Abstract
Paracingulin is a 160-kDa protein localized in the cytoplasmic region of epithelial tight and adherens junctions, where it regulates RhoA and Rac1 activities by interacting with guanine nucleotide exchange factors. Here, we investigate the molecular mechanisms that control the recruitment of paracingulin to cell-cell junctions. We show that paracingulin forms a complex with the tight junction protein ZO-1, and the globular head domain of paracingulin interacts directly with ZO-1 through an N-terminal region containing a conserved ZIM (ZO-1-Interaction-Motif) sequence. Recruitment of paracingulin to cadherin-based cell-cell junctions in Rat1 fibroblasts requires the ZIM-containing region, whereas in epithelial cells removal of this region decreases the junctional localization of paracingulin at tight junctions but not at adherens junctions. Depletion of ZO-1, but not ZO-2, reduces paracingulin accumulation at tight junctions. A yeast two-hybrid screen identifies both ZO-1 and the adherens junction protein PLEKHA7 as paracingulin-binding proteins. Paracingulin forms a complex with PLEKHA7 and its interacting partner p120ctn, and the globular head domain of paracingulin interacts directly with a central region of PLEKHA7. Depletion of PLEKHA7 from Madin-Darby canine kidney cells results in the loss of junctional localization of paracingulin and a decrease in its expression. In summary, we characterize ZO-1 and PLEKHA7 as paracingulin-interacting proteins that are involved in its recruitment to epithelial tight and adherens junctions, respectively.
Keywords: Actin, Cell Adhesion, Cell Junctions, Cytoskeleton, Epithelial Cell, PLEKHA7, ZO-1, Cingulin, p120ctn, Paracingulin
Introduction
Tight junctions (TJ)2 and adherens junctions (AJ) are key elements of the apical junctional complex of vertebrate epithelial cells and are of fundamental importance in the morphogenesis and homeostasis of epithelial tissues. TJ and AJ carry out several functions: canonical roles in cell-cell adhesion and tissue sorting (AJ), cell polarization and permeability barrier functions (TJ), and signaling functions through pathways that control the organization of the cytoskeleton and the activity of transcription factors (for review, see Refs. 1–10). All of these functions are orchestrated by complexes of transmembrane, cytoplasmic, and cytoskeletal proteins. Transmembrane proteins of TJ are anchored to the actin cytoskeleton by cytoplasmic scaffolding proteins, among which ZO-1 and ZO-2 play a critical role (11–13). The major transmembrane proteins of AJ are E-cadherin and nectin, which are anchored to the actin and microtubule cytoskeletons by distinct cytoplasmic protein complexes (9, 10). The linkage of E-cadherin to microtubules is mediated by a newly characterized protein complex, comprising p120ctn and PLEKHA7 (14).
The formation and maintenance of epithelial junctions require the activity of small GTPases of the Rho family (RhoA, Rac1, and Cdc42), which control the assembly and contractility of the actin cytoskeleton and, ultimately, cell adhesion, cell shape, and morphogenesis (15, 16). Several junction-associated proteins are implicated, directly or indirectly, in the control of the activity of Rho family GTPases in epithelia (8, 16–23). Among these, cingulin and paracingulin (also known as cingulin-like protein 1, CGNL1, or JACOP) (24–27) play an important role in the regulation of RhoA and Rac1 activities (27–30). For example, both cingulin and paracingulin recruit GEF-H1, a RhoA activator, to junctions, resulting in the down-regulation of RhoA activity in confluent monolayers (28, 29, 31). In addition, paracingulin promotes the activation of Rac1 during junction formation upon calcium switch, by recruiting the Rac1 activator Tiam1 to junctions (29).
The junctional targeting of paracingulin is of paramount importance to spatially define its regulation of Rac1 and RhoA activities. However, nothing is known about the molecular interactions that control the recruitment of paracingulin to junctions. Paracingulin shares with cingulin a similar dynamic behavior and domain organization, with globular head and tail domains, and a coiled-coil rod domain, but unlike cingulin, it is localized not only at TJ, but also at AJ (26, 27, 32). Although the TJ protein ZO-1 is required for the efficient recruitment of cingulin to TJ (13, 33, 34), it is not known whether paracingulin interacts with ZO-1 and whether this interaction is important for its recruitment to TJ. Furthermore, the question remains open about any potential molecular mechanism of association of paracingulin with AJ. In this paper we address these questions, and we discover that paracingulin interacts not only with ZO-1, but also with the AJ protein PLEKHA7 (14, 35) and that these molecular interactions play a mechanistic role in the recruitment of paracingulin to junctions.
EXPERIMENTAL PROCEDURES
Antibodies
Polyclonal and monoclonal antibodies against PLEKHA7 were described previously (35). Other antibodies were cingulin (Invitrogen/Zymed Laboratories 36-4401), CGNL1 (rabbit 20983, in-house), ZO-1 (R40-76, a kind gift from D. Goodenough, Harvard Medical School), ZO-2 (Santa Cruz Biotechnology 71-1400), afadin (Sigma A-0224), Myc tag (9E10 clone, in-house), His tag (Santa Cruz Biotechnology), and p120ctn (mouse 15D2 and 8D11, a kind gift from Dr. A. Reynolds, Vanderbilt University). Fluorescein isothiocyanate- and tetramethylrhodamine isothiocyanate-labeled secondary antibodies were from Jackson Immunoresearch Laboratories.
Recombinant Protein Expression and Glutathione S-Transferase (GST) Pulldown Assays
GST fusion protein constructs were obtained by cloning PCR amplicons in the pGEX4T1 vector, in-frame with GST. Purification of GST fusion proteins and GST pulldowns were as described previously (29, 36). Full-length PLEKHA7 and paracingulin were expressed in baculovirus-infected Sf29 insect cells, after cloning full-length cDNAs into the pFastBacHT vector in-frame with the N-terminal His tag, and transfection of cells with bacmid DNA (32, 36). Cells were lysed in LBT buffer (150 mm NaCl, 20 mm Tris-HCl, pH 7.5, 5 mm EDTA, 1% Triton X-100, 1 mm PMSF), and the lysates were centrifuged at 100,000 × g to remove insoluble material, prior to pulldown assays. At least two biological repeats were performed for each experimental set.
Yeast Two-hybrid Screen
The globular head domain of human paracingulin (residues 1–603) was fused C-terminally to LexA in the pB27 vector. This construct was used to screen a human placenta library in the presence of 0.5 mm 3-amino-1,2,4-triazole (Hybrigenics, Paris, France). A high confidence interaction (based on the PIM biological score) (37) was detected with four distinct clones of human ZO-1, containing sequences between nucleotides 5468 and 7049, corresponding to residues 1455–1736 of ZO-1. A very high confidence interaction was detected with five distinct clones of human PLEKHA7, containing sequences between nucleotides 1868 and 2321, corresponding to residues 602–892 of human PLEKHA7.
Cell Culture and Transfection
MDCK cells, Caco-2 cells, and Rat1 fibroblasts were cultured in DMEM (containing pyruvate for Caco-2 cells) supplemented with FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, and 1× minimal essential medium (MEM), nonessential amino acids. MDCK and Rat1 cell lines expressing either full-length or N-terminal deletions (Δ1–110, Δ1–209) of YFP-tagged canine CGNL1 (generated in the vector pTRE2Hyg) were obtained by transfection with Lipofectamine 2000 and selection in hygromicin. mpkCCDc14 cells were cultured as described (35). MDCK cell clones depleted of PLEKHA7 were obtained by transfection of wild-type MDCK cells with the pTER vector, containing an insert designed to target the sequence(s) AACCTGCCAAGTGACTACAAGT and ATCGCAGTCACGAGGATTCCTT. Stable transfectants were generated by selection in zeocin (29), and individual clones were isolated by cloning rings. Stable MDCK cell lines depleted of ZO-1, ZO-2, or p120ctn were gifts of Dr. A. Fanning (University of North Carolina) (13) and Dr. A. Reynolds (Vanderbilt University) (38), respectively.
Immunoprecipitation and Immunoblotting
For immunoprecipitations, cells were washed twice in ice-cold PBS and lysed in coimmunoprecipitation buffer (150 mm NaCl, 20 mm Tris-HCl, pH 7.5, 1% Nonidet P-40, 1 mm EDTA, and complete protease inhibitor) for 15 min at 4 °C. Lysates were clarified by centrifugation for 15 min at 13,000 rpm. Antibodies (5 μl of rabbit serum anti-PLEKHA7, anti-CGN, anti-CGNL1, and preimmune sera, 2 μg of anti-p120ctn 15D2 and 8D11) were coupled with 20 μl of pre-washed G-protein Dynabeads (Invitrogen) (1 h at 4 °C) and then incubated with either mpkCCDc14 or MDCK lysates (16 h at 4 °C). Beads were washed in coimmunoprecipitation, and proteins were eluted by boiling in SDS sample buffer and analyzed by SDS-PAGE and immunoblotting. At least three biological repeats were performed for each experimental set.
Immunofluorescence Microscopy
Cells on coverslips were fixed with cold methanol for 10 min at −20 °C, washed with PBS, incubated with primary antibody (1 h at 30 °C), washed, incubated with secondary antibody (30 min at 37 °C), and mounted with Vectashield medium (Reactolab). Specimens were analyzed with Axiovert S100 or Zeiss 510 META microscopes. For semiquantitative analysis of junctional labeling, we compared the junctional labeling of CGN with afadin, and CGNL1 with E-cadherin, reference proteins whose junctional localization was not affected by either ZO-1 or ZO-2 depletion. Five confocal images for each double-immunolabeled sample were analyzed with ImageJ software. Pixel intensity for each channel was measured in the selected junctional area, and the averaged background signal was subtracted. Relative signal was expressed as a ratio between CGN/afadin and CGNL1/E-cadherin, calculated over the five different images for each sample. At least three biological repeats were performed for each experiment.
Quantitative RT-PCR
Quantitative RT-PCR (qRT-PCR) was used to assess mRNA expression in MDCK cell clones. Total RNA was prepared using the RNeasy mini kit (Qiagen), retrotranscribed using iScript cDNA synthesis kit (Bio-Rad), and analyzed by SYBR Green-based PCR as described (39). The following primers were used: PLEKHA7, forward 5′-ATT GCT CAC CAG CAG AGG TT-3′ and reverse 5′-TTC CAG GCA TTT TCC ATC TC-3′; CGNL1 forward 5′-CTC AAG GAC CTG GAA TAC GAG C-3′ and reverse 5′-TCC GAG AGC AAA TCC GAG TT-3′; Hypoxantine-guanine phosphoribosyltransferase forward 5′-TGG ACA GGA CTG AGC GGC-3′ and reverse 5′-TGA GCA CAC AGA GGG CTA CG-3′. Ct, ΔCt, ΔΔCt, and -fold changes were calculated as described (39).
RESULTS
Paracingulin Interacts with ZO-1 through the ZIM-containing Region of the Globular Head Domain and Is Recruited to ZO-1-containing Junctions in a ZIM-dependent Manner
To ask whether paracingulin can interact with ZO-1, we first examined whether paracingulin and ZO-1 form a complex. Immunoblotting analysis of immunoprecipitates prepared from mouse kidney epithelial cells showed that ZO-1 is detected both in cingulin and paracingulin immunoprecipitates (Fig. 1A), indicating that ZO-1 forms complexes with both proteins.
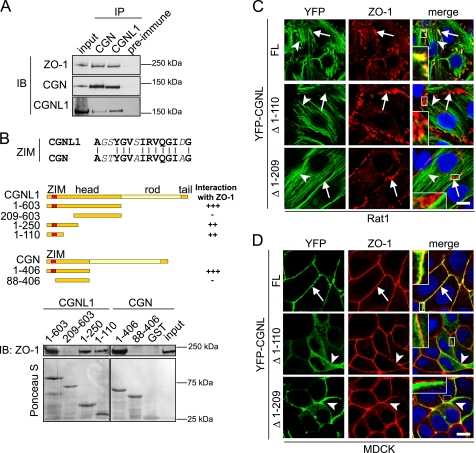
FIGURE 1.
CGNL1 interacts with ZO-1 and is recruited to junctions through the ZIM domain. A, CGNL1 forms a complex with ZO-1 and CGN. Immunoblot (IB) analysis was performed with antibodies against ZO-1, CGN, and CGNL1, of immunoprecipitates (IP) prepared from mpkCCDC14 (mouse kidney) cell lysates, using antibodies against CGN and CGNL1 (a rabbit preimmune serum was used as a negative control). Input was 1/10 of the volume used for the immunoprecipitation. B, ZIM-containing region is required for the interaction between the head region of CGNL1 and ZO-1. Top panel, ZIM sequences (33) of human paracingulin (residues 37–51) and human cingulin (residues 40–54) are aligned, with amino acid identities indicated by cross-bars. Middle panel, diagrams show schematically full-length CGNL1 and CGN (with head, rod, and tail domains), and head GST fusion constructs used in GST pulldowns. The ZIM sequence (in red) is in the N-terminal region of the head domain. The interaction with ZO-1 is scored on the right (interaction: very strong, +++; strong, ++; none detected, −). Bottom panel, ZO-1 immunoblotting (top) and Ponceau S staining (bottom, for normalization of total GST fusion proteins) of GST pulldowns of full-length ZO-1 used GST fusion proteins of CGNL1 and CGN (numbers indicate residues) as bait. Numbers on the right indicate migration of molecular size markers. C, ZIM domain is required for CGNL1 recruitment to AJ in Rat1 fibroblasts. Immunofluorescence microscopy, with anti-ZO-1 antibodies, of Rat1 fibroblasts expressing exogenous YFP-tagged CGNL1 constructs is shown: full-length (FL), deletion of residues 1–110 (Δ1–110), and deletion of residues 1–209 (Δ1–209). All of these constructs target actin stress fibers (arrowheads), but only the full-length CGNL1, which contains the ZIM sequence, colocalizes with ZO-1 at the spot-like AJ (arrows). Scale bar, 10 μm, D, ZIM domain is required for CGNL1 recruitment to TJ in MDCK cells. Immunofluorescence microscopy, with anti-ZO-1 antibodies, of MDCK cells expressing YFP-tagged CGNL1 constructs (see C) is shown. Arrows indicate colocalization with ZO-1. Arrowheads indicate junctional staining that is not colocalized with ZO-1. Scale bar, 10 μm.
The ZIM sequence, which is involved in the junctional recruitment and interaction of cingulin with ZO-1 (33), is present in the N-terminal region of the head domain of paracingulin and shows 75% identity to the homologous sequence of cingulin (Fig. 1B). We asked whether the ZIM-containing region is implicated in the interaction of paracingulin with ZO-1 by carrying out GST pulldown assays (Fig. 1B). Bacterially expressed constructs of the head domain of paracingulin were used as baits to pull down full-length ZO-1, expressed in baculovirus-infected insect cells. As a control, we used bacterially expressed constructs of cingulin. The globular head domains of cingulin and paracingulin interacted strongly with ZO-1 (Fig. 1B) (see also Ref. 25), and removal of the ZIM-containing N-terminal region abolished this interaction (Fig. 1B) (33). A 110-residue N-terminal fragment of the paracingulin head domain, which contains the ZIM sequence, was sufficient to interact with ZO-1 (Fig. 1B). Fragments of the rod domain of paracingulin did not interact with full-length ZO-1 (data not shown). In addition, a C-terminal fragment of human ZO-1 (residues 1455–1736) was found to interact with the globular head domain of paracingulin with a high confidence score, in a yeast two-hybrid screen. In summary, the head domain of paracingulin interacts directly with the C-terminal region of ZO-1, and the ZIM-containing N-terminal region of the head domain of paracingulin is necessary and sufficient for this interaction.
To establish whether the interaction between paracingulin and ZO-1 is physiologically relevant, we tested whether the ZIM-containing region is required for paracingulin recruitment to junctions. To this purpose, we first expressed fluorescently tagged paracingulin in Rat1 fibroblasts. Rat1 fibroblasts do not have TJ and TJ proteins, but form spot-like AJ at sites of cell-cell contact which contain ZO-1, allowing one to examine the interaction of exogenous proteins with ZO-1, independently of TJ (33, 40, 41). Importantly, spot-like AJ of Rat-1 fibroblasts do not contain the AJ protein PLEKHA7, which in these cells is only detectable at centrosomes (data not shown and Ref. 14), in agreement with the notion that PLEKHA7 is confined to belt-like AJ and absent from spot-like AJ (35). Immunofluorescence microscopy showed that in Rat1 fibroblasts full-length paracingulin was colocalized with ZO-1 at sites of cell-cell contact and along actin stress-fibers (Fig. 1C, FL, arrows and arrowheads, and magnified inset). In contrast, two distinct constructs lacking the ZIM sequence localized only along stress fibers (Fig. 1C, Δ1–110 and Δ1–209, arrowheads), and failed to colocalize with ZO-1 (Fig. 1C, Δ1–110 and Δ1–209, arrows, and magnified insets).
Next, we asked whether the ZIM-containing region is required for the localization of paracingulin at junctions of epithelial renal (MDCK) cells that contain belt-like AJ and TJ. Immunofluorescence microscopy showed that exogenous full-length paracingulin was localized at junctions, and the paracingulin labeling significantly colocalized with ZO-1 labeling (Fig. 1D, FL, arrows, and magnified inset). In contrast, the paracingulin constructs lacking the ZIM-containing region were not colocalized with ZO-1 (Fig. 1D, Δ1–110 and Δ1–209, arrowheads, and magnified insets), albeit they were still detected in a junctional localization. This suggested that ZIM-independent protein interactions can recruit paracingulin to junctions in epithelial cells, but not in Rat1 fibroblasts.
Paracingulin Junctional Recruitment Is Decreased in ZO-1-depleted, but Not ZO-2-depleted, Cell Lines
Next, to examine in more detail the role of ZO-1 in the junctional recruitment of paracingulin in epithelial cells, we analyzed the expression and localization of cingulin and paracingulin in MDCK cells depleted of ZO-1. As a control, we used cells depleted of ZO-2. Previous studies showed either a loss or a decrease in the junctional labeling of cingulin in cells devoid or depleted of ZO-1 (13, 34).
Immunoblotting (Fig. 2A) and qRT-PCR analysis (data not shown) showed that the levels of expression of cingulin and paracingulin were not affected by either ZO-1 or ZO-2 depletion. Immunofluorescence microscopy showed decreased junctional labeling for both paracingulin and cingulin in ZO-1-depleted cells (Fig. 2, B and C, and supplemental Fig. 1A, ZO-1 kd, arrowheads), but not in ZO-2-depleted cells (Fig. 2, B and C, and supplemental Fig. 1A, ZO-2 kd, arrows), when cells were grown for 1 or 2 days after plating. However, the junctional labeling for paracingulin, but not cingulin, increased as junctions matured, because 3 days after plating the labeling for junctional paracingulin was almost as strong as wild-type cells (Fig. 2B), suggesting that depletion of ZO-1 results in a delay rather than a block of the junctional recruitment of paracingulin. In contrast, depletion of either ZO-1 or ZO-2 did not affect the junctional localization of AJ markers, such as p120ctn, afadin, and PLEKHA7 (supplemental Fig. 1B) and E-cadherin (data not shown), in agreement with the notion that ZO-1 specifically controls the assembly and function of TJ, but not AJ (13, 42). Thus, ZO-1 is required for the efficient junctional recruitment of cingulin and, to a lesser extent, paracingulin.
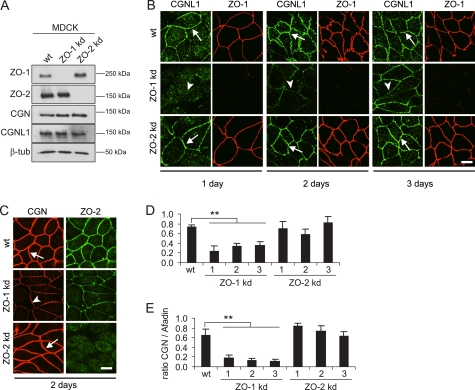
FIGURE 2.
Depletion of ZO-1, but not ZO-2, negatively regulates the junctional recruitment of CGNL1. A, depletion of either ZO-1 or ZO-2 does not affect CGNL1 and CGN protein levels. Immunoblotting analysis of lysates from MDCK cells, either wild-type (wt) or stable lines depleted of either ZO-1 (ZO-1 kd, clone 1), or ZO-2 (ZO-2 kd, clone 1) was performed with antibodies against ZO-1, ZO-2, CGN, CGNL1. β-Tubulin was used as loading control. B, CGNL1 junctional labeling increases at longer times after plating in ZO-1 kd cells. Double immunofluorescence microscopy analysis of MDCK cell lines (wt, ZO-1 kd, and ZO-2 kd) labeled with antibodies against CGNL1 and ZO-1 was performed after 1, 2, or 3 days in culture following plating. Arrows indicate normal junctional labeling, and arrowheads indicate decreased junctional labeling of CGNL1. Scale bar, 10 μm. C, CGN junctional labeling is decreased in ZO-1-, but not ZO-2-depleted cells. Double immunofluorescent microscopy analysis of MDCK cells grown for 2 days after plating (wt, ZO-1 kd, ZO-2 kd) was performed with antibodies against CGN and ZO-2. Arrowheads indicate reduced junctional labeling for CGN. Arrows indicate normal junctional labeling for CGN in wt and ZO-2 kd cells. Scale bar, 10 μm. D and E, semiquantitative analysis of the junctional labeling for CGNL1 (D) and CGN (E) in wt MDCK cells and in different clones of ZO-1 kd cells (clones 1, 2, and 3) and ZO-2 kd cells (clones 1, 2, and 3) was performed. Junctional labeling is expressed as a ratio between labeling of either CGNL1 and E-cadherin (D) or CGN and afadin (E) (see “Experimental Procedures”). D, ratio of CGNL1 to E-cadherin was decreased 57% in ZO-1 kd clones compared with the wild-type (wt = 0.73 ± 0.05; ZO-1 kd clones = 0.32 ± 0.06, p value < 0.001), but not in ZO-2 kd clones (0.71 ± 0.12). E, ratio of CGN to afadin was decreased 80% in ZO-1 kd clones compared with the wild-type (wt = 0.66 ± 0.12; ZO-1 kd clones = 0.15 ± 0.04, p value < 0.001), but not in ZO-2 kd clones (0.74 ± 0.10).
To compare more quantitatively the effect of ZO-1 depletion on the junctional recruitment of paracingulin and cingulin, we measured the intensity of their junctional labeling in different clones of either ZO-1-depleted or ZO-2-depleted cells, using as a reference E-cadherin and afadin. In ZO-1-depleted cells, the labeling for both paracingulin (Fig. 2D) and cingulin (Fig. 2E) was significantly decreased, whereas no significant change was observed in ZO-2-depleted cells. However, whereas cingulin labeling was decreased, on average, by about 77% upon ZO-1 depletion in cells grown for 2 days, the labeling for paracingulin was decreased by about 57%, confirming that ZO-1 has a stronger impact on the junctional recruitment of cingulin than that of paracingulin. Finally, we examined the localization of a ZIM-deleted construct of paracingulin (Δ1–209) in cells depleted either of ZO-1 or ZO-2. Based on the colocalization with occludin and E-cadherin, the full-length paracingulin construct was targeted to both TJ and AJ in wild-type cells and ZO-2-depleted cells, but only to AJ in ZO-1-depleted cells (supplemental Fig. 2A). Deletion of the ZIM-containing region resulted in reduced or no colocalization with occludin, without decreasing the colocalization with E-cadherin (supplemental Fig. 2B). Taken together, these results indicate that the ZIM-containing region specifically targets paracingulin to TJ and is not required for paracingulin recruitment to AJ.
Paracingulin Forms a Complex with PLEKHA7 and p120ctn at Epithelial Adherens Junctions and Interacts Directly with PLEKHA7
To identify novel interacting protein partners that may be important for the recruitment of paracingulin to AJ, we carried out a yeast two-hybrid screen in which the globular head domain of paracingulin was used as a bait to screen a human placenta library. A very high confidence interaction was detected with PLEKHA7, a recently described AJ protein, which interacts with p120ctn (14, 35). To test whether the PLEKHA7-p120ctn complex associates with paracingulin in vivo, we analyzed immunoprecipitates of either paracingulin or PLEKHA7, prepared from lysates of human intestinal epithelial cells. Immunoblot analysis detected PLEKHA7 and p120ctn in paracingulin immunoprecipitates and paracingulin and p120ctn in PLEKHA7 immunoprecipitates (Fig. 3A), demonstrating that the three proteins exist in a complex. The association of paracingulin with p120ctn is specific because it was only detected with an antibody (15D2) that recognizes human p120ctn and not with an antibody (8D11) that does not recognize human p120ctn (Fig. 3B). In addition, depletion of p120ctn resulted in the disappearance of paracingulin from immunoprecipitates (Fig. 3C).
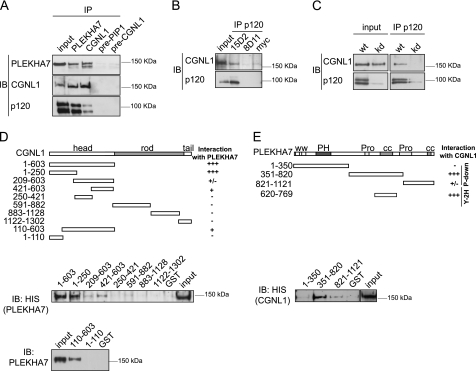
FIGURE 3.
CGNL1 forms a complex with PLEKHA7 and p120ctn and interacts with PLEKHA7 through its globular head domain. A, CGNL1 forms a complex with PLEKHA7. Immunoblotting (IB) analysis was performed (using antibodies against PLEKHA7, CGNL1, and p120ctn) of immunoprecipitates (IP) obtained from lysates of mpkCCDc14 cells, using either anti-PLEKHA7 or anti-CGNL1 antibodies (preimmune sera were used as controls). Note that CGNL1 and p120ctn are detected in the PLEKHA7 immunoprecipitates and PLEKHA7 and p120ctn in CGNL1 immunoprecipitates. B and C, association of CGNL1 with the p120ctn complex is specific. B, immunoblotting analysis was performed (using antibodies against either CGNL1 or p120ctn) of immunoprecipitates from lysates of human intestinal cells (Caco-2), using monoclonal antibodies that bind to (15D2) or do not bind to (8D11) human p120ctn. Note that CGNL1 is specifically coimmunoprecipitated only with the monoclonal anti-p120ctn antibody that recognizes the human isoform (15D2). C, immunoblotting analysis of immunoprecipitates from lysates of either wild-type MDCK cells (wt) or MDCK cell lines depleted of p120ctn (kd), using the 8D11 anti-p120ctn monoclonal antibody is shown. CGNL1 is detected only in immunoprecipitates from wt cells and not in cells depleted of p120ctn. D, CGNL1 interacts with PLEKHA7 through its globular head domain, independently of the ZIM-containing region. Top, schematic diagrams full-length CGNL1 and regions used for the generation of GST fusion proteins used in pulldown experiments. Numbers on the left indicate amino acid residues, and their interaction with full-length PLEKHA7 (for CGNL constructs) by GST pulldown is scored on the right (+++, strong; +, detected; +/−, weak; −, none detected). Bottom, immunoblotting analysis (using either anti-His antibodies or anti-PLEKHA7 antibodies) of GST pulldowns of His-tagged, full-length PLEKHA7 used GST fusions of the indicated constructs. Input was 1/10 of the volume used for each pulldown. E, PLEKHA7 interacts with CGNL1 through its central domain, containing proline-rich and coiled-coil domains. Top, schematic diagrams full-length PLEKHA7, with tryptophane domains (ww), pleckstrin-homology (PH) domain, proline-rich (Pro), coiled-coil (cc) domains, and regions used for the generation of GST fusion proteins from pulldown experiments. Numbers on the left indicate amino acid residues, and their interaction with full-length CGNL1 either by GST pulldown (residues 1–350, 351–820, 821–1121) or yeast two-hybrid screen (Y-2H) is scored on the right. Bottom, immunoblotting analysis was performed (using anti-His antibodies) of GST pulldowns of His-tagged, full-length CGNL1, using GST fusions of the indicated constructs of PLEKHA7.
Next, we used GST pulldown experiments to identify the regions of paracingulin and PLEKHA7 that are involved in their association. The globular head domain of paracingulin and its N-terminal fragment (residues 1–250) interacted strongly with full-length PLEKHA7 (Fig. 3D). Additional fragments of the globular head domain of paracingulin (e.g. residues 209–603 and 421–603) also interacted with PLEKHA7, albeit more weakly (Fig. 3D), whereas fragments of the rod domain of paracingulin did not interact with PLEKHA7 (Fig. 3D). Moreover, the ZIM-containing region of paracingulin (residues 1–110) did not interact with PLEKHA7 (Fig. 3D), indicating that paracingulin interacts with ZO-1 and PLEKHA7 through structurally distinct regions of its head domain. Taken together, these results indicate that the region of the paracingulin head domain that interacts with PLEKHA7 with highest affinity is comprised within residues 110–250. In turn, only the central region of PLEKHA7 (residues 351–820), which comprises a proline-rich domain and a coiled-coil domain, interacted strongly with full-length paracingulin (Fig. 3E). This region overlaps with the PLEKHA7 fragment (residues 602–892) that interacts with paracingulin by the two-hybrid screen assay, suggesting that residues 602–820 comprise the sequences that are sufficient for the interaction of PLEKHA7 with paracingulin. In summary, paracingulin forms a complex with PLEKHA7 and p120ctn, and the globular head domain of paracingulin interacts with the central region of PLEKHA7 in a ZIM-independent manner.
PLEKHA7 but Not p120ctn Is Required to Maintain the Expression and Junctional Association of Paracingulin
PLEKHA7 is exclusively localized at belt-like AJ in epithelial cells (35), and p120ctn was reported to recruit PLEKHA7 to AJ in intestinal cells (14). To test the role of PLEKHA7 and p120ctn in the recruitment of paracingulin to AJ, we analyzed the localization of paracingulin in different stable clones of MDCK cells depleted of either PLEKHA7 or p120ctn.
Depletion of PLEKHA7 by expression of shRNA resulted in an 80–90% decrease in PLEKHA7 protein levels in different stable clones (Fig. 4A), and a dramatic decrease in the immunofluorescent staining for PLEKHA7 at junctions (Fig. 4B). Paracingulin expression was also decreased by about 50%, as determined by immunoblotting and qRT-PCR (Fig. 4A and data not shown), and paracingulin junctional staining was also dramatically decreased (Fig. 4B). This indicated that PLEKHA7 is required for both the expression and the junctional localization of paracingulin. In contrast, the immunofluorescent localizations of p120ctn, ZO-1 (Fig. 4, A and B), E-cadherin, α-catenin, and β-catenin (supplemental Fig. 3, A and B) were not dramatically affected by depletion of PLEKHA7.
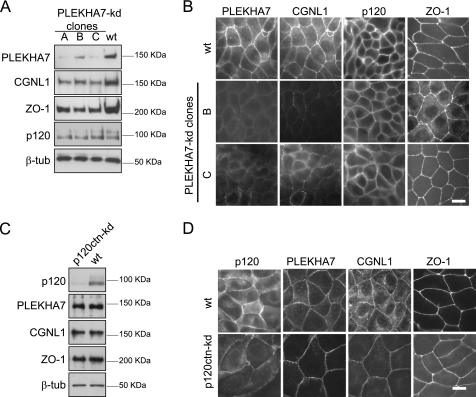
FIGURE 4.
PLEKHA7 is required for CGNL1 recruitment to AJ. A, establishment of PLEKHA7-kd MDCK cell clones. Immunoblotting analysis, using antibodies against PLEKHA7, CGNL1, ZO-1, p120ctn, and β-tubulin of lysates of wild-type MDCK cells and MDCK cell clones (A–C) depleted of PLEKHA7 by expression of shRNA was performed (see “Experimental Procedures”). B, decreased junctional labeling of CGNL1 in PLEKHA7 kd cells. Immunofluorescence microscopy analysis of wild-type MDCK cells and stable MDCK cells depleted of PLEKHA7, was performed using antibodies against PLEKHA7, CGNL1 (double immunofluorescent labeling for PLEKHA7 and CGNL1), p120ctn, and ZO-1. C, immunoblotting analysis, using antibodies against p120ctn, PLEKHA7, CGNL1, ZO-1, and β-tubulin of lysates of wild-type MDCK cells and p120ctn kd cells. D, immunofluorescence microscopy analysis of wild-type MDCK cells and MDCK cells depleted of p120ctn, using antibodies against p120ctn, PLEKHA7, CGNL1 (double immunofluorescent labeling for PLEKHA7 and CGNL1) and ZO-1. Scale bars, 10 μm.
Depletion of p120ctn did not result in changes in the levels of protein expression of PLEKHA7, paracingulin, and ZO-1, or their junctional localization (Fig. 4, C and D), whereas it negatively regulated the expression and the junctional localization of β-catenin and E-cadherin (supplemental Fig. 3, C and D), as previously shown (38). In contrast, depletion of either paracingulin or cingulin did not affect the junctional localization and expression of PLEKHA7 and p120ctn (supplemental Fig. 4). In summary, in MDCK cells PLEKHA7 promotes the expression of paracingulin and its recruitment to AJ, and neither cingulin nor paracingulin affects the junctional recruitment of TJ and AJ proteins (see also Refs. 29, 31, 32).
DISCUSSION
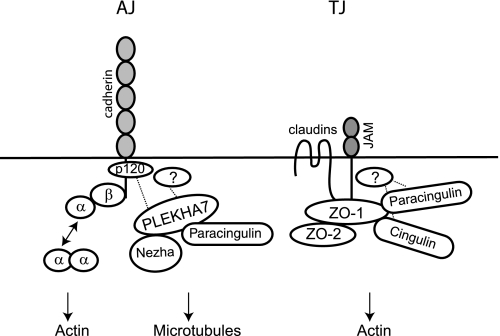
In this paper we address the molecular mechanisms of recruitment of paracingulin to epithelial junctions, and we characterize PLEKHA7 and ZO-1 as interacting partners of paracingulin, which are implicated in the targeting of paracingulin to epithelial AJ and TJ, respectively (Fig. 5).
FIGURE 5.
Molecular interactions of paracingulin at TJ and AJ. Simplified scheme, showing the association of paracingulin with ZO-1 at TJ, and with PLEKHA7 at AJ. Paracingulin and cingulin are partially recruited to TJ through ZO-1 and through additional unknown interactions (dotted lines). ZO-1 is shown as part of a dimer with ZO-2 and interacting with the C-terminal cytoplasmic tail of TJ membrane proteins (claudins and JAM). The TJ-associated protein complex is linked to actin filaments. E-cadherin interacts with actin filaments through different protein complexes, including α-catenin, and linkage to microtubules occurs through the PLEKHA7-nezha complex (14). PLEKHA7 is recruited to AJ through p120ctn in intestinal cells (14) and through additional unknown interactions (dotted lines).
A major new finding reported here is the identification of PLEKHA7 as a protein that interacts with paracingulin and is required for its expression and junctional recruitment. This is a novel function for PLEKHA7, a protein that was independently identified through its binding to the N-terminal domain of p120ctn (14). PLEKHA7 plays a fundamental role in the junctional recruitment of paracingulin in epithelial cells because the presence of ZO-1 at junctions was not sufficient to rescue the expression and junctional localization of paracingulin in PLEKHA7-depleted cells. Interestingly, the p120ctn-PLEKHA7 complex is linked to microtubules through nezha (14), and the junctional accumulation of paracingulin is severely decreased by disruption of the microtubule cytoskeleton (32). This indicates that the integrity of the complex among microtubules, PLEKHA7, and associated proteins is essential to recruit and stabilize paracingulin at AJ. Together with our previous observation that paracingulin forms a complex with E-cadherin (32), the data presented here provide the first biochemical and mechanistic evidence for the reported association of paracingulin with AJ (26).
Paracingulin also interacts with ZO-1, based on immunoprecipitation, in vitro binding, and yeast two-hybrid screen assays. The yeast two-hybrid screen analysis indicates that paracingulin interacts with the C-terminal domain of ZO-1, a region that has been reported to interact with actin, cortactin, protein 4-1, and cingulin (43, 44) and is therefore a key structural module in the interaction of ZO-1 with the actomyosin cytoskeleton. However, the paracingulin-interacting region of ZO-1 (residues 1455–1736) does not overlap with the actin binding region (residues 1152–1371) (45), suggesting that ZO-1 can interact simultaneously with paracingulin and actin. In fact, disruption of actin microfilaments breaks up the linear distribution of paracingulin at junctions without affecting its association with cingulin and ZO-1 (32). Therefore, the integrity of the actin cytoskeleton is not required to maintain the association of paracingulin with the protein complexes at TJ and AJ. Moreover, the localization of exogenous paracingulin at actin stress fibers in transfected Rat1 fibroblasts indicates that paracingulin can interact with the actomyosin cytoskeleton independently of ZO-1. Further studies should address the mechanisms and functional relevance of the interaction of paracingulin with the actin cytoskeleton.
The ZIM-containing, N-terminal region of the globular head domain of paracingulin is a key structural module that controls its recruitment to ZO-1-containing junctions, but not to PLEKHA7-containing AJ. Indeed, in spot-like AJ of Rat1 fibroblasts, which do not contain PLEKHA7, the ZIM-containing region is absolutely required for junctional targeting of paracingulin. However, in epithelial cells, when the ZO-1-paracingulin interaction is inhibited, either through deletion of the ZIM-containing region or through depletion of ZO-1, paracingulin can still be recruited to belt-like AJ junctions. Consistent with this finding, constructs of the globular head domain of paracingulin that lack the ZIM-containing region can still interact with PLEKHA7 in vitro. In addition, in cells depleted of ZO-1, the junctional localization of both cingulin and paracingulin is not abolished, suggesting that redundant ZIM-mediated interactions can recruit these proteins to TJ (Fig. 5). Thus, the impact of ZO-1 on the junctional localization of paracingulin is cell context-dependent, and multiple interactions can recruit paracingulin to TJ, although neither cingulin nor ZO-2 is involved (see also Ref. 32).
In summary, we identify paracingulin as a component of the macromolecular complex that links AJ to microtubules, and we show that PLEKHA7, and, to a lesser extent, ZO-1, are required to recruit paracingulin to epithelial junctions efficiently. Because paracingulin functions as an adaptor protein for guanine nucleotide exchange factors that regulate Rac1 and RhoA activities in epithelial cells (29), these results provide important new information on the molecular mechanisms through which cell-cell junctions spatially regulate the activation of Rho family GTPases.
Supplementary Material
Acknowledgments
We acknowledge the gift of cells and reagents from the collaborators cited in the text, members of the Citi laboratory for comments and discussions, and the National Centres of Competence in Research Imaging Platform for excellent support.
This work was supported by Swiss National Fonds Grants 31003A103637 and 31003A116763 and the State of Geneva.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- TJ
- tight junction(s)
- AJ
- adherens junction(s)
- MDCK
- Madin-Darby canine kidney
- qRT-PCR
- quantitative RT-PCR
- ZIM
- ZO-1 interaction motif.
REFERENCES
- 1. Shin K., Fogg V. C., Margolis B. (2006) Annu. Rev. Cell Dev. Biol. 22, 207–235 [DOI] [PubMed] [Google Scholar]
- 2. Anderson J. M., Van Itallie C. M., Fanning A. S. (2004) Curr. Opin. Cell Biol. 16, 140–145 [DOI] [PubMed] [Google Scholar]
- 3. González-Mariscal L., Tapia R., Chamorro D. (2008) Biochim. Biophys. Acta 1778, 729–756 [DOI] [PubMed] [Google Scholar]
- 4. Perez-Moreno M., Jamora C., Fuchs E. (2003) Cell 112, 535–548 [DOI] [PubMed] [Google Scholar]
- 5. Gumbiner B. M. (2005) Nat. Rev. Mol. Cell Biol. 6, 622–634 [DOI] [PubMed] [Google Scholar]
- 6. McCrea P. D., Gu D., Balda M. S. (2009) Cold Spring Harb. Perspect. Biol. 1, a002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Halbleib J. M., Nelson W. J. (2006) Genes Dev. 20, 3199–3214 [DOI] [PubMed] [Google Scholar]
- 8. Guillemot L., Paschoud S., Pulimeno P., Foglia A., Citi S. (2008) Biochim. Biophys. Acta 1778, 601–613 [DOI] [PubMed] [Google Scholar]
- 9. Takai Y., Miyoshi J., Ikeda W., Ogita H. (2008) Nat. Rev. Mol. Cell Biol. 9, 603–615 [DOI] [PubMed] [Google Scholar]
- 10. Meng W., Takeichi M. (2009) Cold Spring Harb. Perspect. Biol. 1, a002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fanning A. S., Jameson B. J., Jesaitis L. A., Anderson J. M. (1998) J. Biol. Chem. 273, 29745–29753 [DOI] [PubMed] [Google Scholar]
- 12. Umeda K., Ikenouchi J., Katahira-Tayama S., Furuse K., Sasaki H., Nakayama M., Matsui T., Tsukita S., Furuse M., Tsukita S. (2006) Cell 126, 741–754 [DOI] [PubMed] [Google Scholar]
- 13. Van Itallie C. M., Fanning A. S., Bridges A., Anderson J. M. (2009) Mol. Biol. Cell 20, 3930–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meng W., Mushika Y., Ichii T., Takeichi M. (2008) Cell 135, 948–959 [DOI] [PubMed] [Google Scholar]
- 15. Braga V. M., Yap A. S. (2005) Curr. Opin. Cell Biol. 17, 466–474 [DOI] [PubMed] [Google Scholar]
- 16. Samarin S., Nusrat A. (2009) Front. Biosci. 14, 1129–1142 [DOI] [PubMed] [Google Scholar]
- 17. Anastasiadis P. Z., Reynolds A. B. (2001) Curr. Opin. Cell Biol. 13, 604–610 [DOI] [PubMed] [Google Scholar]
- 18. Chen X., Macara I. G. (2005) Nat. Cell Biol. 7, 262–269 [DOI] [PubMed] [Google Scholar]
- 19. Mertens A. E., Rygiel T. P., Olivo C., van der Kammen R., Collard J. G. (2005) J. Cell Biol. 170, 1029–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wittchen E. S., Haskins J., Stevenson B. R. (2003) Mol. Biol. Cell 14, 1757–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Godsel L. M., Dubash A. D., Bass-Zubek A. E., Amargo E. V., Klessner J. L., Hobbs R. P., Chen X., Green K. J. (2010) Mol. Biol. Cell 21, 2844–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wells C. D., Fawcett J. P., Traweger A., Yamanaka Y., Goudreault M., Elder K., Kulkarni S., Gish G., Virag C., Lim C., Colwill K., Starostine A., Metalnikov P., Pawson T. (2006) Cell 125, 535–548 [DOI] [PubMed] [Google Scholar]
- 23. Severson E. A., Lee W. Y., Capaldo C. T., Nusrat A., Parkos C. A. (2009) Mol. Biol. Cell 20, 1916–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Citi S., Sabanay H., Jakes R., Geiger B., Kendrick-Jones J. (1988) Nature 333, 272–276 [DOI] [PubMed] [Google Scholar]
- 25. Cordenonsi M., D'Atri F., Hammar E., Parry D. A., Kendrick-Jones J., Shore D., Citi S. (1999) J. Cell Biol. 147, 1569–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ohnishi H., Nakahara T., Furuse K., Sasaki H., Tsukita S., Furuse M. (2004) J. Biol. Chem. 279, 46014–46022 [DOI] [PubMed] [Google Scholar]
- 27. Guillemot L., Citi S. (2006) in Tight Junctions (Gonzalez-Mariscal L. ed) pp. 54–63, Landes Bioscience-Springer Science, New York [Google Scholar]
- 28. Aijaz S., D'Atri F., Citi S., Balda M. S., Matter K. (2005) Dev. Cell 8, 777–786 [DOI] [PubMed] [Google Scholar]
- 29. Guillemot L., Paschoud S., Jond L., Foglia A., Citi S. (2008) Mol. Biol. Cell 19, 4442–4453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Terry S. J., Zihni C., Elbediwy A., Vitiello E., Leefa Chong San I. V., Balda M. S., Matter K. (2011) Nat. Cell Biol. 13, 159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guillemot L., Citi S. (2006) Mol. Biol. Cell 17, 3569–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paschoud S., Yu D., Pulimeno P., Jond L., Turner J. R., Citi S. (2011) Mol. Membr. Biol. 28, 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. D'Atri F., Nadalutti F., Citi S. (2002) J. Biol. Chem. 277, 27757–27764 [DOI] [PubMed] [Google Scholar]
- 34. Umeda K., Matsui T., Nakayama M., Furuse K., Sasaki H., Furuse M., Tsukita S. (2004) J. Biol. Chem. 279, 44785–44794 [DOI] [PubMed] [Google Scholar]
- 35. Pulimeno P., Bauer C., Stutz J., Citi S. (2010) PLoS One 5, e12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Citi S., D'Atri F., Cordenonsi M., Cardellini P. (2001) in Cell Cell Interactions (Fleming T. P. ed), 2nd Ed., pp. 153–176, IRL Press, Oxford [Google Scholar]
- 37. Rain J. C., Selig L., De Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schächter V., Chemama Y., Labigne A., Legrain P. (2001) Nature 409, 211–215 [DOI] [PubMed] [Google Scholar]
- 38. Dohn M. R., Brown M. V., Reynolds A. B. (2009) J. Cell Biol. 184, 437–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guillemot L., Hammar E., Kaister C., Ritz J., Caille D., Jond L., Bauer C., Meda P., Citi S. (2004) J. Cell Sci. 117, 5245–5256 [DOI] [PubMed] [Google Scholar]
- 40. Van Itallie C. M., Anderson J. M. (1997) J. Cell Sci. 110, 1113–1121 [DOI] [PubMed] [Google Scholar]
- 41. Bordin M., D'Atri F., Guillemot L., Citi S. (2004) Mol. Cancer Res. 2, 692–701 [PubMed] [Google Scholar]
- 42. Ooshio T., Kobayashi R., Ikeda W., Miyata M., Fukumoto Y., Matsuzawa N., Ogita H., Takai Y. (2010) J. Biol. Chem. 285, 5003–5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Citi S. (2001) in Tight Junctions (Cereijido M., Anderson J. eds), 2nd Ed., pp 231–264, CRC Press, Boca Raton, FL [Google Scholar]
- 44. Fanning A. S., Anderson J. M. (2009) Ann. N.Y. Acad. Sci. 1165, 113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fanning A. S., Ma T. Y., Anderson J. M. (2002) FASEB J. 16, 1835–1837 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.