Abstract
The bacterial Rcs phosphorelay is a stress-induced defense mechanism that controls the expression of numerous genes, including those for capsular polysaccharides, motility, and virulence factors. It is a complex multicomponent system that includes the histidine kinase (RcsC) and the response regulator (RcsB) and also auxiliary proteins such as RcsF. RcsF is an outer membrane lipoprotein that transmits signals from the cell surface to RcsC. The physiological signals that activate RcsF and how RcsF interacts with RcsC remain unknown. Here, we report the three-dimensional structure of RcsF. The fold of the protein is characterized by the presence of a central 4-stranded β sheet, which is conserved in several other proteins, including the copper-binding domain of the amyloid precursor protein. RcsF, which contains four conserved cysteine residues, presents two nonconsecutive disulfides between Cys74 and Cys118 and between Cys109 and Cys124, respectively. These two disulfides are not functionally equivalent; the Cys109–Cys124 disulfide is particularly important for the assembly of an active RcsF. Moreover, we show that formation of the nonconsecutive disulfides of RcsF depends on the periplasmic disulfide isomerase DsbC. We trapped RcsF in a mixed disulfide complex with DsbC, and we show that deletion of dsbC prevents the activation of the Rcs phosphorelay by signals that function through RcsF. The three-dimensional structure of RcsF provides the structural basis to understand how this protein triggers the Rcs signaling cascade.
Keywords: Bacteria, Crystal Structure, Disulfide, Protein Folding, Protein Structure, Escherichia coli, Oxidative Folding, Periplasm, Protein-disulfide Isomerase, Thioredoxin
Introduction
The envelope of Gram-negative bacteria is a complex macromolecular structure that serves as a permeability barrier protecting bacterial cells from ever-changing environmental conditions (1–3). To maintain envelope integrity, Gram-negative bacteria have evolved complicated signaling systems that allow them to react to a range of envelope stresses by modulating the expression of specific genes. Several envelope stress response systems have been described in Escherichia coli, including the Rcs phosphorelay (4, 5).
The Rcs signaling cascade was first identified by its role in the transcriptional regulation of genes involved in the biosynthesis of colanic acid in E. coli (6). Colanic acid is a capsule polysaccharide that is produced by bacteria in response to envelope perturbations, osmotic stress, and growth on surfaces (7). Production of colanic acid leads to the formation of a mucoid layer on the bacterial cell surface, which results in a distinctive mucoid phenotype (see Fig. 4). In addition to its role in controlling capsule synthesis, the Rcs phosphorelay is required for normal biofilm formation, contributes to antibiotic resistance, and regulates virulence-associated structures involved in motility and host recognition (8–10).
FIGURE 4.
Deletion of dsbC prevents the induction of the Rcs phosphorelay by deletion of mdoG. A, mdoG, dsbC, dsbD, mdoG dsbC, and mdoG dsbD mutant strains were grown on M63 minimal medium for 2 days at room temperature. Similar results were obtained for the mdoH mutants (data not shown). B, mdoG dsbC strain was complemented with either pBAD33, pBAD33::dsbC, pBAD33::dsbCCXXS, or pBAD33::dsbCSXXS and grown on M63 minimal medium for 2 days at room temperature. Like DsbCSXXS, DsbCCXXS keeps the chaperone activity but cannot function as reductase.
The Rcs phosphorelay is a complex signaling pathway that includes three proteins, RcsC, RcsD, and RcsB (9, 11). RcsC is an inner membrane sensor protein that presents an N-terminal histidine kinase domain and a C-terminal receiver domain (12). In response to environmental signals, RcsC is predicted to autophosphorylate a conserved histidine residue located in the kinase domain. The phosphoryl group is then transferred to a conserved aspartate residue present in the receiver domain (9, 13). RcsD, which is also located in the inner membrane, contains a histidine-containing phosphotransmitter domain (Hpt), which mediates phosphoryl transfer between the receiver domain of RcsC and an aspartate residue located in the receiver domain of RcsB, a cytoplasmic response regulator (14). Phosphorylation of RcsB allows this cytoplasmic protein to bind to target promoters on the chromosomal DNA via a DNA-binding domain (9, 15).
In addition to RcsC, RcsD, and RcsB, the Rcs phosphorelay involves auxiliary proteins that are required for the normal functioning of the system (9, 11). In E. coli, the best characterized auxiliary proteins are RcsA and RcsF. RcsA is an unstable cytoplasmic protein that belongs to the same family of transcriptional regulators as RcsB (16). RcsA interacts with phosphorylated RcsB to form a heterodimer that binds to the promoters of a subset of Rcs-regulated genes, including those involved in capsule synthesis (17, 18).
RcsF is a small lipoprotein (14 kDa) anchored in the outer membrane and oriented toward the periplasm (19). RcsF was first identified by its ability to induce the Rcs phosphorelay when overexpressed (20), an activation that is dependent on RcsC (21). The function of RcsF is to transduce signals that this protein perceives in the outer part of the envelope to RcsC (9). For instance, treatments that alter the integrity of the outer membrane, such as exposure to cationic antimicrobial peptides (22), or that target the peptidoglycan layer, like exposure to lysozyme (23) or β-lactam antibiotics (10), activate the Rcs phosphorelay. This induction has been shown to be dependent on RcsF (19). Moreover, mutations that lead to defects in the envelope, such as mutations in the lipopolysaccharide synthesis pathway or in mdoG and mdoH, two genes that are involved in the synthesis of membrane-derived oligosaccharides, also activate the Rcs cascade in an RcsF-dependent manner (19, 24).
Despite the important role played by the Rcs phosphorelay in monitoring envelope integrity, in biofilm formation, and in bacterial virulence, we still have a poor understanding of the molecular mechanisms that activate the system and control its correct functioning. In particular, the physiological signals that turn on the Rcs phosphorelay remain unknown, and how the outer membrane protein sensor RcsF transfers environmental signals to RcsC is not understood.
The Rcs phosphorelay has been mostly studied using bacterial genetics. To elucidate how this system works and to understand the molecular mechanisms that control its activity, biochemical and structural data are required.
Here, we report the three-dimensional crystal structure of the sensor protein RcsF. The protein fold is characterized by the presence of a central four-stranded β sheet. Two nonconsecutive disulfides, including one cross-strand disulfide, are present in the structure. We show that formation of these disulfides involves the periplasmic protein-disulfide isomerase DsbC. Our work provides the structural and biochemical basis that should facilitate the understanding of the mechanism used by RcsF to transmit signals across the periplasm.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
The bacterial strains used in this study are listed in supplemental Table S1. All the strains are derivatives of the E. coli K-12 strain MC4100 (25). Cells were grown aerobically in either LB or M63 minimal media. Unless otherwise indicated, M63 minimal medium was supplemented with 0.2% glucose, vitamins, 1 mm MgSO4, and a mixture of amino acids. When necessary, growth media were supplemented with either chloramphenicol (25 μg/ml), kanamycin (50 μg/ml), or ampicillin (200 μg/ml). All alleles were moved by bacteriophage P1 transduction using standard procedures (26). The resistance cassette was removed from the chromosome by using the pCP20 plasmid encoding the FLP recombinase (27).
Construction of the Expression Plasmids
The genomic DNA of E. coli MC4100 was used as a template for the PCR. The primers are listed in supplemental Table S2. The PCR amplifications were performed using Pfu DNA polymerase (Fermentas), and samples were subjected to 25 cycles of 30 s denaturation at 95 °C, 30 s annealing at 55 °C, and 3 min elongation at 68 °C, followed by 5 min at 68 °C. The rcsF gene, without the sequence corresponding to the signal peptide and the lipobox, was amplified using rcsF Fw1 as forward primer (containing an NcoI site, CCATGG) and rcsF Rv1 as reverse primer (containing an XhoI site, CTCGAG) (see supplemental Table S2 for primers list). The PCR product was analyzed by electrophoresis on a 1% agarose gel and stained with ethidium bromide. It was then purified, double-digested, and ligated into the plasmid pET28a (Novagen) for expression of the protein with a C-terminal His tag and no N-terminal His tag (the sequence of the recombinant protein is provided supplemental Table S2). For expression of RcsF in the periplasm using the OmpA signal sequence, the rcsF gene, without the sequence corresponding to the first 16 residues, was amplified using rcsF Fw2 (containing a KasI site, GGCGCC) as forward primer and rcsF Rv2 (containing an NcoI site, CCATGG) as reverse primer. The amplification product was then cloned into the pASK-IBA16 vector (IBA BioTAGnology®). The truncated version of rcsF starting at residue Pro48 (RcsFtrunc) was also cloned into the pASK-IBA16 vector, after PCR amplification using the primers rcsFtrunc Fw and rcsF Rv2. The resulting plasmids (pET28a::rcsF, pASK-IBA16::rcsF, and pASK-IBA::rcsFtrunc) were first transformed into E. coli XLIBlue competent cells for sequencing. pET28a::rcsF was then transformed into Rosetta-Gami BL21 (New England Biolabs)-competent cells for expression of RcsF in the oxidizing cytoplasm of that strain. pASK-IBA16::rcsF and pASK-IBA16::rcsFtrunc were transformed in different MC4100 background for phenotypic characterization.
Site-directed Mutagenesis
The QuikChange mutagenesis kit (Stratagene) was used for the generation of the RcsF mutants in which the cysteine residues are replaced by serines. The plasmid pASK-IBA16::rcsF was used as a template. The forward and reverse primers that were used are listed in supplemental Table S2. The PCR amplification reaction was performed using Pfu DNA polymerase, and samples were subjected to 20 cycles of 30 s denaturation at 95 °C, 30 s annealing at 55 °C, and 9 min elongation at 68 °C, followed by a 11-min step at 68 °C. The DpnI restriction enzyme was used to digest the parental supercoiled double-stranded DNA. The plasmids harboring the sequence of the mutated proteins were transformed into E. coli XLIBlue competent cells for sequencing and then in PL262 (rcsF::kan) competent cells for phenotypic characterization.
Expression and Purification of the Recombinant RcsF Protein
Rosetta-Gami BL21 cells transformed with the pET28a::rcsF expression plasmid were grown at 37 °C in 4 liters of LB medium containing ampicillin (200 mg/liter) and kanamycin (50 mg/liter) to an A600 of 0.6. Isopropyl 1-thio-β-d-galactopyranoside (0.5 mm) was then added to induce expression during 3 h with shaking. The cells were harvested by centrifugation, and the cell pellet was resuspended in 20 ml of a buffer containing NaPi, pH 8.0, 100 mm and NaCl 0.3 m (Buffer A). The cells were then disrupted using a French press. After centrifugation (40 min, 18,000 × g), the supernatant was diluted three times and loaded onto a Ni-NTA6 chromatography column equilibrated with Buffer A. Elution was performed by applying a linear gradient of imidazole (from 30 to 300 mm) in Buffer A, and the eluted fractions were analyzed by SDS-PAGE containing 16% (v/v) acrylamide. The protein was ≈99% homogeneous after this step.
Crystallization, Data Collection, and Processing
Before crystallization, the protein solution at a concentration of 11 mg/ml was mixed with 5-amino-2,4,6-triiodoisophthalic acid (I3C) neutralized with LiOH to reach a concentration of 20 mm. All attempts to grow crystals in the absence of I3C were completely unsuccessful. Furthermore, the replacement of I3C with 5-amino-2,4,6-tribromoisophthalic acid (the same compound with three bromo atoms instead of the three iodine atoms) did not produce any crystals. The incorporation of the I3C compound known as the “magic triangle” (28) initially aimed at providing a way to obtain phases by single wavelength anomalous dispersion, but it turned out to be an essential ligand for crystal growth. Crystals for data collection were grown by vapor diffusion using the hanging drop method at 18 °C. The well solution (500 μl) consisted of 1.8 m ammonium sulfate, 0.1 m sodium acetate, pH 4.5, and 0.02% (w/v) sodium azide. The drop was formed by mixing 1 μl of the protein + I3C mixture with 1 μl of the well solution. Crystals appeared after several weeks.
Before data collection, crystals were soaked for a few seconds in a solution similar to the mother liquor but containing 20% (w/v) glycerol as cryo-protectant and flash-cooled at 100 K. Data were collected on beamline X12 at EMBL, DESY. Two data sets were collected. The first data set was collected at a long wavelength with a high level of redundancy, to obtain a good anomalous signal from the iodine atoms for phasing using the single wavelength anomalous dispersion method. For the second data set, a shorter wavelength was used and was allowed to reach a higher resolution for the refinement. The diffraction images were processed and merged with the XDS program package (29). The crystals are trigonal, space group P3121, with one molecule in the asymmetric unit. Statistics of data collection and processing are given in Table 1.
TABLE 1.
Data collection and refinement statistics
| Data set 1 (structure solution) | Data set 2 (refinement) | |
|---|---|---|
| Wavelength | 1.54975 Å | 0.977910 Å |
| Space group | P3121 | P3121 |
| Unit cell | ||
| a | 56.14 Å | 55.71 Å |
| c | 61.81 Å | 61.68 Å |
| Resolution range | ||
| Overall (ov) | 19.1 to 2.46 | 19.0 to 2.0 |
| Highest shell (hs) | 2.52 to 2.46 | 2.20 to 2.0 |
| No. of reflections | ||
| Total | 87,651 | 27,634 |
| Unique | 7,889 | 7,786 |
| Friedel pairs | Not merged | Merged |
| Completeness (ov/hs) | 99.4/97.4% | 99.6/99.6% |
| Rmerge (ov/hs) | 0.083/0.377 | 0.082/0.522 |
| 〈I/σ(I)〉 (ov/hs) | 21.9/6.0 | 11.5/2.7 |
| Refinement | ||
| R-factor (ov/hs) | 0.196/0.240 | |
| Rfree (ov/hs) | 0.258/0.330 | |
| Root mean square deviation from ideality | ||
| Bonds | 0.022 Å | |
| Angles | 2.16° | |
| No. of water molecules | 46 | |
| Mean B values | ||
| Protein | 30.3 Å2 | |
| Water | 37.4 Å2 | |
| Sulfate | 35.0 Å2 | |
| I3C | 34.1 Å2 | |
The structure was solved by the single wavelength anomalous dispersion method applied to data set 1 using the Auto-Rickshaw procedure (30). The resulting model was refined against the high resolution data (data set 2) using REFMAC5 (31) of the CCP4 suite (32) and was manually completed and corrected using COOT (33). The final structure contains residues 48–131 of the protein chain, 46 water molecules, 1 sulfate ion, and 1 I3C molecule. Some statistics related to the refinement are given in Table 1.
Coordinates and Structure Factors
Atomic coordinates and structure factors have been deposited in the PDB with accession code 2y1b.
Trapping and Purification of the DsbC-RcsF Complex
A 1-liter LB culture of the dsbC strain carrying pBAD33::dsbCCXXS was grown at 25 °C to an A600 of 0.6. Expression was induced for 5 h by the addition of 0.2% l-arabinose. Proteins were precipitated using trichloroacetic acid (TCA) (10%) and resuspended in 25 ml of NaPi, pH 8, 100 mm, NaCl 300 mm, SDS 0.3%, urea 8 m, and iodoacetamide 100 mm to prevent any further disulfide bond rearrangement. The lysate was centrifuged at 15,000 × g for 45 min. After incubation (overnight at room temperature) of the cleared mixture with 2 ml of Ni-NTA resin (Qiagen), a 1-ml column was packed with the protein-Ni-NTA resin complex and was washed thoroughly using NaPi, pH 8, 100 mm, NaCl 300 mm, and SDS 0.3%. Elution of the complexes involving DsbC CXXS was performed using a step gradient from 30 to 300 mm imidazole. Only one fraction eluted from the column, which was concentrated 10-fold and analyzed by SDS-PAGE with or without DTT. After electrophoresis, the proteins were transferred to a nitrocellulose membrane and probed with an anti-RcsF antibody (1:3000) produced from a rabbit immunized with the purified protein (Eurogentec). Anti-rabbit IgG (Sigma) was used as the secondary antibody at a 1:5000 dilution. Thermo Scientific Pierce ECL Western blotting substrate and Fuji film were used to visualize the protein bands.
Mass Spectrometry
Intact mass measurement of RcsF was done by diluting the purified protein with 50:50 acetonitrile/water containing 0.1% formic acid to an approximate concentration of 5 μm. The sample was introduced by off-line infusion using a capillary electrospray at 1.5 μl/min. An LTQ XL mass spectrometer (LTQ XL, Thermo Fisher Scientific) was used to acquire mass spectra from m/z 400 to 2000 in centroid mode. Electrospray source conditions such as “source fragmentation” voltage and the tube lens voltage were optimized to help desolvation but without fragmenting the intact protein. Default values were used for most other data acquisition parameters. The resulting spectra were averaged up to 200 scans and were deconvoluted using ProMass software (Thermo Fisher Scientific). We determined the mass of the protein used for crystallization to verify the sequence of the expression product.
In Vivo Expression Levels of RcsF
The bacterial strains were grown in minimal medium at 30 °C to an A600 of 0.8, and 1-ml samples were taken. Proteins were precipitated with 10% ice-cold TCA and centrifuged at 13,000 × g for 1 min. The pellets were washed with acetone, dried, and resuspended with 5× Laemmli buffer (SDS 10%, bromophenol blue 0.05%, glycerol 61%, Tris-HCl, pH 6.8, 300 mm). Standardized samples were analyzed by SDS-PAGE under nonreducing conditions. After electrophoresis, proteins were transferred to a nitrocellulose membrane and probed with an anti-RcsF antibody.
RESULTS
Three-dimensional Structure of RcsF
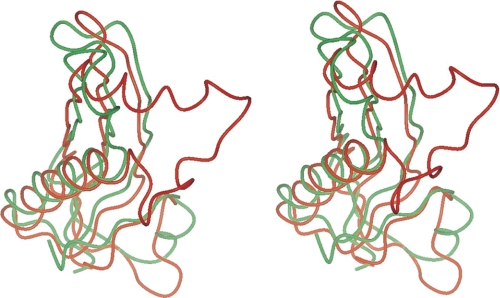
Here, we report the three-dimensional crystal structure of the RcsF protein sensor. The overall fold is illustrated in Fig. 1 and is characterized by the presence of a central four-stranded β sheet. One face of the β sheet is covered by a long α helix (α2), whereas the short helix α1 appears on the other side of the β sheet. After a search for a similar fold in the PDB using the DALI server (34), the best structural alignment corresponded to a protein of unknown function from the Gram-positive bacteria Bacillus cereus (PDB code 1vr4). 75 Cα atoms were superposed with a root mean square distance of 1.8 Å. A stereoscopic view of the superposition of the Cα traces of RcsF and code 1vr4 is shown in Fig. 2. The central β sheet and the α2 helix are well conserved. Two main differences are observed as follows: the α1 helix of RcsF has disappeared and is replaced by a loop in 1vr4 and the loop between β2 and α2 in RcsF is replaced in 1vr4 by an additional helix followed by a much longer loop. Other structural homologues of RcsF include a selenium-binding protein from Methanococcus vannielii (PDB code 2jz7), dodecin from Halobacterium salinarum (PDB code 2vx9-A), and the copper-binding domain of the Alzheimer peptide precursor protein (PDB code 2fma-A).
FIGURE 1.
Overall fold of the RcsF molecule. The protein is colored from green (N terminus) to pink (C terminus). A, topological diagram on which the disulfide bonds are represented by a yellow line. The limits of the secondary structure elements are indicated. B, ribbon diagram in a more or less similar orientation. The side chains of the cysteine residues taking part in the disulfide bonds are drawn as balls and sticks. A was prepared with Topdraw (54) and B with CCP4mg (55).
FIGURE 2.
Fold of RcsF is conserved in other proteins. Stereoscopic view of a superposition of the Cα traces of RcsF (green) and PDB entry 1vr4 (red). The orientation is similar to that of Fig. 1B. This figure was prepared with CCP4mg (55).
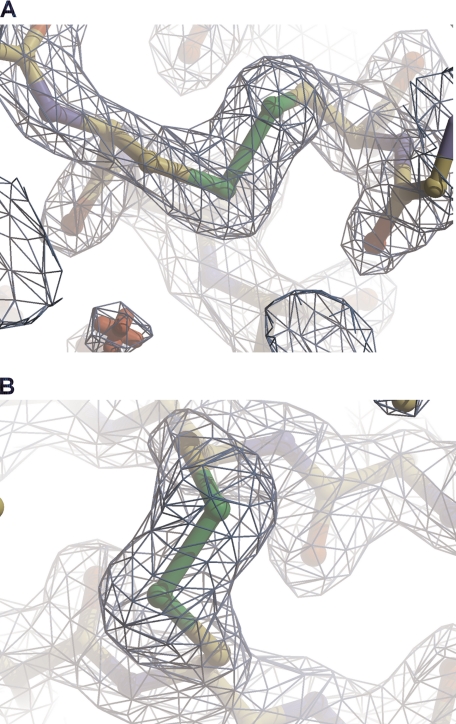
Two disulfide bonds are observed in the structure. The first one between Cys74 and Cys118 links a residue in the β2 strand to a residue located in the β3-β4 loop. The second disulfide bond between Cys109 and Cys124 is a cross-strand disulfide (CSD) that links the β3 and β4 strands. Both disulfide bonds are illustrated in the electron density in Fig. 3.
FIGURE 3.
RcsF presents two nonconsecutive disulfides. The regions of the disulfide bond between Cys74 and Cys118 (A) and between Cys109 and Cys124 (B) are shown. The (2mFo − DFc) electron density map is contoured at a level of 1.5 σ. This figure was prepared with COOT (33).
I3C Ligand
As explained under “Experimental Procedures,” the presence of the I3C ligand was essential for crystallization, suggesting that a strong interaction exists between this ligand and the protein. Indeed, we observed a close contact (3.05 Å) between one of the iodine atoms and the carbonyl oxygen atom of Pro82. This interaction is nearly linear with a bond angle of 170.6° around the iodine atom and is usually described as “halogen bonding” (35, 36). On the aromatic ring of the I3C molecules, the two positions in ortho relative to this iodine atom are occupied by electron-attracting carboxylate groups that favor the appearance of a positive region on the outermost portion of the halogen surface. This phenomenon is known as a σ-hole and is responsible for the interaction with a nucleophile like the oxygen atom of a carbonyl group. One of the carboxylate groups is hydrogen-bonded to Ser84 Oγ.
N-terminal Segment of RcsF Is Disordered
The protein that was used for crystallization was expressed as a soluble protein containing a C-terminal His tag and lacking the first 16 residues corresponding to the signal peptide and the lipobox (remember that RcsF is a lipoprotein whose lipid moiety is anchored in the outer membrane). However, the observed electron density begins at residue Pro48, suggesting that about 30 residues in the N-terminal part of the protein are completely disordered. The integrity of the sequence of the purified protein was confirmed using mass spectrometry to rule out any nonspecific proteolytic cleavage of the protein during expression and purification. The measured average mass was 13,609 ± 2 Da, which is in good agreement with a theoretical average mass of 13,612 Da taking into account the complete processing of the N-terminal methionine.
We then decided to determine whether the portion of RcsF that is not visible in the structure is important for the activity of the protein. It has been shown recently that expression of RcsF as a protein soluble in the periplasm constitutively induces the Rcs phosphorelay (22). Therefore, we decided to study the ability of a truncated version of the protein (RcsFtrunc), corresponding to residues Pro48 to Lys134, to induce the Rcs phosphorelay when expressed in the periplasm, using strain mucoidity as a read-out. As explained in the Introduction, induction of the Rcs system leads to colanic acid production, which results in a distinctive mucoid phenotype. As indicated in Table 2, we observed that expression of RcsFtrunc leads to a mucoid phenotype comparable with that observed when wild-type RcsF is expressed unanchored in the periplasm (supplemental Fig. S1). Thus, we can conclude from this experiment that the fraction of the polypeptide chain that is disordered in the crystal is not required for signaling, at least when RcsF is expressed as a soluble protein in the periplasm.
TABLE 2.
Mucoid phenotype of strains expressing RcsF cysteine to serine mutants
The cysteine to serine RcsF mutants were expressed as soluble proteins in the periplasm of an rcsF deletion strain (PL262). Strains were grown on LB medium (containing ampicillin) for 2 days at room temperature.
| Expressed protein | Mucoid phenotype | |
|---|---|---|
| wtRcsF | CCCC | +++ |
| RcsFtrunc | CCCC | +++ |
| RcsFC74S | SCCC | ++ |
| RcsFC109S | CSCC | − |
| RcsFC118S | CCSC | +++ |
| RcsFC124S | CCCS | − |
| RcsFC74S/C118S | SCSC | +++ |
| RcsFC109/C124S | CSCS | − |
Cys109–Cys124 Disulfide Is Required for Assembly
A striking feature of the RcsF structure is the presence of two nonconsecutive disulfides. We wanted to determine whether these disulfides are required for activation of the Rcs phosphorelay. We generated single cysteine to serine mutants and tested whether expression of the mutants in the periplasm leads to a mucoid phenotype (Table 2 and supplemental Fig. S2). We observed that expression of RcsFC74S and RcsFC118S, in which only the Cys109–Cys124 disulfide remains, leads to a mucoid phenotype. In contrast, expression of the RcsFC109S and RscFC124S mutants, in which only the Cys74–Cys118 disulfide can be formed, does not activate the production of capsule polysaccharides (Table 2). Similar results were obtained when the cysteine pairs Cys74–Cys118 and Cys109–Cys124 were mutated; only expression of an RcsF mutant, in which Cys74 and Cys118 are replaced by serines (this mutant conserves the Cys109–Cys124 disulfide), resulted in a mucoid phenotype (Table 2). The mutated proteins in which the Cys109–Cys124 disulfide cannot be formed were not detected by Western blot analysis, in contrast to the mutants lacking the Cys74–Cys118 disulfide (data not shown). This suggests that the inability of the mutant proteins lacking the Cys109–Cys124 disulfide to activate the phosphorelay is due to a folding defect. Thus, these experiments suggest that the Cys109–Cys124 disulfide is important for the assembly of an active RcsF in the cell envelope, whereas the Cys74–Cys118 disulfide seems to be dispensable.
dsbC Deletion Prevents Signaling through RcsF
In E. coli, disulfides are introduced into periplasmic proteins by the oxidoreductase DsbA (37). The dependence of RcsF on DsbA for oxidative folding has already been reported by Kadokura et al. (38) and was not explored further in this work. DsbA is a powerful oxidant that rapidly reacts with unfolded proteins to oxidize them (37, 39). When disulfides need to be formed between cysteine residues that are not consecutive in the sequence, as is the case in RcsF, DsbA may introduce non-native disulfides, which leads to protein misfolding and proteolysis. These non-native disulfides are corrected by DsbC, a soluble dimeric protein that functions as a protein-disulfide isomerase in the periplasm (39, 40). Each monomer of DsbC possesses a thioredoxin domain harboring a catalytic CXXC motif. The cysteine residues of this motif are kept reduced in the periplasm by DsbD, an inner membrane protein that catalyzes the transfer of reducing equivalents from the cytoplasmic thioredoxin system across the membrane to DsbC (41). These reducing equivalents maintain DsbC in a reduced state in the periplasm, allowing the protein to function as an isomerase/reductase (see below). Several DsbC substrates with multiple cysteines have already been identified; they include four periplasmic enzymes (42, 43) as well as the essential β-barrel protein LptD (44).
The presence of two disulfides formed between cysteines that are not consecutive in the sequence of RcsF suggested that the assembly of this protein involves DsbC. To test this hypothesis, we searched to determine whether deletion of dsbC affects signaling via RcsF. As explained in the Introduction, it has been reported that mutations that alter the integrity of the cell envelope, such as mdoG and mdoH, induce the Rcs phosphorelay in an RcsF-dependent manner (19). We generated double mutants mdoH dsbC and mdoG dsbC to determine whether deletion of dsbC alters the mucoid phenotype of single mutants mdoH and mdoG. We found that whereas the mdoG and mdoH strains appear mucoid on minimal media, in agreement with reported data, the double mutants do not (Fig. 4A). This indicates that deletion of dsbC prevents the induction of the Rcs phosphorelay via RcsF and suggests that RcsF assembly involves DsbC.
The nonmucoid phenotype of the double mutants could result from the absence of the isomerase activity of DsbC or from the absence of the chaperone activity of DsbC, which has been described in vitro (45). To test these two hypotheses, we studied the phenotype of mdoG dsbD and mdoH dsbD double mutants. In strains lacking dsbD, DsbC is not recycled and is mostly found in the oxidized state (46), which prevents the protein from functioning as an isomerase/reductase but does not affect the chaperone activity. We found that the double mutants mdoG dsbD and mdoH dsbD are less mucoid than the single mutants mdoG and mdoH (Fig. 4A), suggesting that it is the isomerase activity of DsbC that is important for RcsF assembly. However, in contrast to the deletion of dsbC, the deletion of dsbD did not completely abolish the production of colanic acid, which prevented us from clearly discriminating between the two hypotheses. Thus, to firmly establish the involvement of the isomerase activity of DsbC in RcsF assembly, we compared the ability of wild-type DsbC and of a DsbC mutant that lacks the isomerase activity but keeps the chaperone activity (DsbCSXXS) to restore the mucoid phenotype of the double mutant mdoG dsbC. As shown in Fig. 4B, we observed that only expression of wild-type DsbC was able to restore the induction of the Rcs phosphorelay in the double mutant, indicating that it is the isomerase activity of DsbC that is involved in the assembly of RcsF.
RcsF Interacts with DsbC in Vivo
This prompted us to further investigate the interaction between RcsF and DsbC. As explained above, the catalytic site of DsbC is maintained in a reduced state in the oxidizing environment of the periplasm, which is critical to allow DsbC to react with non-native disulfides. The first cysteine of the CXXC catalytic motif performs a nucleophilic attack on an incorrect disulfide, which results in the formation of an unstable mixed disulfide complex between DsbC and the substrate protein. There are two ways to resolve this mixed disulfide, either by attack of another cysteine of the misfolded protein, which results in the formation of a more stable disulfide in the substrate and the release of reduced DsbC, or by attack of the other cysteine of the CXXC motif. In this second case, DsbC is released in the oxidized state and functions as a true reductase.
To confirm that DsbC is involved in the assembly of RcsF, we expressed a mutated version of DsbC presenting a CXXS catalytic site (DsbCCXXS) in a dsbC strain. Replacing the second cysteine of the catalytic site by a serine is a favorite approach to trap the complexes formed between the proteins from the thioredoxin family with their substrates (47, 48). As explained above, the C-terminal cysteine of the CXXC motif is the residue that performs a nucleophilic attack on the mixed disulfides formed between DsbC and its substrates. Thus, replacing this cysteine by a serine stabilizes the mixed disulfides and prevents DsbC from functioning as a reductase (Fig. 5A). After expression of the mutant protein in the periplasm of a dsbC deletion strain, the proteins were precipitated with TCA, and all free cysteines were alkylated with iodoacetamide to prevent any further disulfide bond rearrangements. The proteins were resuspended in a buffer containing 0.3% SDS and no reductant to maintain DsbCCXXS covalently bound to its potential substrate proteins. DsbCCXXS was then purified under denaturing conditions using a His tag present at the C terminus of the protein. Only one peak eluted from the column when an imidazole gradient was applied. DsbC was the most abundant protein in that peak, but other proteins of higher molecular weight were also present (data not shown). As the purification was performed under denaturing conditions, nonspecific binding to the column was minimal, and the bands co-eluting with DsbC likely correspond to complexes between DsbC and other proteins. To determine whether RcsF was trapped in complex with DsbC, the fractions were analyzed by Western blot using an anti-RcsF antibody. Although a band corresponding to the DsbC-RcsF complex could not be clearly identified (see under “Discussion”), we observed that addition of DTT led to the appearance of a band recognized by the anti-RcsF antibody and migrating at the size expected for RcsF (∼14 kDa) (Fig. 5B). This indicates that RcsF forms a mixed disulfide complex with DsbC, and thus that RcsF interacts with DsbC in vivo.
FIGURE 5.
Identification of RcsF trapped in complex with DsbC. A, the first cysteine residue of the CXXC motif of DsbC performs a nucleophilic attack on an incorrect disulfide, which results in the formation of a mixed disulfide complex. To trap DsbC substrates, the C-terminal cysteine was replaced by a serine. This mutant protein is still able to react with non-native disulfides but lacks the second cysteine of the catalytic motif required to resolve the mixed disulfide complex. B, complexes formed between DsbCCXXS and its substrates were purified using Ni-NTA resin. The samples were analyzed by SDS-PAGE without or with DTT. RcsF was detected by Western blot using an anti-RcsF antibody. The band marked by an asterisk corresponds to RcsF that was released from the mixed disulfide complex with DsbC upon DTT addition.
Deletion of dsbC Decreases the Levels of RcsF in the Cell Envelope
We then sought to determine whether the deletion of dsbC affects the assembly of RcsF in vivo by comparing the levels of RcsF in wild-type and dsbC strains. As shown in Fig. 6, we observed that deletion of dsbC decreases the abundance of RcsF indicating that the absence of DsbC impairs the correct assembly of RcsF in the cell envelope. Deletion of dsbD also led to decreased RcsF levels, although to a lesser extent than the deletion of dsbC, which is in agreement with the mild mucoid phenotype exhibited by the mdoG dsbD mutant.
FIGURE 6.
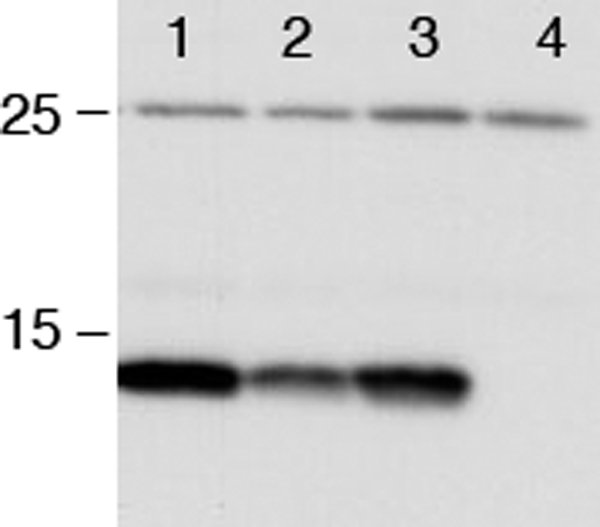
In vivo expression levels of RcsF. The RcsF expression level was determined in different backgrounds (line 1, wild type; line 2, dsbC; line 3, dsbD; line 4, rcsF). Cells were grown to exponential phase at 30 °C in M63 minimal medium, normalized by absorbance, and precipitated by TCA. RcsF was detected by Western blot using an anti-RcsF antibody.
DISCUSSION
The Rcs phosphorelay plays an important role in the response of bacteria to envelope stress by controlling the transcription of many genes, including the cell division gene ftsZ, the catalase katE, and those involved in capsule synthesis, biofilm formation, and virulence factors (9). About 30 years of research on the Rcs phosphorelay have led to the identification of the major components of this system and the characterization of the activation cascade. However, surprisingly little is known about the molecular mechanisms that activate the system and fine-tune its activity in response to changes in the integrity of the cell envelope. One of the most fascinating questions is how the outer membrane lipoprotein RcsF is activated and how, upon activation, it transmits the signal to the histidine kinase sensor RcsC, which is localized in the inner membrane. Biochemical and structural data are required to elucidate the details of the molecular mechanisms that led to the activation of this complicated multicomponent system and that allow signal transduction from the cell envelope to the cytoplasm.
Here, we report the structure of RcsF. The fold of the protein is characterized by a central four-stranded β sheet, sandwiched by two α helices. A similar fold has already been described in several proteins. Representative examples include dodecin, a small (7 kDa) riboflavin-binding protein found in prokaryotes that assembles to hollow sphere-like dodecameric complexes (49, 50), and the Alzheimer peptide precursor protein whose copper-binding domain is a structural homologue to RcsF (51).
Previous work by Kadukora et al. (38) led to the identification of RcsF as a substrate for the thiol-oxidase DsbA, which indicated that at least two of the four cysteine residues of RcsF are disulfide-bonded. This is nicely confirmed by the structure of RcsF, in which two disulfides can be observed. Interestingly, these two disulfides are formed between cysteine residues that are not consecutive in the sequence, Cys74–Cys118 and Cys109–Cys124. This suggested that the assembly of RcsF involves the periplasmic protein-disulfide isomerase DsbC. The involvement of DsbC in the formation of the RcsF disulfides was confirmed by a combination of genetic and biochemical evidence. 1) We showed that deletion of dsbC prevents the induction of the Rcs phosphorelay in mdoG and mdoH mutants, in which Rcs activation had been shown to occur through RcsF. 2) By using complementation experiments with wild-type DsbC or with a DsbC variant lacking the catalytic cysteine residues, we showed that it is the isomerase activity of DsbC that is important for RcsF assembly. 3) We showed that dsbC deletion leads to decreased levels of RcsF in the outer membrane. Furthermore, we observed that RcsF copurifies under nonreducing and denaturing conditions with a DsbC variant lacking the resolving cysteine of the CXXC. Addition of DTT to reduce the mixed disulfide linking DsbC to its target proteins led to the release of RcsF, which provides good evidence that these two proteins interact in the periplasm. The disulfide-bonded DsbC-RcsF complex (40 kDa) could not be clearly identified on the Western blot probably because of an altered migration and transfer of the RcsF-DsbC complex and of a decreased accessibility of the RcsF epitope(s) when RcsF is bound to the isomerase.
We also tested the functional role of the two nonconsecutive disulfides, which allowed us to conclude that the two disulfides of RcsF are not functionally equivalent; the Cys109–Cys124 disulfide seems particularly important for the stability of the protein as attested by the fact that removal of this disulfide impairs the assembly of RcsF more significantly than removal of the Cys74–Cys118 disulfide. Importantly, similar results were obtained independently by Prouvost (56) on Erwinia chrysantemi RcsF. In a recent publication, Farris et al. (22) generated mutants of Salmonella enterica serovar Typhimurium RcsF presenting cysteine to serine point mutations. They tested whether those RcsF mutants could activate the Rcs phosphorelay in response to polymyxin B, a cationic antimicrobial peptide, using a wza::lacZ transcriptional reporter (the wza gene is regulated by the Rcs system). In contrast to our results showing that RcsF mutants lacking the Cys74–Cys118 disulfide are still able to signal, they report that no β-galactosidase activity was observed for any of the four point mutants. This discrepancy could result from the fact that they used S. enterica serovar Typhimurium RcsF as well as a different reporter of Rcs activation.
Recently, Singh et al. (52) determined the disulfide connectivity of RcsF to test a new mass spectrometry analysis method that they had designed. They found one disulfide between Cys74 and Cys124. They deduced from that result that the second disulfide of RcsF connects Cys109–Cys118. They specifically searched their mass spectrometry data for this disulfide and found it. The structural and genetic data that we report indicate that, in contrast to what they found, RcsF contains two nonconsecutive disulfides between Cys74–Cys118 and Cys109–Cys124. To determine whether other oxidized forms of RcsF were present in the sample that was used for crystallization, we analyzed the RcsF protein preparation by LC-MS/MS. Because RcsF has four cysteines, there are six different disulfides that can be formed. We found all the possible cysteine pairs in the sample (data not shown). However, using an internal standard, we quantified the various cysteine pairs and found that the Cys74–Cys118 and Cys109–Cys124 disulfides, which correspond to those observed in the structure of the native protein, represent more than 90% of the disulfide population. This suggests that the RcsF protein preparation that was used for crystallization is contaminated by low amounts of non-native RcsF presenting incorrect disulfides. Singh et al. (52) do not report any quantification results, which limits the interpretation of the data. However, it is likely that the disulfides that they identified correspond to those of a minor, non-native form of RcsF.
A striking feature of the disulfide connecting Cys109 and Cys124 is that it links two adjacent antiparallel β strands and thus occurs in a secondary structure that is already stabilized by hydrogen bonds. The Cys109–Cys124 disulfide belongs to the category of the cross-strand disulfides (see the review by Wouters et al. (53) and references therein). CSD are relatively unusual. However, more and more structures presenting CSD have been deposited in the PDB over the past 10 years. For instance, CSD are found in the N-terminal domain (α domain) of DsbD, in the HIV-1 primary receptor CD4, and in ribonucleotide reductase. In most cases, CSD do not function as structure stabilizers but appear to play the role of a redox switch. Further studies are required to determine whether the CSD observed in RcsF also plays a functional redox role.
Supplementary Material
Acknowledgments
We thank Asma Boujtat and Steve Calberson for technical assistance and Matthieu Depuydt for critical reading of the manuscript. We thank the European Community for Access to Research Infrastructure Action of the Improving Human Potential Programme to the EMBL Hamburg Outstation, Contract RII3-CT-2004-506008.
This work was supported by the Interuniversity Attraction Pole Programme-Belgian Science Policy, Network P6/05 (to J.-F. C.), and by grants from the Fonds de la Recherche Scientifique-Fonds National de la Recherche Scientifique (to J.-F. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2, Figs. S1 and S2, and additional references.
The atomic coordinates and structure factors (code 2y1b) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Ni-NTA
- nickel-nitrilotriacetic acid
- I3C
- 5-amino-2,4,6-triiodoisophthalic acid
- PDB
- Protein Data Bank
- CSD
- cross-strand disulfide.
REFERENCES
- 1. Bos M. P., Robert V., Tommassen J. (2007) Annu. Rev. Microbiol. 61, 191–214 [DOI] [PubMed] [Google Scholar]
- 2. Leverrier P., Vertommen D., Collet J. F. (2010) Proteomics 10, 771–784 [DOI] [PubMed] [Google Scholar]
- 3. Ruiz N., Kahne D., Silhavy T. J. (2006) Nat. Rev. Microbiol. 4, 57–66 [DOI] [PubMed] [Google Scholar]
- 4. Ruiz N., Silhavy T. J. (2005) Curr. Opin. Microbiol. 8, 122–126 [DOI] [PubMed] [Google Scholar]
- 5. MacRitchie D. M., Buelow D. R., Price N. L., Raivio T. L. (2008) Adv. Exp. Med. Biol. 631, 80–110 [DOI] [PubMed] [Google Scholar]
- 6. Gottesman S., Trisler P., Torres-Cabassa A. (1985) J. Bacteriol. 162, 1111–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Whitfield C. (2006) Annu. Rev. Biochem. 75, 39–68 [DOI] [PubMed] [Google Scholar]
- 8. Huang Y. H., Ferrières L., Clarke D. J. (2006) Res. Microbiol. 157, 206–212 [DOI] [PubMed] [Google Scholar]
- 9. Majdalani N., Gottesman S. (2005) Annu. Rev. Microbiol. 59, 379–405 [DOI] [PubMed] [Google Scholar]
- 10. Laubacher M. E., Ades S. E. (2008) J. Bacteriol. 190, 2065–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clarke D. J. (2010) Future Microbiol. 5, 1173–1184 [DOI] [PubMed] [Google Scholar]
- 12. Stout V., Gottesman S. (1990) J. Bacteriol. 172, 659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clarke D. J., Joyce S. A., Toutain C. M., Jacq A., Holland I. B. (2002) J. Bacteriol. 184, 1204–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takeda S., Fujisawa Y., Matsubara M., Aiba H., Mizuno T. (2001) Mol. Microbiol. 40, 440–450 [DOI] [PubMed] [Google Scholar]
- 15. Stout V. (1994) Res. Microbiol. 145, 389–392 [DOI] [PubMed] [Google Scholar]
- 16. Stout V., Torres-Cabassa A., Maurizi M. R., Gutnick D., Gottesman S. (1991) J. Bacteriol. 173, 1738–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wehland M., Bernhard F. (2000) J. Biol. Chem. 275, 7013–7020 [DOI] [PubMed] [Google Scholar]
- 18. Kelm O., Kiecker C., Geider K., Bernhard F. (1997) Mol. Gen. Genet. 256, 72–83 [DOI] [PubMed] [Google Scholar]
- 19. Castanié-Cornet M. P., Cam K., Jacq A. (2006) J. Bacteriol. 188, 4264–4270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gervais F. G., Drapeau G. R. (1992) J. Bacteriol. 174, 8016–8022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hagiwara D., Sugiura M., Oshima T., Mori H., Aiba H., Yamashino T., Mizuno T. (2003) J. Bacteriol. 185, 5735–5746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Farris C., Sanowar S., Bader M. W., Pfuetzner R., Miller S. I. (2010) J. Bacteriol. 192, 4894–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Callewaert L., Vanoirbeek K. G., Lurquin I., Michiels C. W., Aertsen A. (2009) J. Bacteriol. 191, 1979–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Majdalani N., Heck M., Stout V., Gottesman S. (2005) J. Bacteriol. 187, 6770–6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Casadaban M. J. (1976) J. Mol. Biol. 104, 541–555 [DOI] [PubMed] [Google Scholar]
- 26. Miller J. (1992) A Short Course in Bacterial Genetics, pp. 221–222, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27. Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beck T., Krasauskas A., Gruene T., Sheldrick G. M. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 1179–1182 [DOI] [PubMed] [Google Scholar]
- 29. Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 30. Panjikar S., Parthasarathy V., Lamzin V. S., Weiss M. S., Tucker P. A. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 449–457 [DOI] [PubMed] [Google Scholar]
- 31. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 32. Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 33. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Holm L., Rosenström P. (2010) Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Politzer P., Lane P., Concha M. C., Ma Y., Murray J. S. (2007) J. Mol. Model 13, 305–311 [DOI] [PubMed] [Google Scholar]
- 36. Politzer P., Murray J. S., Clark T. (2010) Phys. Chem. Chem. Phys. 12, 7748–7757 [DOI] [PubMed] [Google Scholar]
- 37. Depuydt M., Messens J., Collet J. F. (2011) Antioxid. Redox. Signal., in press [DOI] [PubMed] [Google Scholar]
- 38. Kadokura H., Tian H., Zander T., Bardwell J. C., Beckwith J. (2004) Science 303, 534–537 [DOI] [PubMed] [Google Scholar]
- 39. Bardwell J. C., McGovern K., Beckwith J. (1991) Cell 67, 581–589 [DOI] [PubMed] [Google Scholar]
- 40. Hiniker A., Bardwell J. C. (2003) Biochemistry 42, 1179–1185 [DOI] [PubMed] [Google Scholar]
- 41. Katzen F., Beckwith J. (2000) Cell 103, 769–779 [DOI] [PubMed] [Google Scholar]
- 42. Vertommen D., Depuydt M., Pan J., Leverrier P., Knoops L., Szikora J. P., Messens J., Bardwell J. C., Collet J. F. (2008) Mol. Microbiol. 67, 336–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Berkmen M., Boyd D., Beckwith J. (2005) J. Biol. Chem. 280, 11387–11394 [DOI] [PubMed] [Google Scholar]
- 44. Denoncin K., Vertommen D., Paek E., Collet J. F. (2010) J. Biol. Chem. 285, 29425–29433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen J., Song J. L., Zhang S., Wang Y., Cui D. F., Wang C. C. (1999) J. Biol. Chem. 274, 19601–19605 [DOI] [PubMed] [Google Scholar]
- 46. Rietsch A., Bessette P., Georgiou G., Beckwith J. (1997) J. Bacteriol. 179, 6602–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Depuydt M., Leonard S. E., Vertommen D., Denoncin K., Morsomme P., Wahni K., Messens J., Carroll K. S., Collet J. F. (2009) Science 326, 1109–1111 [DOI] [PubMed] [Google Scholar]
- 48. Collet J. F., Messens J. (2010) Antioxid. Redox. Signal. 13, 1205–1216 [DOI] [PubMed] [Google Scholar]
- 49. Grininger M., Staudt H., Johansson P., Wachtveitl J., Oesterhelt D. (2009) J. Biol. Chem. 284, 13068–13076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bieger B., Essen L. O., Oesterhelt D. (2003) Structure 11, 375–385 [DOI] [PubMed] [Google Scholar]
- 51. Kong G. K., Adams J. J., Harris H. H., Boas J. F., Curtain C. C., Galatis D., Masters C. L., Barnham K. J., McKinstry W. J., Cappai R., Parker M. W. (2007) J. Mol. Biol. 367, 148–161 [DOI] [PubMed] [Google Scholar]
- 52. Singh P., Shaffer S. A., Scherl A., Holman C., Pfuetzner R. A., Larson Freeman T. J., Miller S. I., Hernandez P., Appel R. D., Goodlett D. R. (2008) Anal. Chem. 80, 8799–8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wouters M. A., Fan S. W., Haworth N. L. (2010) Antioxid. Redox. Signal. 12, 53–91 [DOI] [PubMed] [Google Scholar]
- 54. Bond C. S. (2003) Bioinformatics 19, 311–312 [DOI] [PubMed] [Google Scholar]
- 55. Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 56. Prouvost A. F. (2008) Rôle du Périplasme dans la Perception par la Bactérie de son Environnement: Utilisation des β-Galactanes par Erwinia chrysanthemi: Voie de Signalisation du Système Rcs dans la Virulence d'Erwinia chrysanthemi. Doctoral dissertation, Universite des Sciences et Technologies de Lille, Lille, France [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.