Abstract
Plant cytochrome P450s are involved in the production of over a hundred thousand metabolites such as alkaloids, terpenoids, and phenylpropanoids. Although cytochrome P450 genes constitute one of the largest superfamilies in plants, many of the catalytic functions of the enzymes they encode remain unknown. Here, we report the identification and functional characterization of a cytochrome P450 gene in a new subfamily of CYP71, CYP71BJ1, involved in alkaloid biosynthesis. Co-expression analysis of putative cytochrome P450 genes in the Catharanthus roseus transcriptome identified candidate genes with expression profiles similar to known terpene indole alkaloid biosynthetic genes. Screening of these candidate genes by functional expression in Saccharomyces cerevisiae yielded a unique P450-dependent enzyme that stereoselectively hydroxylates the alkaloids tabersonine and lochnericine at the 19-position of the aspidosperma-type alkaloid scaffold. Tabersonine, which can be converted to either vindoline or 19-O-acetylhörhammericine, represents a branch point in alkaloid biosynthesis. The discovery of CYP71BJ1, which forms part of the pathway leading to 19-O-acetylhörhammericine, will help illuminate how this branch point is controlled in C. roseus.
Keywords: Cytochrome P450, Enzyme Catalysis, Hydroxylase, Metabolism, Plant, Alkaloid, Biosynthesis
Introduction
Cytochrome P450 enzymes (P450s)5 play a key role in the development and survival of plants (1). P450s participate in the biosynthesis of a wide variety of compounds throughout primary and specialized metabolism, including fatty acids, terpenoids, phenylpropanoids, cyanogenic glucosides, glucosinolates, and alkaloids. Notably, plants have an unusually large number of P450 genes compared with prokaryotes and other eukaryotic organisms. Furthermore, plant P450s are typically stringently substrate-specific enzymes that catalyze highly regio- and stereoselective transformations (2, 3). The catalytic functions of most plant P450s are unknown. Identifying a P450 that catalyzes a specific biosynthetic transformation poses a challenge due to the homology shared by P450 genes and the typical lack of correlation between primary structure and catalytic function (4).
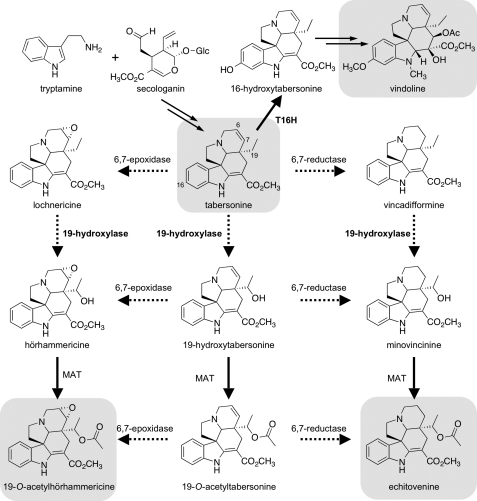
The medicinal plant Catharanthus roseus is currently the sole source of two anticancer agents subject to widespread clinical use, the bisindole alkaloids vinblastine and vincristine (5). In C. roseus, the alkaloid tabersonine can be transformed into the bisindole precursor vindoline, in aerial organs, or 19-O-acetylhörhammericine in roots (see Fig. 1) (5, 6). Although hydroxylation at the 16-position of tabersonine ultimately leads to vindoline, tabersonine can be converted by a P450-dependent 6,7-epoxidase to form lochnericine (7–9) or a putative P450-dependent 19-hydroxylase to form 19-hydroxytabersonine (10). Both lochnericine and 19-hydroxytabersonine are proposed intermediates in 19-O-acetylhörhammericine biosynthesis (Fig. 1), and minovincinine 19-hydroxy-O-acetyltransferase is the only gene in this pathway for which a cognate cDNA has been isolated and characterized (6). Although alkaloid biosynthesis in C. roseus involves several biotransformations known or predicted to be P450-dependent (11), only five C. roseus P450s have been functionally characterized, namely flavonoid 3′,5′-hydroxylase (CYP75A8), cinnamate 4-hydroxylase (CYP73A4), geraniol 10-hydroxylase (CYP76B6), secologanin synthase (CYP72A1), and tabersonine 16-hydroxylase (T16H; CYP71D12). Only the latter three participate in alkaloid biosynthesis (12–16). These C. roseus P450 genes were characterized functionally using various techniques, including in vitro substrate specificity assays and cDNA library screening. Although such traditional approaches have proven effective for the functional elucidation of many P450 genes, no versatile approach exists to identify diverse P450s, particularly those carrying out more specialized functions (4). Recently, co-expression analysis has been used effectively to guide the functional characterization of P450 genes in Arabidopsis thaliana (17, 18).
FIGURE 1.
Proposed biosynthesis of tabersonine-derived alkaloids in C. roseus. The major alkaloids vindoline, 19-O-acetylhörhammericine, and echitovenine, as well as the tabersonine precursor, are highlighted in gray. Secologanin and tryptamine are biosynthetic precursors of tabersonine. In aerial parts of the plant, tabersonine is hydroxylated by T16H as the first committed step in vindoline biosynthesis. In roots, tabersonine can be decorated via several pathways to make alkaloids such as 19-O-acetylhörhammericine and echitovenine. Most of the biosynthetic steps leading toward these root-derived alkaloids are uncharacterized at the genetic level (dashed arrows).
Here, we describe the use of a C. roseus expressed sequence tag collection generated by transcriptome sequencing and a corresponding expression profile data set to facilitate the identification and characterization of a unique P450. Hierarchical clustering of gene expression data identified P450 candidates that are co-expressed with genes known to be involved in alkaloid biosynthesis. We developed a whole cell assay to demonstrate that yeast cell cultures expressing one of these P450 gene candidates produce a hydroxylated product in the culture medium when supplemented with tabersonine. This enzyme, CYP71BJ1, is the first member of a new plant P450 subfamily and appears to be a tabersonine/lochnericine 19-hydroxylase.
EXPERIMENTAL PROCEDURES
Chemicals
Tabersonine was a generous gift from Viresh Rawal (University of Chicago, Chicago, IL). Unless otherwise noted, all other chemicals were obtained from Sigma-Aldrich.
Sequence and Expression Data
The C. roseus transcriptome and expression mapping data were generated by the Medicinal Plant Genomics Consortium at Michigan State University. Separate cDNA libraries were constructed from mRNA from three C. roseus tissues (seedlings, seedlings treated with methyl jasmonate, and suspension culture cells treated with yeast extract) using the Illumina mRNA-seq kit (Illumina, San Diego, CA) and sequenced (single end, 36 bp) on the Illumina Genome Analyzer II platform at the Michigan State University Research Technology Support Facility. A custom Perl script was used to remove reads with low complexity and low quality regions. Reads with a base with a quality score <20 or with >2 N bases were considered low quality and removed. The low complexity threshold was defined as a read composed of >85% of a single nucleotide. Transcript assemblies were constructed using the Velvet (version 0.7.61)/Oases (version 0.1.17) de novo transcriptome assembler (19). The hash length for the initial Velvet assembly was 27, and the minimum transcript length cut-off used for Oases was 200 bp. The final assembly contained 33,992 transcripts with an N50 of 780 bp. The longest transcript from each contig was selected as the representative transcript, and reads from the individual libraries were mapped (up to two mismatches permitted) to the assembled transcriptome using Bowtie (20). Expression values (reads per kb transcript per million mapped reads) were calculated with a custom Perl script.
Hierarchical Clustering (Co-expression) Analysis
Expression levels of P450 candidates (as designated by pfam analysis), and known alkaloid biosynthetic genes, in control and elicited seedlings, and cell suspension cultures were subjected to hierarchical clustering analysis using CLUSTER (version 3.0) for Mac. The resulting dendrogram was viewed with Treeview (21). The expression mapping data and complete Treeview cluster are reported in the supplemental data. All P450 sequences used in this analysis have been deposited to GenbankTM.
Plant Material
C. roseus hairy root cultures were grown according to Morgan et al. (10, 22), and C. roseus seedlings were grown and elicited with methyl jasmonate as reported previously (11).
Gene Cloning
A T16H gene, codon-optimized for expression in Saccharomyces cerevisiae was synthesized (GenScript, Piscataway, NJ) with BamHI and XhoI restriction sites for directional cloning into pYeDP60. The CYP71BJ1 ORF was amplified from elicited seedling cDNA (6 days after elicitation) using a sense primer (5′-AAA AAA GGA TCC ATG TTG TCT TCA TTG AAA GAT-3′) containing an engineered BamHI restriction site (underlined) and an antisense primer (5′-AAA AAA GAA TTC TTA AAA AAT GGT AAC CGG AGT TG-3′) containing an EcoRI restriction site. The apparent full-length ORF for candidate “Locus_41747” was amplified using a sense primer (5′-AAA AAA GGA TCC ATG AAC TTC TCT CTC ACC TCT CCC ATT TTC C-3′) containing an engineered BamHI restriction site (underlined) and start codon (boldface) and an antisense primer (5′-AAA AAA GAA TTC TTA GTT TCC TTC AAC TAC AGT TGA GAT GCT AGG-3′) containing an EcoRI restriction site. The full-length ORF for CYP81Z1 was amplified using a sense primer (5′-AAA AAA GGA TCC ATG GAG GTT TCC TTT TTC TAC ACC TC-3′) containing an engineered BamHI restriction site (underlined) and an antisense primer (5′- AAA AAA CCC GGG TTA TGG GTT ATT TTC CAT-3′) containing an SmaI restriction site. Using directional cloning, the candidate P450 ORFs were cloned into pYeDP60, sequenced, and transformed into S. cerevisiae WAT11 (harboring the integrated A. thaliana P450 reductase ATR1) cells (22).
The 16-hydroxytabersonine 16-O-methyltransferase (16OMT; GenBankTM accession no. EF444544.1) ORF was amplified from elicited seedling cDNA using a sense primer (5′- AAT AAA GGA TCC ATG GAT GTT CAA TCT GAG G-3′) containing a BamHI restriction site and an antisense primer (5′-ATT TTA TTT TCT CGA GTC AAG GAT AAA CCT CAA TG-3′) containing an XhoI restriction site. Using directional cloning, the ORF was cloned into pYES3-CT and sequenced. Using lithium acetate, the 16OMT-pYES3-CT construct was transformed into S. cerevisiae WAT11 cells already containing the T16H construct (23).
Yeast Strain, Growth, and Whole Cell Assay for Activity
S. cerevisiae WAT11 was used as the host strain for the expression candidate P450 genes and the T16H gene in pYeDP60. S. cerevisiae harboring the desired plasmid were grown, and protein expression was induced as reported previously (22). To functionally characterize CYP71BJ1, a 5-ml S. cerevisiae WAT11 cell culture expressing this ORF was grown for 5 h after induction at 28 °C, at which point, the cell culture was supplemented with 147 μm of tabersonine (from a 73.3 mm dimethyl sulfoxide stock solution). After 24 h, the cells were removed by centrifugation, and the medium was extracted with ethyl acetate and analyzed by LC-MS.
Isolation of Enzymatic Product
Two 500-ml cultures of S. cerevisiae WAT11 cells harboring the CYP71BJ1 construct were grown and induced, and after 5 h of protein expression, the medium was supplemented with 147 μm tabersonine. Cells were removed 24 h later by centrifugation at 3810 × g for 15 min. The combined medium was then extracted with 1 liter of ethyl acetate three times and concentrated. The CYP71BJ1 enzymatic product was purified by column chromatography. A mobile phase of hexanes:ethyl acetate (1:1) was used for the first column, and dichloromethane:methanol (99.5:0.5) was used for the second column. 1H NMR, 13C NMR, COSY, and 1H,13C HSQC spectra were recorded on a Bruker 400 MHz spectrometer. Exact mass measurements were made on a Bruker Daltonics APEXIV 4.7 Tesla Fourier Transform Ion Cyclotron Resonance Mass Spectrometer with electrospray ionization.
(R)-19-hydroxytabersonine
1H NMR (CDCl3): δ 8.91 (1H, s), 7.27 (1H, d, J = 6.9), 7.15 (1H, t, J = 7.7), 6.89 (1H, t, J = 7.5), 6.82 (1H, d, J = 7.8), 5.92 (1H, dd, J = 4.9, 10.2), 5.80 (1H, d, J = 9.9), 3.79 (3H, s), 3.47 (1H, dd, J = 4.9, 15.9), 3.30–3.35 (1H, m), 3.23 (1H, d, J = 16.2), 3.06 (1H, t, J = 7.0), 2.89 (1H, dd, J = 15.4), 2.78 (1H, s), 2.73–2.79 (1H, m), 2.49 (1H, d, J = 15.4), 2.07–2.14 (1H, m), 1.83–1.87 (1H, m), 1.55 (1H, d, J = 3.3), 0.88 (3H, d, J = 6.4); 13C NMR (CDCl3): d 168.54, 166.38, 143.03, 137.71, 129.44, 127.81, 126.31, 121.54, 120.93, 109.37, 91.26, 67.00, 66.62, 55.57, 51.29, 51.02, 50.10, 46.31, 43.90, 27.57, 17.36; ESI-MS(C21H24N2O3+) m/z calculated: 353.1860 [M+H]+, found: 353.1848 [M+H]+. The UV absorbance maxima are observed at 229, 296, and 331 nm.
Isolation of Lochnericine and Hörhammericine
C. roseus hairy roots (5 g) were macerated in 50 ml of methanol followed by sonication for 1 h. After filtration, the methanol extract was analyzed on a Beckman Coulter System Gold 125 HPLC equipped with a model 168 photodiode array detector at 330 nm. Analytes were separated on a Lichrosorb reverse phase column (Select B, 25 cm × 4.0 mm column, 5-μm particle size) using 20–50% acetonitrile:water (0.1% trifluoroacetic acid) for 20 min. Preparative HPLC of lochnericine and hörhammericine was performed on a Beckman Coulter System Gold equipped with a 125 solvent module, and a 166P detector at 330 nm using the same gradient. Analytes were separated on a reverse-phase column (Grace Vydac 2.2 cm × 25 cm, 10-μm particle size). The absorbance maxima and exact mass spectrometry data for lochnericine and hörhammericine are listed in the supplemental data (24, 25).
Preparation of Microsomes, Kinetic Assays, and Substrate Specificity Assays
Microsomes enriched with CYP71BJ1 or T16H were isolated using the high density procedure according to Pompon et al. (22). The total microsomal protein content was determined using a bichinchoninic acid assay by Pierce. CYP71BJ1 assays were prepared in a final volume of 100 μl containing 55 μg of microsomal protein, 1 mm NADPH, 4 mm dithiothreitol, 100 μm ajmaline internal standard, and 100 mm sodium phosphate buffer (pH 7.0). Assays were initiated by the addition of 30 μm substrate unless otherwise noted. The following substrates were assayed with microsomes: tabersonine, yohimbine, catharanthine, dihydrotabersonine, norharmane, lochnericine, 16-hydroxytabersonine (3 μm), 16-methoxytabersonine (300 nm), and vindolinine (5 μm). Dihydrotabersonine was prepared as reported by Liscombe et al. (26). Yeast cultures co-expressing T16H and 16OMT enzymes were supplemented with tabersonine (147 μm) to produce 16-hydroxytabersonine and 16-methoxytabersonine. Assays lacking microsomes or NADPH served as negative controls. All assays were incubated at 30 °C for 1 h or overnight, after which 10% of the assay volume was quenched with 1 ml of HPLC-grade methanol. Assays were clarified by centrifugation for 5 min in a microcentrifuge and then analyzed by LC-MS. Ultra-performance liquid chromatography and MS analyses were performed according to Liscombe et al. (26).
Steady State Enzyme Kinetics
A dimethyl sulfoxide stock (7.3 mm) of tabersonine was prepared and then diluted with water to a final concentration of 5.9 mm. Serial dilutions of this stock were used for enzyme assays. Enzyme assays (0.1-ml reaction volume) contained 1.2 mg of CYP71BJ1 enriched microsomes, 1 μm ajmaline (for an internal standard), 1 mm NADPH in sodium phosphate buffer (100 mm, pH 7.0), and 4 mm dithiothrietol. Reactions were initiated by the addition of tabersonine (290 nm, 550 nm, 570 nm, 590 nm, 1.2 μm, 2.3 μm, 4.8 μm, 5.3 μm, 6.2 μm, 9.2 μm) and incubated at 30 °C. Time points were chosen such that the rate of product formation was linear, to ensure accurate measure of initial rates (between 3 and 15 min after initiation of the reaction). The reactions were analyzed by LC-MS and values of Vmax and Km were estimated using nonlinear fitting (OriginPro; version 7.0, Northampton, MA). The errors reported are based on the 95% confidence of the S.D. of three independent experiments.
Inhibition Assays
Aqueous stocks of the suicide inhibitor aminobenzotriazole (ABT) ranging from 64 mm to 250 μm were used for inhibition assays. A concentration of 9.2 μm of tabersonine substrate in the presence of 0, 25 μm, 100 μm, 400 μm, 1.6 mm, and 6.4 mm ABT was used. The errors reported are based on the 95% confidence of the S.D. of three independent experiments.
RESULTS
Cluster Analysis and Molecular Cloning
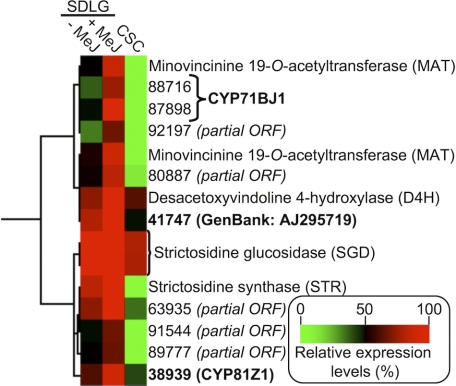
To identify genes encoding cytochrome(s) P450 involved in the oxygenation of tabersonine in C. roseus, we performed hierarchical clustering analysis with expression mapping data. Expression profiles of known alkaloid biosynthetic genes (tryptophan decarboxylase, geraniol 10-hydroxylase, loganic acid methyltransferase, secologanin synthase, strictosidine synthase, strictosidine glucosidase, T16H, 16OMT, 2,3-dihydro-3-hydroxytabersonine-N-methyltransferase, desacetoxyvindoline-4-hydroxylase, deacetylvindoline acetyltransferase, and minovincinine 19-hydroxy-O-acetyltransferase (5, 6, 26)) and putative P450 genes were included in the analysis. The results of hierarchical clustering were visualized as a heat map, a section of which is presented in Fig. 2, showing the clusters that we focused on in this study (supplemental data). Contigs 92197, 80887, 63935, 91544, and 89777 all represented partial ORFs. However, two contigs (88716 and 87898) that clustered with minovincinine 19-hydroxy-O-acetyltransferase (Fig. 2), which acetylates the 19-hydroxyl group of minovincinine and hörhammericine (Fig. 1) (6), were determined by manual sequence analysis to represent the same gene transcript and could be assembled to form the full-length ORF encoding CYP71BJ1 (supplemental Fig. S1). Two other apparent full-length ORFs (41747 and CYP81Z1) were also present in the cluster and were selected for further characterization. The gene candidates were cloned from methyl jasmonate-elicited seedling cDNA, cloned into the yeast expression vector pYeDP60, and expressed in S. cerevisiae WAT11 cells, a yeast strain optimized for plant P450 protein expression (22).
FIGURE 2.
Hierarchical clustering analysis to examine putative P450-dependent enzyme-encoding genes co-expressed with minovincinine O-acetyltransferase. The sub-cluster that became the focus of this study is shown. Putative full-length candidates (shown in boldface) were selected for further characterization. SDLG, aseptically grown seedlings; −MeJ, without methyl jasmonate treatment; +MeJ, treated with methyl jasmonate; CSC, cell suspension culture.
Protein Expression and Whole Cell Assay
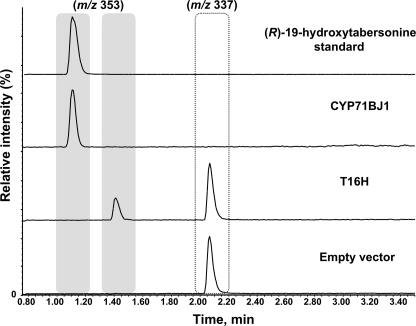
We developed a whole cell yeast assay to functionally characterize P450s. As a positive control, we cloned a synthetic, yeast codon-optimized version of the previously characterized T16H gene (16) into the pYeDP60 vector and transformed the resulting T16H-pYeDP60 construct into yeast. Protein expression was induced in a 5-ml yeast culture for 5 h, at which point the medium was supplemented with tabersonine. In 24 h, 16-hydroxytabersonine was detected in the yeast medium as evidenced by LC-MS analysis (Fig. 3). With the success of this positive control, we used this assay to detect products derived from tabersonine in yeast cultures expressing CYP71BJ1, 41747, and CYP81Z1 candidate P450s.
FIGURE 3.
LC-MS selected ion chromatograms (m/z 337 and m/z 353) of the medium of tabersonine-supplemented yeast cell cultures expressing CYP71BJ1, T16H, or empty pYeDP60 vector.
Yeast cultures that expressed CYP71BJ1 were supplemented with tabersonine, and after 24 h, the tabersonine starting material was completely converted to a more hydrophilic compound with a mass consistent with hydroxylation or epoxidation (Fig. 3). Notably, the CYP71BJ1 product exhibited a retention time distinct from that observed for the T16H product, 16-hydroxytabersonine (Fig. 3). No enzymatic product was observed when tabersonine was incubated with yeast cultures harboring the empty pYeDP60 plasmid (Fig. 3) or those expressing the other candidate P450s, 41747 and CYP81Z1.
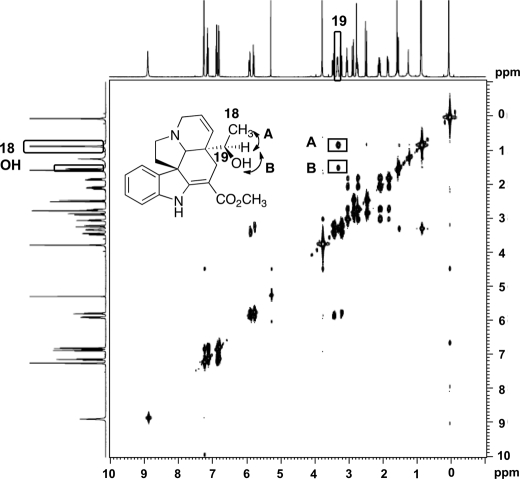
We isolated and purified the tabersonine-derived CYP71BJ1 product from 1 liter of yeast culture supplemented with 147 μm tabersonine. The enzymatic product was identified as (R)-19-hydroxytabersonine, as evidenced by high resolution mass spectrometry, 1H NMR, 13C NMR, COSY (Fig. 4), and HSQC NMR (supplemental Figs. S2–S4). The COSY spectrum shows a cross-peak between the doublet at 1.55 ppm corresponding to the proton in the 19-hydroxyl group and the doublet at 2.78 ppm corresponding to the proton at C19 (Fig. 4). Furthermore, we observed a cross-peak between the protons at C18 (0.88 ppm) and the proton at the 19-position (Fig. 4).
FIGURE 4.
COSY correlations for 19-hydroxylation of tabersonine isolated from yeast culture expressing CYP71BJ1. A represents the cross-peak between the proton at C19 and the 3 protons at C18. B represents the cross-peak between the proton in the 19-hydroxyl group and the proton at C19.
Substrate Specificity Analysis
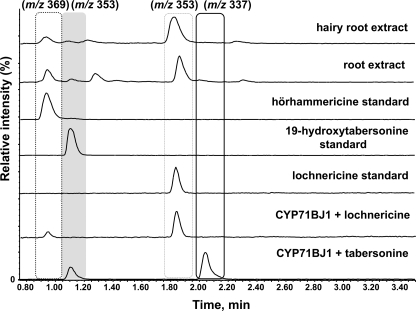
Microsomes were isolated from yeast cultures expressing CYP71BJ1 and assayed with a variety of alkaloids representing different terpene indole alkaloid skeletons to determine the substrate scope of the enzyme (supplemental Fig. S5). Assays lacking NADPH cofactor or CYP71BJ1-enriched microsomes served as negative controls. Although neither lochnericine nor hörhammericine are available commercially, both are known to be present in hairy root cultures and in the roots of mature C. roseus plants (9, 10, 25). We could isolate small quantities of lochnericine, a potential physiological substrate for CYP71BJ1 (Fig. 1), from hairy roots. Although we could not obtain quantities sufficient for characterization by NMR, the isolated standard exhibited the correct exact mass and UV signature (24). Moreover, a microsomal fraction enriched in CYP71BJ1 converted this isolated compound to a product with an exact mass and UV signature consistent with hörhammericine (Fig. 5 and supplemental Fig. S6 and Table S1) (24, 25). Only tabersonine and lochnericine were transformed by CYP71BJ1.
FIGURE 5.
Selected ion chromatograms from LC-MS chromatograms of enriched CYP71BJ1 microsomes assayed with tabersonine (m/z 337) and lochnericine (m/z 353) and C. roseus mature root and hairy root extracts containing lochnericine and hörhammericine (m/z 369) alkaloids.
Enzyme Kinetics and Oxygenase Inhibition
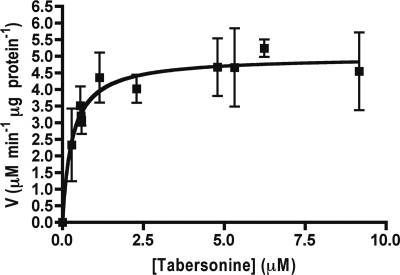
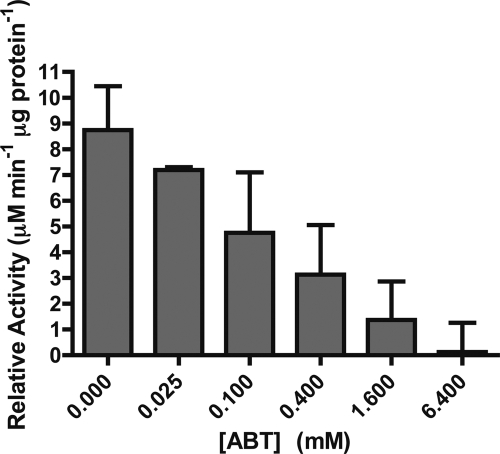
CYP71BJ1-enriched microsomes assayed with tabersonine demonstrate Michaelis-Menten kinetics with an apparent Km of 300 ± 50 nm and Vmax of 5.0 ± 0.2 μm min−1 μg−1 (Fig. 6). We also observed reduced activity when increasing concentrations of the ABT-P450 monooxygenase suicide inhibitor was added to the assay. CYP71BJ1 activity decreased to <1% when 6.4 mm ABT is present (Fig. 7).
FIGURE 6.
Steady state Michaelis-Menten kinetics derived from initial rates of CYP71BJ1-enriched microsomes with tabersonine. Error bars represent the 95% confidence of the S.D. from three independent experiments.
FIGURE 7.
ABT inhibition of CYP71BJ1 19-hydroxylase activity with tabersonine. Error bars represent the 95% confidence of the S.D. from three independent experiments.
DISCUSSION
Using co-expression analysis, followed by functional characterization with a whole cell yeast assay, we identified a role for C. roseus CYP71BJ1 in alkaloid biosynthesis. Co-expression data shows that CYP71BJ1 has an expression pattern similar to minovincinine 19-hydroxy-O-acetyltransferase, an enzyme that acetylates the 19-position of hörhammericine or minovincinine to form 19-O-acetylhörhammericine or echitovenine, respectively. Formation of hörhammericine from tabersonine requires the catalytic action of two enzymes, tabersonine 6,7-epoxidase and a 19-hydroxylase. Biochemical evidence strongly suggests that both of these enzymes are P450-dependent (10, 27). Heterologous expression and subsequent incubation of CYP71BJ1 with tabersonine revealed that this enzyme stereoselectively catalyzes hydroxylation of tabersonine at C19. Large scale whole cell assays allowed isolation of milligram quantities of the enzymatic product for complete structural characterization.
Kutney et al. (9) hypothesize that hörhammericine biosynthesis proceeds either through a 6,7-epoxidation of tabersonine to yield lochnericine, which is subsequently hydroxylated at the 19-position. Alternatively, hydroxylation at the 19-position can occur directly on tabersonine to yield 19-hydroxytabersonine, which then undergoes a 6,7-epoxidation (9, 10). NMR characterization of the CYP71BJ1 enzymatic product revealed that the absence of the 6,7-epoxide in lochnericine does not alter the stereoselectivity of the hydroxylation; the 1H NMR chemical shifts of the enzymatic product of CYP71BJ1 and tabersonine match those of the previously reported (R)-19-hydroxytabersonine (28, 29). Thus, the order of biosynthetic steps involved in hörhammericine biosynthesis is still ambiguous because CYP71BJ1 accepts lochnericine and tabersonine in vitro. The kinetics of CYP71BJ1 were measured only with tabersonine since limited amounts of lochnericine could be isolated from C. roseus hairy root cultures. As a result, it is not clear whether CYP71BJ1 quantitatively prefers tabersonine to lochnericine in vitro. However, the low Km value for tabersonine suggests that 19-hydroxylation of tabersonine could occur in planta. Both lochnericine and 19-hydroxytabersonine are observed in C. roseus, suggesting that both compounds are plausible biosynthetic intermediates. To further explore the qualitative substrate preference of this enzyme, we subjected microsomes containing CYP71BJ1 to a competition assay. Microsomes were incubated with a 1:1 mixture of tabersonine and lochnericine (55 μm concentration each). After 15 h of incubation, LC-MS analysis indicated that although both substrates were turned over by the enzyme, lochnericine appeared to be the preferred substrate (supplemental Fig. S7). Morgan and Shanks (10) observed a concomitant reduction in hörhammericine and accumulation of lochnericine in C. roseus hairy roots after treatment with the P450 suicide inhibitor ABT. As such, the ABT sensitivity of CYP71BJ1 provides some further support for the involvement of CYP71BJ1 in hörhammericine biosynthesis in vivo, perhaps favoring a biosynthetic route via lochnericine. Further studies to disrupt the function of CYP71BJ1 in plant tissue such as RNAi in hairy roots might resolve these questions and provide further insight into the physiological role of CYP71BJ1.
The substrate scope of CYP71BJ1 seems to be controlled partially by the presence of the 2,3-double bond because dihydrotabersonine is not an accepted substrate. Importantly, the vindoline precursors 16-hydroxytabersonine and 16-O-methoxytabersonine are not hydroxylated by CYP71BJ1. Because hydroxylation at the 16-position is the first step in the conversion of tabersonine to vindoline, we speculate that the substrate specificity of CYP71BJ1 may play a role in preventing the 19-hydroxylation of intermediates destined for vindoline biosynthesis. Silencing of CYP71BJ1 in C. roseus may therefore improve the metabolic flux toward the commercially valuable vindoline-derived bisindole alkaloids.
In summary, we report that CYP71BJ1, the first member of a new CYP71 subfamily, stereoselectively hydroxylates lochnericine and tabersonine at the 19-position. We are currently assaying more P450 candidates with tabersonine to detect 6,7-epoxidase activity to further understand how vindoline and 19-O-acetylhörhammericine biosynthesis are regulated.
Supplementary Material
Acknowledgments
We thank David Nelson for assigning the CYP family of CYP71BJ1. We acknowledge the assistance of Greg Fedewa for preparation of cDNA libraries and Brieanne Vaillancourt for sequencing of the cDNA libraries.
This work was supported in part by grants from the National Institutes of Health (GM074820 and RC2GM092521). Funding for the MIT-DCIF Avance (DPX) 400 MHz NMR spectrometer was provided by the National Institutes of Health (Award 1S10RR13886-01).

The on-line version of this article (available at http://www.jbc.org) contains supplemental data, Table S1, and Figs. S1–S7.
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) HQ901597, JF266591, and JF266590.
The Illumina mRNA-seq reads are available in the NCBI Sequence Read Archive under study accession no. SRP005953.
- P450
- cytochrome P450
- contig
- group of overlapping clones
- 16OMT
- 16-hydroxytabersonine 16-O-methyltransferase
- T16H
- tabersonine 16-hydroxylase
- ABT
- aminobenzotriazole.
REFERENCES
- 1. Nielsen K. A., Moller B. L. (2005) in Cytochrome P450: Structure, Mechanism, and Biochemistry (de Montellano P. R. ed.), 3rd Ed., pp. 553–583, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 2. Jensen K., Møller B. L. (2010) Phytochemistry 71, 132–141 [DOI] [PubMed] [Google Scholar]
- 3. Nelson D. R., Schuler M. A., Paquette S. M., Werck-Reichhart D., Bak S. (2004) Plant Physiol. 135, 756–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mizutani M., Ohta D. (2010) Annu. Rev. Plant Biol. 61, 291–315 [DOI] [PubMed] [Google Scholar]
- 5. O'Connor S. E., Maresh J. J. (2006) Nat. Prod. Rep. 23, 532–547 [DOI] [PubMed] [Google Scholar]
- 6. Laflamme P., St-Pierre B., De Luca V. (2001) Plant Physiol. 125, 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Furuya T., Sakamoto K., Iida K., Asada Y., Yoshikawa T., Sakai S., Aimi N. (1992) Phytochemistry 31, 3065–3068 [DOI] [PubMed] [Google Scholar]
- 8. Shanks J. V., Bhadra R., Morgan J., Rijhwani S., Vani S. (1998) Biotechnol. Bioeng. 58, 333–338 [DOI] [PubMed] [Google Scholar]
- 9. Kutney J. P., Choi L. S., Kolodziejczyk P., Sleigh S. K., Stuart K. L., Worth B. R., Kurz W. G., Chatson K. B., Constabel F. (1980) Phytochemistry 19, 2589–2595 [Google Scholar]
- 10. Morgan J. A., Shanks J. V. (1999) Phytochemistry 51, 61–68 [DOI] [PubMed] [Google Scholar]
- 11. Facchini P. J., De Luca V. (2008) Plant J. 54, 763–784 [DOI] [PubMed] [Google Scholar]
- 12. Kaltenbach M., Schröder G., Schmelzer E., Lutz V., Schröder J. (1999) Plant J. 19, 183–193 [DOI] [PubMed] [Google Scholar]
- 13. Hotze M., Schröder G., Schröder J. (1995) FEBS Lett. 374, 345–350 [DOI] [PubMed] [Google Scholar]
- 14. Collu G., Unver N., Peltenburg-Looman A. M., van der Heijden R., Verpoorte R., Memelink J. (2001) FEBS Lett. 508, 215–220 [DOI] [PubMed] [Google Scholar]
- 15. Irmler S., Schröder G., St-Pierre B., Crouch N. P., Hotze M., Schmidt J., Strack D., Matern U., Schröder J. (2000) Plant J. 24, 797–804 [DOI] [PubMed] [Google Scholar]
- 16. Schröder G., Unterbusch E., Kaltenbach M., Schmidt J., Strack D., De Luca V., Schröder J. (1999) FEBS Lett. 458, 97–102 [DOI] [PubMed] [Google Scholar]
- 17. Ehlting J., Sauveplane V., Olry A., Ginglinger J. F., Provart N. J., Werck-Reichhart D. (2008) BMC Plant Biol. 8, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matsuno M., Compagnon V., Schoch G. A., Schmitt M., Debayle D., Bassard J. E., Pollet B., Hehn A., Heintz D., Ullmann P., Lapierre C., Bernier F., Ehlting J., Werck-Reichhart D. (2009) Science 325, 1688–1692 [DOI] [PubMed] [Google Scholar]
- 19. Zerbino D. R., Birney E. (2008) Genome Res. 18, 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009) Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saldanha A. J. (2004) Bioinformatics 20, 3246–3248 [DOI] [PubMed] [Google Scholar]
- 22. Pompon D., Louerat B., Bronine A., Urban P. (1996) Methods Enzymol. 272, 51–64 [DOI] [PubMed] [Google Scholar]
- 23. Gietz R. D., Woods R. A. (2002) Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 24. Hisiger S., Jolicoeur M. (2007) Phytochem. Rev. 6, 207–234 [Google Scholar]
- 25. Abraham D. J., Farnsworth N. R., Loub W. D., Blomster R. N. (1969) J. Org. Chem. 34, 1575–1576 [Google Scholar]
- 26. Liscombe D. K., Usera A. R., O'Connor S. E. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 18793–18798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rodriguez S., Compagnon V., Crouch N. P., St-Pierre B., De Luca V. (2003) Phytochemistry 64, 401–409 [DOI] [PubMed] [Google Scholar]
- 28. Langlois N., Andriamialisoa R. Z. (1979) J. Org. Chem. 44, 2468–2471 [DOI] [PubMed] [Google Scholar]
- 29. Kohl W., Witte B., Hofle G. (1981) Z. Naturforsch. B 36, 1153–1162 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.