for OxR1 paired with the same β-arrestins (Table 2). BRET EC50 values for OxR1 proximity to WT β-arrestins treated with OxB could not be obtained, as sufficiently high concentrations of OxB could not be tested (Table 2). This indicates substantially lower potency than at OxR2, even if statistical analysis is precluded. OxB BRET EC50 values for OxR2 proximity to β-arrestins were not significantly different than respective EC50 values from OxA dose-response analyses.
The β-arrestin proximity potency shift observed between the different OxR subtypes activated by OxB (Table 2) appears to reflect that observed for inositol phosphate production (Table 1), which in turn reflects published differences in OxB binding affinity at the two receptors (4). Therefore, for the remainder of the study, we focused our attention on investigating potential differences between the OxR subtypes when activated by OxA, which has similar binding affinity (4), and inositol phosphate production potency (Table 1) at both receptors.
eBRET Monitoring of OxA-induced OxR-β-Arrestin Proximity
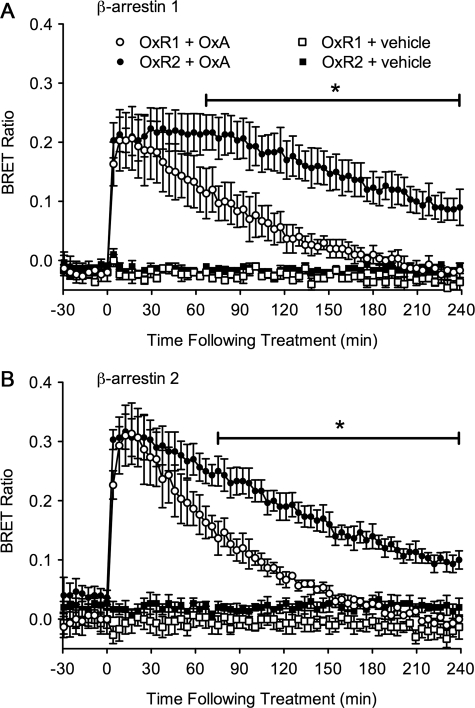
Following treatment with 1 μm OxA, COS-7 cells transfected with BRET-tagged pairs of OxR and β-arrestin subtypes elicited a robust initial increase in BRET response (Fig. 1, A and B). The response was similarly observed for all OxR-β-arrestin combinations, although slightly heightened for OxR interactions with β-arrestin 2. However, subsequent kinetics differed between OxR subtypes. OxR2-expressing cells displayed a significantly more stable BRET signal with both β-arrestins in comparison with OxR1 over 4 h of continual stimulation with OxA. A significant divergence between OxR subtype kinetic profiles was observed after 67 min for β-arrestin 1 (p < 0.05; Fig. 1A), and 75 min for β-arrestin 2 (p < 0.05; Fig. 1B) and continued over the rest of the 4-h measurement period.
FIGURE 1.
eBRET kinetic data for OxA-induced OxR proximity to β-arrestin 1 and 2. To assess EGFP-tagged OxR1 and OxR2 proximity to Rluc-tagged β-arrestin 1 (A) and β-arrestin 2 (B), a basal BRET signal was established in the absence of ligand for 30 min. Cells were then treated with either vehicle or OxA and monitored in real time for up to 4 h. Results are presented as the mean BRET ratio ± S.E. of at least three independent experiments. *, p < 0.05 between OxR subtypes treated with OxA.
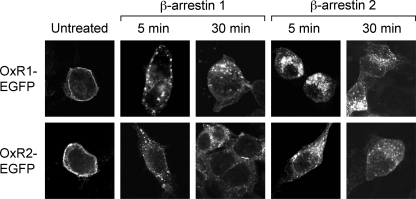
Confocal Microscopy Indicates That Both OxR Subtypes Internalize into Endosomes following OxA Treatment
HEK293 cells transfected with OxR1-EGFP or OxR2-EGFP exhibited fluorescence on the cell surface indicative of receptor expression at the plasma membrane (Fig. 2). Following treatment with 1 μm OxA for either 5 or 30 min, similar punctate fluorescence was observed throughout the cytosol of cells containing all combinations of EGFP-tagged OxRs and untagged β-arrestins, consistent with similar internalization into endosomes (Fig. 2).
FIGURE 2.
Confocal microscopy showing redistribution of OxRs following OxA treatment. OxR1-EGFP or OxR2-EGFP was visualized in HEK293 cells overexpressing untagged β-arrestin 1 or 2. The distribution of EGFP-tagged OxRs is shown in untreated cells compared with those treated with OxA for 5 or 30 min as indicated.
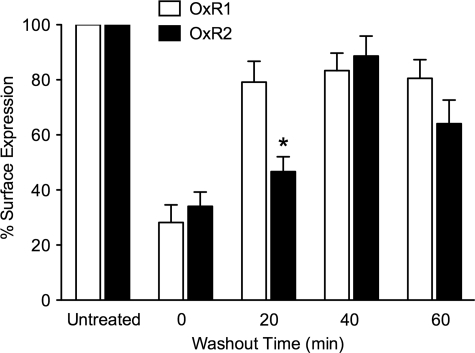
OxR Subtypes Recycle at Different Rates following OxA Treatment
Recycling of the receptor back to the cell surface was monitored after treatment with OxA for 60 min and subsequent washing out of agonist for specified time periods (Fig. 3). After immediate cell fixation following agonist treatment, both stably expressing cell lines exhibited a similar decrease in cell surface HA immunoreactivity consistent with the similar levels of receptor internalization observed with confocal microscopy. However, after a 20-min washout period, a significantly lower percentage of HA-OxR2 was detected at the cell surface compared with HA-OxR1 (Fig. 3). This difference was not observed for washout periods of 40 min and greater (Fig. 3).
FIGURE 3.
OxR recycling rates following OxA treatment. HA-OxR1 and HA-OxR2 stable cell lines were treated for 60 min with OxA, following which cells were either washed and fixed immediately (0 min washout) or washed and incubated in media for varying time periods before fixing. Data are presented as the percentage of receptors at the cell surface relative to untreated samples (mean ± S.E.) from three independent experiments. *, p < 0.05 compared with OxR1 at same time point.
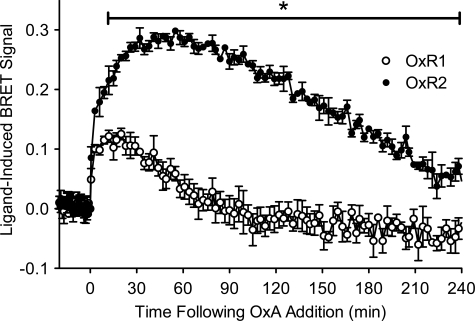
β-Arrestin 2-Ubiquitin eBRET Kinetics Differ between OxR Subtypes
Upon OxA treatment of cells co-expressing BRET-tagged β-arrestin 2 and ubiquitin constructs, in addition to an untagged OxR subtype, a robust increase in BRET signal was observed for both receptors (Fig. 4). The maximal BRET signal was significantly higher for cells expressing OxR2 compared with OxR1 after 12 min of OxA stimulation and remained so over the 4-h measurement period (Fig. 4). In addition, the time at which the peak BRET signal for OxR2 was reached was later than for OxR1. The BRET signal for OxR2 was relatively sustained over the 4-h measurement period. In contrast, the OxR1 signal was transient, returning to and remaining at base line after ∼75 min of ligand stimulation (Fig. 4).
FIGURE 4.
eBRET kinetic data for OxA-induced β-arrestin 2-ubiquitin proximity in the presence of OxR1 or OxR2. Kinetic profiles of changes in proximity between Rluc8-tagged β-arrestin 2 and Venus-tagged ubiquitin in the presence of untagged OxR1 or OxR2 transiently transfected in HEK293FT cells. Cells were treated with OxA and measured over a 4-h period. Data are expressed as mean ± S.E. of three independent experiments; *, p < 0.05 between OxR1 and OxR2 from 12 min up to 4 h post-agonist stimulation.
OxR Subtypes Display Different ERK1/2 Phosphorylation Kinetics
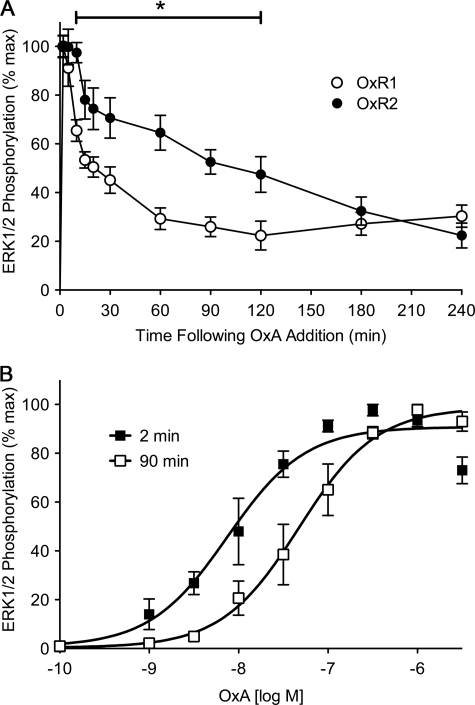
Using the homogeneous cell-based SureFire assay, ERK1/2 phosphorylation was monitored over a period of 4 h following stimulation with a maximal dose of OxA in cells stably transfected with either OxR subtype (Fig. 5A). Maximal levels of phosphorylated ERK1/2 (pERK1/2) were observed 2 min after agonist stimulation, and all subsequent measurement data were normalized to this maximal value. Relative levels of pERK1/2 were similar at 5 min post-agonist stimulation; however, a significantly lower level of pERK1/2 was observed for OxR1 compared with OxR2 at 10 min and up to 2 h following sustained agonist stimulation (Fig. 5A). Kinetically, OxR2-expressing cells exhibited a more stable, prolonged pERK1/2 profile compared with OxR1. pERK1/2 levels in OxR1-expressing cells dropped to ∼30% of maximal levels after 60 min and were similar throughout the remainder of the 4-h time course, whereas OxR2 exhibited a more gradual decrease over the same time period (Fig. 5A).
FIGURE 5.
ERK1/2 phosphorylation data for OxR1 and OxR2 stably transfected in HEK293 cells. A, stably transfected HEK293 cells were treated with OxA and measured over a 4-h period. Data were normalized to time-matched vehicle treatments and are expressed as a percentage of the maximal response induced at 2 min post-agonist treatment. Data are expressed as mean ± S.E. of four independent experiments. *, p < 0.05 between OxR1 and OxR2 from 10 to 120 min post-agonist stimulation. B, dose-response data were collected at 2 and 90 min post OxA treatment of OxR2-expressing cells. Data were expressed as a percentage of the maximal response induced at the time point. Data are expressed as mean ± S.E. of four independent experiments.
OxR2 Exhibits a Time-dependent Shift in ERK1/2 Phosphorylation Potency
OxA-induced ERK1/2 phosphorylation dose-response data were generated at two distinct time points, 2 and 90 min post-agonist stimulation. For OxR1 at 90 min, the ERK1/2 phosphorylation exhibited insufficient dose dependence to generate meaningful data, and so comparison of the 2- and 90-min time points could not be made. However, for OxR2 a significantly higher dose of OxA was required at 90 min (EC50 ± S.E. of 47.3 ± 12.3 nm) compared with 2 min (EC50 ± S.E. of 7.5 ± 1.3 nm) to achieve a similar proportion of pERK1/2 (p < 0.05 evaluated using logEC50 values; Fig. 5B).
DISCUSSION
Potency of β-Arrestin Recruitment to OxRs
The potencies of β-arrestin recruitment to both OxRs were surprisingly low, being 2 orders of magnitude lower than observed for inositol phosphate production. These potencies are lower than we have observed for other GPCRs interacting with β-arrestins using BRET, including angiotensin II receptor type 1 (37), thyrotropin-releasing hormone receptor (40), and chemokine receptors CCR5 and CXCR4 (42). Investigations of the role of receptor phosphorylation in receptor-β-arrestin interactions, and subsequent receptor internalization, have taken advantage of the properties of mutant β-arrestins (43–46). Mutant β-arrestins (R169E and R170E) do not require receptor phosphorylation for activation (47) and were included in the study to assess whether receptor phosphorylation was a limiting factor for the potency of β-arrestin recruitment to OxRs in these cells. Overall, this would appear not to be the case as the only statistically significant difference when comparing WT and mutant β-arrestins was with OxA acting on the combination of β-arrestin 1 and OxR2 in COS-7 cells (2.7-fold shift). These data suggest that either nonlimiting amounts of GRK were present or that GRK phosphorylation may not be required for β-arrestin binding. However, as a previous study observed that elimination of a particular GRK phosphorylation motif in OxR1 resulted in a loss of OxR1 internalization and β-arrestin co-localization (48), it is likely that GRK phosphorylation is required for OxR-β-arrestin binding.
Temporal Differences between OxR Subtypes with Respect to β-Arrestin Recruitment
Our eBRET data imply that although both OxRs are capable of associating with both β-arrestins following OxA activation, OxR2 appears to have a more stable interaction with β-arrestins that is sustained significantly longer compared with OxR1, with this being reflected in the BRET signal from populations of many individual receptor-β-arrestin interactions in many cells. The small but significant difference in BRET EC50 values between OxR subtypes, at least in COS-7 cells (Table 2), is also consistent with OxR2 interacting with β-arrestins more avidly than OxR1.
The role of GRK phosphorylation in β-arrestin binding to activated GPCRs has been discussed previously (20, 49–51), the stability of which has been found to be positively correlated with the composition and quantity of serine/threonine residue clusters in the C-tail of receptors (49, 51). Sequence analysis of amino acid residues in the C-tail of OxR subtypes indicates that OxR1 has two putative GRK-specific phosphorylation sites (17, 48, 52, 53), only one of which appears to play a significant role in β-arrestin interaction (48). In contrast, OxR2 contains three of these motifs (17). Thus, the potential for more extensive phosphorylation of the OxR2 subtype following agonist binding may contribute to the more stable interaction observed between OxR2 and β-arrestins compared with OxR1.
It should be noted that other intracellular serine/threonine residues outside the C-tail of GPCRs could also play a role in β-arrestin recruitment. Neuropeptide Y receptor subtype 5 exhibits a robust interaction with β-arrestin 2 measured using BRET despite possessing a relatively short C-tail with few phosphorylation sites (54). However, Y receptor subtype 5 has an extended third intracellular loop with phosphorylation sites (54). In addition, membrane association of the C-tail by palmitoylation can also affect β-arrestin-binding stability (55–57). However, both OxRs possess similar putative palmitoylation sites in the proximal C-tail and GRK phosphorylation sites in the third intracellular loop.
OxR1 Recycles More Rapidly than OxR2
Results from studies of receptor-β-arrestin coupling using chimeric receptors in which the C-tails of GPCRs were swapped indicated that, at least for the receptors investigated, the rate of β-arrestin-dependent GPCR recycling and resensitization directly correlated with the presence or absence of highly conserved serine/threonine clusters within the C-tail of the receptor (58, 59). The fate of the receptor-β-arrestin complex is often correlated with the strength and stability of this interaction, which is itself typically dependent upon the amino acid composition of the receptor C-tail (19, 53, 59). Work with the somatostatin receptor subtypes, for example, showed that receptor degradation, recycling, and up/down-regulation were all determined, to an extent, by receptor phosphorylation and β-arrestin coupling (60). From our study, it would appear that the higher rate of OxR1 dissociation from either form of β-arrestin, despite a strong initial interaction (as evidenced by the eBRET kinetic assays), is associated with more rapid recycling back to the plasma membrane compared with OxR2.
OxA-induced β-Arrestin-Ubiquitin Proximity Provides Further Insights into OxR Complex Stability
Covalent modification of receptors and their binding partners appears to be critical for regulating the activity of many activated GPCRs (61). Ubiquitination seems unique in its ability to potentially govern multiple facets of receptor function, including internalization, compartmentalization, signaling, and degradation of the receptor complex, depending on the stability and secondary structure of ubiquitin linkages to target proteins (31). β-Arrestin ubiquitination may be influential in regulating some of these receptor-mediated events with certain GPCRs (25, 32, 62). Furthermore, loss-of-function mutation of β-arrestin with respect to receptor-specific β-arrestin ubiquitination may result in the inability of some receptor complexes to internalize into endosomes (24, 32) and alter the ability of β-arrestin to scaffold and compartmentalize pERK1/2 signaling complexes (24). Characterization of the temporal aspects of β-arrestin-ubiquitin proximity is potentially informative for investigating the stability of receptor complexes and the influence it may exert on β-arrestin-mediated activities. Using BRET, the kinetics of β-arrestin-ubiquitin proximity has been explored for two GPCRs, β2-adrenergic receptor and vasopressin receptor 2, that represent class A and B β-arrestin-binding receptors, respectively (39). There is some evidence for enzymes that co-regulate ubiquitination and deubiquitination events associating with the receptor complex in a class-dependent manner to allow stable ubiquitination of vasopressin receptor 2 and more transient ubiquitination of β2-adrenergic receptors upon agonist stimulation (62). Similarly, chimeric β2-adrenergic receptors and vasopressin receptor 2 with swapped C-tails appear to exhibit β-arrestin ubiquitination stability dependent on the origin of the C-tail (33).
In this study, OxR2-expressing cells display a more stable, prolonged BRET signal for energy transfer between tags on β-arrestin 2 and ubiquitin that is kinetically similar to that of β-arrestin recruitment to this receptor, whereas OxR1-expressing cells display a more transient BRET signal indicating more transient receptor-arrestin-ubiquitin complex formation compared with OxR2. Note that we have not directly assessed either β-arrestin or receptor ubiquitination per se, and although most of the BRET signal is presumed to result from β-arrestin interaction with ubiquitin, as these are the BRET-tagged proteins in the complex, it cannot be excluded that a component of the signal results from ubiquitin interacting directly with the OxR or other proteins in the macromolecular complex. This would bring the Venus tag on ubiquitin into close proximity to the Rluc8 tag on the β-arrestin that is also interacting with the receptor complex. Either way, the observations that more sustained ubiquitination correlates with increased stability of the receptor-β-arrestin complex are consistent with previous findings (33, 39, 62) and support our observations from the eBRET assays monitoring receptor-β-arrestin proximity and the recycling assays.
Correlation with ERK1/2 Phosphorylation
We observed significantly elevated and sustained pERK1/2 following activation of OxR2 compared with OxR1 at time points typically associated with non-G protein-mediated ERK1/2 phosphorylation (34, 63). The elevated level of pERK1/2 for OxR2 at the later time points compared with OxR1 is consistent with OxR2 forming a more stable secondary signaling complex that is likely to involve β-arrestin. Consequently, this may also indicate that OxR2 scaffolds and maintains pERK1/2 in the cytosol rather than allowing nuclear translocation as has been observed for non-G protein-mediated ERK1/2 signaling (64–66). Indeed, the possibility of non-G protein-mediated ERK1/2 signaling has been observed in a study involving chemical inhibition of the G protein-mediated pERK1/2 pathway for OxR2 (67). Furthermore, significant differences in OxA potency at OxR2 when comparing 2- and 90-min stimulation time points also indicate that ERK1/2 activation occurs via different mechanisms at the different time points. Indeed, the observed ERK1/2 activation that is presumed to be predominantly via a non-G protein-mediated pathway occurs at higher doses, consistent with the lower potency of receptor-β-arrestin proximity compared with G protein-mediated signaling.
Do Both OxRs Fit the Current Classification for β-Arrestin Usage?
GPCRs exhibiting weak, transient β-arrestin interactions with an apparent preference for β-arrestin 2 tend to be deemed class A receptors, although GPCRs that form strong, stable interactions with both β-arrestins tend to be denoted class B receptors for β-arrestin usage (59). Results from the eBRET kinetic studies indicate that both OxRs can associate robustly with both β-arrestins, superficially making them both candidates for class B classification according to β-arrestin usage (52, 59). Indeed, previous confocal microscopy studies have shown β-arrestin 2 co-localized with OxR1 in acidic endosomes following OxA stimulation (48). The class B assignment is also supported by the combination of our confocal microscopy and ELISA data showing that both OxRs are internalized to similar degrees. Furthermore, although OxR1 displays a more transient association than OxR2, the profiles are similar for both β-arrestins. However, the rapid recycling observed with OxR1 does not fit the class B categorization well and neither does the relatively weak and transient ubiquitination profile. Interestingly, such unusual behavior resembles that observed with somatostatin sst2A receptor that was classified uneasily as class B for β-arrestin usage as it formed stable complexes with both β-arrestins, yet was rapidly resensitized and recycled to the plasma membrane without detectable receptor ubiquitination (60). Bradykinin 2 receptor is another notable example as it appears to interact strongly with β-arrestin 2 when bradykinin is present (42, 68), but it recycles rapidly following agonist washout (68), similar to our observations with OxR1. Moreover, the functional differences revealed by the pERK1/2 kinetic profiles further highlight the differences between the OxR subtypes. It has been considered previously (60, 68) and our data further indicate that the taxonomy of GPCRs according to β-arrestin usage may need to be refined to discriminate between receptors that interact strongly with both β-arrestins yet exhibit divergent kinetic profiles. Perhaps a class C for β-arrestin usage is required, as suggested by Simaan et al. (68), for bradykinin 2 receptor, with OxR1 and sst2A receptor fitting into this category. Furthermore, our findings indicate that temporal characterization of the receptor-arrestin-ubiquitin complex may help to differentiate potential members of the different classes, with the ultimate aim of improving our understanding of receptor systems in vitro so that they can be correlated more effectively to physiological and pathological roles in vivo.
In conclusion, the orexin neuroendocrine system is an intriguing physiological entity with vast potential as a target for therapeutic intervention, particularly as it appears to play a unique and fundamental role in regulating and integrating so many key biological systems from sleep-wake to metabolism. A greater understanding of the mechanisms regulating its function is crucial if this pharmacological potential is to be tapped.
Acknowledgments
We thank Ruth Seeber for expertise in generating cDNA fusion constructs. We acknowledge the facilities and scientific and technical assistance of the Australian Microscopy and Microanalysis Research Facility at the Centre for Microscopy, Characterization, and Analysis, University of Western Australia, a facility funded by the University, State, and Commonwealth Governments.
This work was supported in part by the National Health and Medical Research Council of Australia via Project Grant 404087 (to K. A. E. and K. D. G. P.) and Fellowships 212064 and 353709 (to K. A. E. and K. D. G. P., respectively).
- OxA
- orexin A
- OxB
- orexin B
- OxR
- orexin receptor
- B2R
- bradykinin 2 receptor
- BRET
- bioluminescence resonance energy transfer
- C-tail
- C-terminal tail
- eBRET
- extended BRET
- EGFP
- enhanced GFP
- GPCR
- G protein-coupled receptor
- GRK
- G protein-coupled receptor kinase
- pERK1/2
- phosphorylated ERK1/2
- Rluc
- Renilla luciferase.
REFERENCES
- 1. Saper C. B., Scammell T. E., Lu J. (2005) Nature 437, 1257–1263 [DOI] [PubMed] [Google Scholar]
- 2. Sakurai T. (2007) Nat. Rev. Neurosci. 8, 171–181 [DOI] [PubMed] [Google Scholar]
- 3. Tsujino N., Sakurai T. (2009) Pharmacol. Rev. 61, 162–176 [DOI] [PubMed] [Google Scholar]
- 4. Sakurai T., Amemiya A., Ishii M., Matsuzaki I., Chemelli R. M., Tanaka H., Williams S. C., Richardson J. A., Kozlowski G. P., Wilson S., Arch J. R., Buckingham R. E., Haynes A. C., Carr S. A., Annan R. S., McNulty D. E., Liu W. S., Terrett J. A., Elshourbagy N. A., Bergsma D. J., Yanagisawa M. (1998) Cell 92, 573–585 [DOI] [PubMed] [Google Scholar]
- 5. de Lecea L., Kilduff T. S., Peyron C., Gao X., Foye P. E., Danielson P. E., Fukuhara C., Battenberg E. L., Gautvik V. T., Bartlett F. S., 2nd, Frankel W. N., van den Pol A. N., Bloom F. E., Gautvik K. M., Sutcliffe J. G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 322–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heinonen M. V., Purhonen A. K., Makela K. A., Herzig K. H. (2008) Acta Physiol. 192, 471–485 [DOI] [PubMed] [Google Scholar]
- 7. Date Y., Ueta Y., Yamashita H., Yamaguchi H., Matsukura S., Kangawa K., Sakurai T., Yanagisawa M., Nakazato M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 748–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gurevich V. V., Gurevich E. V. (2006) Pharmacol. Ther. 110, 465–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hanyaloglu A. C., von Zastrow M. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 537–568 [DOI] [PubMed] [Google Scholar]
- 10. Vrecl M., Anderson L., Hanyaloglu A., McGregor A. M., Groarke A. D., Milligan G., Taylor P. L., Eidne K. A. (1998) Mol. Endocrinol. 12, 1818–1829 [DOI] [PubMed] [Google Scholar]
- 11. van Koppen C. J., Jakobs K. H. (2004) Mol. Pharmacol. 66, 365–367 [DOI] [PubMed] [Google Scholar]
- 12. Rasmussen T. N., Novak I., Nielsen S. M. (2004) Eur. J. Biochem. 271, 4366–4374 [DOI] [PubMed] [Google Scholar]
- 13. Pals-Rylaarsdam R., Gurevich V. V., Lee K. B., Ptasienski J. A., Benovic J. L., Hosey M. M. (1997) J. Biol. Chem. 272, 23682–23689 [DOI] [PubMed] [Google Scholar]
- 14. Hislop J. N., Caunt C. J., Sedgley K. R., Kelly E., Mundell S., Green L. D., McArdle C. A. (2005) J. Mol. Endocrinol. 35, 177–189 [DOI] [PubMed] [Google Scholar]
- 15. Heding A., Vrecl M., Hanyaloglu A. C., Sellar R., Taylor P. L., Eidne K. A. (2000) Endocrinology 141, 299–306 [DOI] [PubMed] [Google Scholar]
- 16. Bhatnagar A., Willins D. L., Gray J. A., Woods J., Benovic J. L., Roth B. L. (2001) J. Biol. Chem. 276, 8269–8277 [DOI] [PubMed] [Google Scholar]
- 17. Marchese A., Paing M. M., Temple B. R., Trejo J. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 601–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luttrell L. M., Lefkowitz R. J. (2002) J. Cell Sci. 115, 455–465 [DOI] [PubMed] [Google Scholar]
- 19. Reiter E., Lefkowitz R. J. (2006) Trends Endocrinol. Metab. 17, 159–165 [DOI] [PubMed] [Google Scholar]
- 20. Dromey J. R., Pfleger K. D. (2008) Endocr. Metab. Immune Disord. Drug Targets 8, 51–61 [DOI] [PubMed] [Google Scholar]
- 21. Luttrell L. M., Gesty-Palmer D. (2010) Pharmacol. Rev. 62, 305–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rajagopal S., Rajagopal K., Lefkowitz R. J. (2010) Nat. Rev. Drug Discov. 9, 373–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pouysségur J., Volmat V., Lenormand P. (2002) Biochem. Pharmacol. 64, 755–763 [DOI] [PubMed] [Google Scholar]
- 24. Shenoy S. K., Lefkowitz R. J. (2005) J. Biol. Chem. 280, 15315–15324 [DOI] [PubMed] [Google Scholar]
- 25. Shenoy S. K., McDonald P. H., Kohout T. A., Lefkowitz R. J. (2001) Science 294, 1307–1313 [DOI] [PubMed] [Google Scholar]
- 26. Hislop J. N., Henry A. G., Marchese A., von Zastrow M. (2009) J. Biol. Chem. 284, 19361–19370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ciechanover A. (2005) Nat. Rev. Mol. Cell Biol. 6, 79–87 [DOI] [PubMed] [Google Scholar]
- 28. Hislop J. N., von Zastrow M. (2011) Traffic 12, 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haglund K., Dikic I. (2005) EMBO J. 24, 3353–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dikic I., Dötsch V. (2009) Nat. Struct. Mol. Biol. 16, 1209–1210 [DOI] [PubMed] [Google Scholar]
- 31. Pickart C. M., Fushman D. (2004) Curr. Opin. Chem. Biol. 8, 610–616 [DOI] [PubMed] [Google Scholar]
- 32. Shenoy S. K., Barak L. S., Xiao K., Ahn S., Berthouze M., Shukla A. K., Luttrell L. M., Lefkowitz R. J. (2007) J. Biol. Chem. 282, 29549–29562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shenoy S. K., Lefkowitz R. J. (2003) J. Biol. Chem. 278, 14498–14506 [DOI] [PubMed] [Google Scholar]
- 34. Ahn S., Shenoy S. K., Wei H., Lefkowitz R. J. (2004) J. Biol. Chem. 279, 35518–35525 [DOI] [PubMed] [Google Scholar]
- 35. Shenoy S. K., Drake M. T., Nelson C. D., Houtz D. A., Xiao K., Madabushi S., Reiter E., Premont R. T., Lichtarge O., Lefkowitz R. J. (2006) J. Biol. Chem. 281, 1261–1273 [DOI] [PubMed] [Google Scholar]
- 36. Kroeger K. M., Hanyaloglu A. C., Seeber R. M., Miles L. E., Eidne K. A. (2001) J. Biol. Chem. 276, 12736–12743 [DOI] [PubMed] [Google Scholar]
- 37. Pfleger K. D., Dromey J. R., Dalrymple M. B., Lim E. M., Thomas W. G., Eidne K. A. (2006) Cell. Signal. 18, 1664–1670 [DOI] [PubMed] [Google Scholar]
- 38. Dantuma N. P., Groothuis T. A., Salomons F. A., Neefjes J. (2006) J. Cell Biol. 173, 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perroy J., Pontier S., Charest P. G., Aubry M., Bouvier M. (2004) Nat. Methods 1, 203–208 [DOI] [PubMed] [Google Scholar]
- 40. Kocan M., See H. B., Seeber R. M., Eidne K. A., Pfleger K. D. (2008) J. Biomol. Screen. 13, 888–898 [DOI] [PubMed] [Google Scholar]
- 41. Lee H. J., Mun H. C., Lewis N. C., Crouch M. F., Culverston E. L., Mason R. S., Conigrave A. D. (2007) Biochem. J. 404, 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. See H. B., Seeber R. M., Kocan M., Eidne K. A., Pfleger K. D. (2011) Assay Drug Dev. Technol. 9, 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hanyaloglu A. C., Vrecl M., Kroeger K. M., Miles L. E., Qian H., Thomas W. G., Eidne K. A. (2001) J. Biol. Chem. 276, 18066–18074 [DOI] [PubMed] [Google Scholar]
- 44. Celver J. P., Lowe J., Kovoor A., Gurevich V. V., Chavkin C. (2001) J. Biol. Chem. 276, 4894–4900 [DOI] [PubMed] [Google Scholar]
- 45. Gray J. A., Bhatnagar A., Gurevich V. V., Roth B. L. (2003) Mol. Pharmacol. 63, 961–972 [DOI] [PubMed] [Google Scholar]
- 46. Pan L., Gurevich E. V., Gurevich V. V. (2003) J. Biol. Chem. 278, 11623–11632 [DOI] [PubMed] [Google Scholar]
- 47. Gurevich V. V., Dion S. B., Onorato J. J., Ptasienski J., Kim C. M., Sterne-Marr R., Hosey M. M., Benovic J. L. (1995) J. Biol. Chem. 270, 720–731 [DOI] [PubMed] [Google Scholar]
- 48. Milasta S., Evans N. A., Ormiston L., Wilson S., Lefkowitz R. J., Milligan G. (2005) Biochem. J. 387, 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. DeWire S. M., Ahn S., Lefkowitz R. J., Shenoy S. K. (2007) Annu. Rev. Physiol. 69, 483–510 [DOI] [PubMed] [Google Scholar]
- 50. Pfleger K. D., Dalrymple M. B., Dromey J. R., Eidne K. A. (2007) Biochem. Soc. Trans. 35, 764–766 [DOI] [PubMed] [Google Scholar]
- 51. Ribas C., Penela P., Murga C., Salcedo A., García-Hoz C., Jurado-Pueyo M., Aymerich I., Mayor F., Jr. (2007) Biochim. Biophys. Acta 1768, 913–922 [DOI] [PubMed] [Google Scholar]
- 52. Oakley R. H., Laporte S. A., Holt J. A., Barak L. S., Caron M. G. (2001) J. Biol. Chem. 276, 19452–19460 [DOI] [PubMed] [Google Scholar]
- 53. Prossnitz E. R. (2004) Life Sci. 75, 893–899 [DOI] [PubMed] [Google Scholar]
- 54. Berglund M. M., Schober D. A., Statnick M. A., McDonald P. H., Gehlert D. R. (2003) J. Pharmacol. Exp. Ther. 306, 147–156 [DOI] [PubMed] [Google Scholar]
- 55. Charest P. G., Bouvier M. (2003) J. Biol. Chem. 278, 41541–41551 [DOI] [PubMed] [Google Scholar]
- 56. Gehret A. U., Hinkle P. M. (2010) Biochem. J. 428, 235–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ponimaskin E., Dumuis A., Gaven F., Barthet G., Heine M., Glebov K., Richter D. W., Oppermann M. (2005) Mol. Pharmacol. 67, 1434–1443 [DOI] [PubMed] [Google Scholar]
- 58. Oakley R. H., Laporte S. A., Holt J. A., Barak L. S., Caron M. G. (1999) J. Biol. Chem. 274, 32248–32257 [DOI] [PubMed] [Google Scholar]
- 59. Oakley R. H., Laporte S. A., Holt J. A., Caron M. G., Barak L. S. (2000) J. Biol. Chem. 275, 17201–17210 [DOI] [PubMed] [Google Scholar]
- 60. Tulipano G., Stumm R., Pfeiffer M., Kreienkamp H. J., Höllt V., Schulz S. (2004) J. Biol. Chem. 279, 21374–21382 [DOI] [PubMed] [Google Scholar]
- 61. Torrecilla I., Tobin A. B. (2006) Curr. Pharm. Des. 12, 1797–1808 [DOI] [PubMed] [Google Scholar]
- 62. Shenoy S. K., Modi A. S., Shukla A. K., Xiao K., Berthouze M., Ahn S., Wilkinson K. D., Miller W. E., Lefkowitz R. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6650–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Werry T. D., Christopoulos A., Sexton P. M. (2006) Curr. Pharm. Des. 12, 1683–1702 [DOI] [PubMed] [Google Scholar]
- 64. Tohgo A., Pierce K. L., Choy E. W., Lefkowitz R. J., Luttrell L. M. (2002) J. Biol. Chem. 277, 9429–9436 [DOI] [PubMed] [Google Scholar]
- 65. Defea K. (2008) Br. J. Pharmacol. 153, S298–S309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cervantes D., Crosby C., Xiang Y. (2010) Circ. Res. 106, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tang J., Chen J., Ramanjaneya M., Punn A., Conner A. C., Randeva H. S. (2008) Cell. Signal. 20, 1651–1661 [DOI] [PubMed] [Google Scholar]
- 68. Simaan M., Bédard-Goulet S., Fessart D., Gratton J. P., Laporte S. A. (2005) Cell. Signal. 17, 1074–1083 [DOI] [PubMed] [Google Scholar]