Abstract
Survivin is a multifunctional protein with essential roles in cell division and inhibition of apoptosis, but the molecular underpinnings of its cytoprotective properties are poorly understood. Here we show that homozygous deletion of the aryl hydrocarbon receptor-interacting protein (AIP), a survivin-associated immunophilin, causes embryonic lethality in mice by embryonic day 13.5–14, increased apoptosis of Ter119−/CD71− early erythropoietic progenitors, and loss of survivin expression in its cytosolic and mitochondrial compartments in vivo. In import assays using recombinant proteins, AIP directly mediated the import of survivin to mitochondria, thus enabling its anti-apoptotic function, whereas a survivin 1–141 mutant that does not bind AIP was not imported to mitochondria and failed to inhibit apoptosis. AIP-directed mitochondrial import of survivin did not affect cell division, was independent of the organelle transmembrane potential, did not require the chaperone Heat Shock Protein 90 (Hsp90), and was inhibited by cytosolic factor(s) present in normal cells. shRNA knockdown of the mitochondrial import receptor Tom20 abolished mitochondrial import of survivin and sensitized tumor cells to apoptosis, whereas silencing of Tom70 had no effect. Therefore, an AIP-Tom20 recognition contributes to cell survival in development and cancer by mediating the mitochondrial import of survivin.
Keywords: Apoptosis, Cancer Tumor Promoter, Chaperone Chaperonin, Mitochondria, Mitochondrial Transport
Introduction
Members of the inhibitor of apoptosis (IAP)3 gene family are multifunctional proteins (1) that participate in cell survival, cell proliferation, and a plethora of signal transduction pathways (2). These properties are commonly exploited in cancer (2), where aberrant IAP expression is frequently associated with deregulated cell survival (3), resistance to conventional or targeted therapies (4), and enhanced metastatic dissemination in vivo (5). This phenotype is exemplified by survivin (6), an IAP with essential roles in cell division and inhibition of apoptosis aberrantly overexpressed in virtually every human tumor in vivo (7). Although it is still unclear which of the two main functions of survivin is predominantly exploited in cancer, evidence from transgenic models points to a pivotal role of survivin cytoprotection in the growth of epithelial (8) and hematopoietic (9) malignancies, potentially via deregulation of stem cell-like compartment(s) in vivo (10).
As far as mechanistic requirements, recent evidence has implicated a pool of survivin localized to mitochondria as specifically earmarked to inhibit apoptosis (11). Several models have been proposed as to how mitochondrial survivin may function in cytoprotection, including sequestration of proapoptotic Smac (12), inhibition of its release from mitochondria (13), or, alternatively, assembly of a phosphorylation-regulated complex with X-chromosome-linked inhibitor of apoptosis protein (XIAP) in the cytosol (14) that enhances XIAP stability and synergistically inhibits caspases (14, 15). Although a mitochondrial pool of survivin has been almost exclusively detected in tumor, as opposed to normal cells (11), and directly linked to disease progression in vivo (14), survivin lacks a recognizable mitochondrial import sequence, and the molecular underpinnings of its localization to mitochondria, and thus anti-apoptotic functions, have not been elucidated.
Preprotein import to mitochondria is a stepwise process that involves the translocase of outer membrane (TOM) complex located on the outer mitochondrial membrane (16, 17). The TOM complex contains at least seven subunits, including Tom40, 22, 7, 6, and 5 and two major receptor molecules, Tom20 and Tom70 (18). Reconstitution experiments of mitochondrial import reactions in vitro have led to a working model in which Tom70 recognizes preproteins with an internal mitochondrial targeting signal, in a process contributed by cytosolic chaperones, including Hsp70 and Hsp90 (19). Conversely, preproteins containing an amino-terminal, cleavable mitochondrial targeting sequence are preferentially translocated via the Tom20 receptor, with the cytosolic immunophilin, aryl hydrocarbon receptor-interacting protein (AIP, also called XAP-2) (20, 21) assisting in this process (22). Intriguingly, biochemical evidence indicates that survivin associates with Hsp90 (23) as well as AIP (24) through non-overlapping recognition sites. Although these interactions have been linked to enhanced survivin stability against proteasomal degradation in vivo (23, 24), their potential role in subcellular trafficking, including mitochondrial localization, has not been determined.
In this study, we investigated mechanisms of survivin localization to mitochondria and their potential impact on apoptosis inhibition, especially in tumors. We found that an AIP-Tom20 recognition provides for the main mechanism of survivin import to mitochondria and that this pathway exerts an essential prosurvival role during erythropoietic development in vivo.
EXPERIMENTAL PROCEDURES
Cell Lines and Antibodies
An AIP gene-trapped embryonic stem cell line, RRI002, was obtained from the Mutant Mouse Regional Resource Center. RRI002 cells contain a disrupted AIP locus in which a β-galactosidase reporter gene substitutes the last COOH terminus 300 amino acids of AIP. In RRI002 cells, the inserted β-galactosidase gene is expressed under the control of the endogenous AIP promoter. Cervical carcinoma HeLa, breast adenocarcinoma MCF-7, and human embryonic kidney HEK293T cells were obtained from the ATCC and maintained in culture as recommended by the supplier. Antibodies against AIP (Novus Biologicals), survivin (Novus Biologicals), Tom20 (Santa Cruz Biotechnology), Tom70 (Novus Biologicals), COX-IV (Clontech), Smac (Pro-Sci), β-actin (Sigma), and Hsp90 (BD Biosciences) were used. The Hsp90 ATPase inhibitor geldanamycin (GA) was obtained from LC Laboratories (Woburn, MA).
Generation of AIP Knockout Mice
All experiments involving animals were approved by an Institutional Animal Care and Use Committee. To generate chimeric AIP knockout mice, gene-trapped RRI002 embryonic stem cells were microinjected into mouse blastocysts by the Transgenic Animal Modeling Core at the University of Massachusetts Medical School, and male chimeric mice were bred with C57Bl/6 female mice. For genotyping experiments, genomic DNA was isolated from mouse tail using a DNeasy tissue kit (Qiagen) according to the manufacturer's instructions. A primer set comprising AIP-forward 5′-GTTATGTACCACTTCTGCTAGGAGC-3′ and AIP-reverse 5′-TGCAGCGTCCGAAAGTGGAA-3′ was used to amplify the wild-type AIP allele, with generation of a product of 800 nt (Fig. 1A). A primer set comprising β-galactosidase-reverse 5′-CCTCTTCGCTATTACGCCAGC-3′ in combination with the AIP-forward primer was used to amplify the gene-trapped allele with generation of a product of 1000 nt (Fig. 1A). Amplification reactions were performed using Platinum TaqDNA polymerase (Invitrogen) according to the manufacturer's instructions. PCR products were separated by electrophoresis on 1% agarose gels, stained with ethidium bromide, and visualized under ultraviolet light. Backcross mating with C57Bl/6 was performed for more than 10 generations to rederive the genetic background.
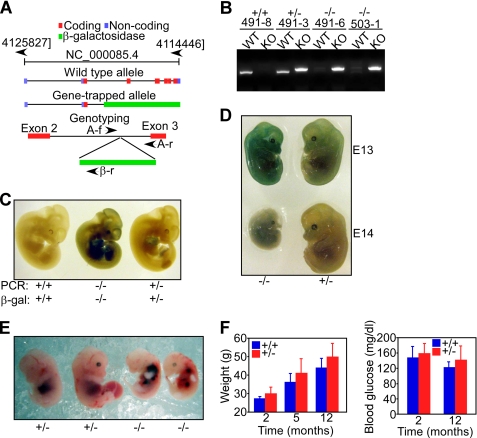
FIGURE 1.
Characterization of AIP knockout embryos. A, disruption of the AIP locus by gene trapping. The position of coding, non-coding, and untranslated regions in the AIP gene, and the position of the β-galactosidase gene insertion in the AIP locus are shown. The position and orientation of the various primers (AIP-forward (A-f), AIP-reverse (A-r), and β-galactosidase-reverse (β-r)) utilized for genotyping is indicated. B, PCR genotyping of AIP+/+, AIP+/−, and AIP−/− mice. KO, knockout; WT, wild type. C, β-galactosidase staining and PCR analysis of the indicated AIP mouse genotypes. D, morphology of AIP−/− or AIP+/− embryos at the indicated gestational age (E). E, hemorrhages in AIP−/− embryos at E12.5. F, wild-type (+/+) or AIP+/− mice were monitored for changes in overall body weight or blood glucose concentrations at the indicated time intervals. Data are mean ± S.E. of individual animal groups (weight analysis, AIP+/+, n = 11; AIP−/−, n = 22; glucose level analysis, AIP+/+, n = 5; AIP−/−, n = 9).
Plasmid Construction
A cDNA encoding the phosphate carrier gene (PiC) was purchased from Invitrogen, amplified with primers 5′-AAAAAGGATCCAGGAGGATGTTCTCGTCCGTAGC-3′ (forward) and 5′-AAAAACTCGAGCTACTCAGTTAACCCAAGCTTCTTCTTC-3′ (reverse), and inserted into pcDNA 3.0 (Invitrogen) after digestion with BamHI and XhoI. A cDNA encoding the mitochondrial import receptor Tom20 was amplified with primers 5′-AAAAAGAATTCAGAGCTGGGCTTTCCAAGTTACC-3′ (forward) and 5′-AAAAACTCGAGTCATTCCACATCATCTTCAGCCA-3′ (reverse). A cDNA encoding the mitochondrial import receptor Tom70 was amplified with primers 5′-AAAAAGAATTCCTTGATAGAGCCCAAGCAGCC-3′ (forward) and 5′-AAAAACTCGAGTTATAATGTTGGTGGTTTTAATCCGTA-3′ (reverse). The amplified PCR products were digested with EcoRI and XhoI and directionally inserted in the pGEX-4T plasmid (Amersham Biosciences). Plasmid constructs encoding pGEX-Hsp90 (23), pGEX-Survivin, pGEX-Survivin 1–141, pcDNA-Survivin, pcDNA Survivin 1–141, pGEX-AIP, pcDNA-AIP, and pcDNA-Smac-FLAG have been described previously (24).
Recombinant Protein Expression
BL21-CodonPlus-RIL Escherichia coli (Stratagene) were transformed with the various cDNA constructs in pGEX-4T and grown to A600 ∼0.8, and recombinant protein expression was induced in the presence of 0.2 mm isopropyl 1-thio-β-d-galactopyranoside (American Bioanalytical) for 5 h at 30 °C. Cells were harvested by centrifugation at 6000 × g for 20 min, and the bacterial pellet was suspended in 50 mm Tris (pH 7.4), 150 mm NaCl, 5 mm MgCl2, and 1 mm DTT and lysed by sonication. After removal of insoluble material by centrifugation at 15,000 × g for 30 min twice, the resulting soluble fractions were mixed with glutathione-agarose (Sigma) and incubated for 4 h at 4 °C. Bound proteins were recovered by centrifugation at 1000 × g for 1 min and washed three times in 50 mm Tris (pH 7.4), 500 mm NaCl, 5 mm MgCl2, and 1 mm DTT. In some experiments, the GST frame was cleaved from the expressed recombinant protein by incubation with 10 units/ml thrombin (Sigma) for 16 h at 4 °C, followed by neutralization in p-aminobenzamidine-agarose (Sigma) for 1 h at 4 °C twice. Protein concentration was determined by a protein assay reagent (Bio-Rad) using bovine serum albumin (Sigma) as a standard.
In Vitro Pull-down and Immunoprecipitation
Glutathione-agarose beads (Sigma) linked to various recombinant proteins were incubated in H buffer containing 20 mm Hepes (pH 7.7), 75 mm KCl, 0.1 mm EDTA, 2.5 mm MgCl2, 1 mg/ml BSA, and 0.05% Nonidet P-40 for 5 min at 22 °C. Protein A-Sepharose beads (Amersham Biosciences) were washed in IP buffer containing 50 mm Tris (pH 7.4), 150 mm NaCl, 1% Triton X-100, 0.5% Nonidet P-40, plus a protease inhibitor mixture (Roche). Aliquots of 35S-labeled proteins were produced using a TNT quick-coupled transcription/translation system (Promega) in the presence of [35S]methionine (Amersham Biosciences). Cell extracts were prepared by incubating PBS-washed cells in IP buffer for 1 h at 4 °C, followed by centrifugation at 13,000 × g for 10 min to remove insoluble material. For pull-down experiments, aliquots of bacterially expressed recombinant proteins, 35S-labeled proteins, or cell extracts were incubated with the conjugated beads for 16 h at 4 °C, washed in H-buffer or IP buffer five times, and then bound material was separated by SDS gel electrophoresis and analyzed by autoradiography or Western blotting (24).
Gene Silencing
siRNA CCAUGACAGACGAAGAGAA targeting AIP was purchased from Dharmacon (characterized previously (24)). An siRNA directed to Tom20 (ON-TARGET plus, SMARTpool L-006487-01-0005) was used. shRNA clones targeting the mitochondrial import receptor Tom20 or Tom70 were derived from a microRNA-adapted shRNA library (shRNAmir) and purchased from Open Biosystems. The shRNA constructs are inserted into the pGIPZ lentiviral vector containing a GFP reporter gene. For transfection experiments, control non-targeting siRNA or the various siRNAs were combined with oligofectamine (Invitrogen) in OPTI-MEM I medium (Invitrogen) according to the manufacturer's instructions. Cells were washed in OPTI-MEM I and incubated with the siRNA/oligofectamine mixture for 4 h, reconstituted with complete medium containing 10% FBS, and maintained in culture for an additional 48 h. Plasmid transfections were carried out using Lipofectamine reagent (Invitrogen) according to the manufacturer's instructions. Transfected cultures were confirmed for protein expression or knockdown by Western blotting after 24 h and processed for functional experiments.
Generation of Stably Transfected Cell Lines
Wild-type HA-tagged survivin or HA-tagged survivin 1–141 mutant cDNA characterized previously (24) was cloned in the BamHI and XhoI sites of the pcDNA5/FRT/TO plasmid (Invitrogen). Each expression vector was cotransfected with pOG44 (Invitrogen) into Flp-In T-Rex-293 cells (Invitrogen) using Lipofectamine (Invitrogen). Stable cell lines were selected in the presence of 15 μg/ml blasticidin plus 200 μg/ml hygromycin B, and conditional expression of HA-reactive survivin was determined after tetracycline induction (Tet-On system), by Western blotting. To generate cell lines with stable knockdown of Tom20 or Tom70, plasmid DNA expressing non-targeting or Tom20- or Tom70-directed shRNA (Open Biosystems) was transfected into HeLa cells using Lipofectamine. Cultures were selected in the presence of 4 μg/ml puromycin, and individual clones with high levels of GFP expression by fluorescence microscopy were characterized for differential silencing of Tom20 or Tom70 by Western blotting. Independently established clones were processed for functional experiments.
Mitochondrial Isolation
HeLa cells were detached by treatment with trypsin/EDTA (Gibco-BRL) and suspended in complete DMEM medium (Gibco-BRL). After centrifugation, the cell pellets were washed once in ice-cold PBS (pH 7.4), and mitochondrial and cytosolic fractions were isolated using a mitochondria isolation kit (Pierce) according to the manufacturer's instructions and as reported in published protocols (14). In some experiments, mitochondria were isolated from normal mouse liver or E11 embryos using a mitochondria isolation kit (Sigma), and samples from normal or tumor cell types were analyzed for mitochondrial preprotein import (see below).
Mitochondrial Import Assay
In vitro mitochondrial import assays were performed as described previously (19). Briefly, 35S-labeled recombinant proteins were diluted in one volume of MCS buffer containing 500 mm sucrose, 80 mm potassium acetate, 20 mm HEPES-KOH (pH 7.5), 5 mm magnesium acetate, and two volumes of MC buffer (250 mm sucrose, 80 mm potassium acetate, 20 mm HEPES-KOH (pH 7.5), 5 mm magnesium acetate). 50 μl of diluted 35S-labeled proteins were mixed with 30 μg of freshly isolated mitochondria, and the final volume was adjusted to 200 μl with MC buffer. The import reaction was continued for 20 min at 30 °C and terminated by cooling of the samples on ice. Mitochondria were reisolated by centrifugation, suspended in MC buffer in the presence of 50 μg/ml proteinase K for 10 min, and mixed with 1 mm PMSF (Calbiochem) to terminate the proteolytic reaction. Mitochondria were collected by centrifugation, and imported proteins were separated by SDS gel electrophoresis, followed by autoradiography. In some experiments, the mitochondrial uncoupler valinomycin (1 μm) or the Hsp90 inhibitor GA (2 μm) was added to the import reaction before analysis of protein import into mitochondria. As control, changes in mitochondrial membrane potential in the presence or absence of valinomycin (15 μm) were analyzed in HeLa cells by JC-1 staining and flow cytometry and quantified as a decrease in the green/red fluorescence ratio. The mitochondrial uncoupler carboxyl cyanide 3-chlorophenyl hydrazone (CCCP, 50 μm) was used as a control for these experiments.
Analysis of Apoptosis
Wild-type (+/+) or AIP knockout (−/−) embryos were harvested at E12.5, and fetal liver cells were mechanically dissociated by pipetting in Iscove modified Dulbecco's medium containing 20% FBS, 2 mm l-glutamine, and 100 μm β-mercaptoethanol. Single cell suspensions were prepared by filtration through 70-μm cell strainers. Reticulocytes were lysed by brief osmotic shock, and nucleated cells were counted. Analysis of erythroid differentiation was carried out by double labeling for the erythroid-specific marker Ter119 and non-erythroid CD71 (transferrin receptor) using multiparametric flow cytometry as described (25). This protocol identifies several distinct subpopulations of erythroid progenitors prior to the onset of erythropoietin dependence with potential for formation of erythroid colonies (CFU-e). These cells mature into a Ter119−/CD71− phenotype comprising erythropoietin-dependent CFU-e (26). For analysis of cell viability, Ter119- and CD71-stained fetal liver cells were analyzed for reactivity with annexin V by flow cytometry. For quantification of apoptosis, AIP+/+ or AIP−/− fetal liver cell populations were analyzed by dual color labeling for annexin V/DAPI and multiparametric flow cytometry. Alternatively, wild-type (+/+), heterozygous (+/−), or knockout (−/−) AIP embryos were collected at E11, subcellularly fractionated in mitochondrial (5 μg) and cytosolic (30 μg) extracts, and analyzed for differential protein expression by Western blotting. In other experiments, HEK293T cells stably transfected with wild-type survivin or the survivin 1–141 truncated mutant were stimulated with 2 μg/ml Tet to induce the expression of the various recombinant proteins, incubated with increasing concentrations of taxol (0–1 μm, Sigma) for 24 h, and analyzed for changes in cell viability by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide as described. Alternatively, HeLa cells carrying stable shRNA knockdown of Tom20 or Tom70 were treated with the broad-spectrum apoptotic stimulus staurosporine (0.5 μm) and analyzed for cell viability by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide after 24 h.
Immunofluorescence Microscopy
HEK293T stable transfectants were incubated with Tet to induce the expression of HA-tagged survivin or the survivin 1–141 mutant. Cells were washed in PBS (pH 7.4) and fixed in 4% paraformaldehyde for 10 min at 22 °C. After permeabilization with 0.1% Triton X-100 in PBS (pH 7.4) for 5 min, cells were blocked in 3% BSA in PBS containing 0.2% Tween 20 for 1 h at 22 °C and incubated with an antibody to α-tubulin (Sigma) for 18 h at 4 °C. After washes, cells were incubated with Alexa Fluor 594-conjugated secondary anti-mouse reagent (Molecular Probes), followed by Alexa Fluor 488-labeled antibody to HA, and analyzed on an Axioplan 2 fluorescence microscope (Zeiss). Images were processed using Photoshop CS2.
Statistical Analysis
Data were analyzed using the unpaired t test on a GraphPad software package for Windows (Prism 4.0). A p value of 0.05 was considered as statistically significant.
RESULTS
AIP Regulation of Cell Survival in Vivo
We began this study by investigating a potential developmental role of AIP in tissue integrity in vivo. For these experiments, we generated AIP knockout mice (27, 28) using gene-trap technology in which a β-galactosidase gene inserted in the AIP locus disrupts the entire coding sequence of AIP downstream of exon 2 (Fig. 1A). In genotyping experiments (Fig. 1B), PCR products of 800 nt or, conversely, 1000 nt, uniquely identified the wild-type AIP allele or the β-galactosidase-containing disrupted allele, respectively (Fig. 1B). Consistent with the expression of β-galactosidase under the control of the endogenous AIP promoter, AIP−/− embryos strongly stained for β-galactosidase expression by immunohistochemistry (Fig. 1C). In agreement with recent findings (27, 28), homozygous deletion of AIP was embryonic lethal, with loss of viability and morphologic involution of 100% of AIP−/− embryos by E13.5–14 (Fig. 1D). Additional features of AIP−/− embryos examined at E12.5 included decreased vascular structures in the yolk sac, reduced liver size, and frequent hemorrhages (Fig. 1E). Conversely, mice with heterozygous deletion of AIP were born at expected rates, were fertile, and exhibited no changes in overall body weight or blood glucose levels over a 12-month period, compared with AIP+/+ mice (Fig. 1F). At variance with a recent report (28), histologic examination of brain samples collected from AIP+/− mice was unremarkable, with no evidence of increased incidence of pituitary tumors, compared with wild-type mice (not shown).
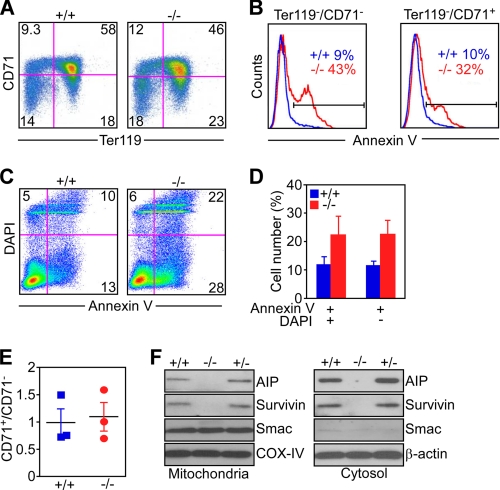
To determine whether lethality of AIP−/− embryos was associated with developmental defects in cell survival, we next examined fetal liver erythropoiesis using an established multiparametric flow cytometry protocol that quantifies the differential expression of CD71 and Ter119 markers in erythroblast populations at progressive stages of differentiation (26). Compared with wild-type liver cells, AIP−/− embryos exhibited a significant decrease in the more differentiated erythroblast populations, characterized by de novo expression of Ter119 (29) (Fig. 2A). Consequently, AIP−/− liver cells were enriched in the more primitive, Ter119− erythroblast populations (Fig. 2A), retaining CFU-e ability (29). To determine whether the block in erythroblast differentiation observed in AIP−/− embryos was due to defective cell survival, we next carried out annexin V staining of differentiating liver cell populations. In these experiments, primitive Ter119−/CD71− erythroblasts from AIP−/− embryos exhibited considerably increased reactivity for annexin V by flow cytometry, compared with their wild-type counterparts (Fig. 2B). In addition, Ter119−/CD71+ erythroid populations, which are highly dependent on erythropoietin as a survival factor, remained intensely annexin V-positive in AIP−/− embryos by flow cytometry (Fig. 2B, p = 0.0032). To independently validate genuine induction of apoptosis under these conditions, we next performed dual labeling experiments for annexin V and DAPI analyzed by multiparametric flow cytometry. Consistent with the data reported above, AIP−/− fetal liver cells exhibited increased apoptosis, compared with wild-type populations (Fig. 2C), with a higher number of annexin V-positive cells, regardless of DAPI reactivity (Fig. 2D). Finally, we asked whether the loss of erythroblast populations in AIP−/− embryos solely reflected increased apoptosis (Fig. 2, B–D) or was also potentially contributed by cell cycle defects. In these experiments, the ratio between CD71+/CD71− liver cells was indistinguishable in wild-type or AIP−/− embryos (Fig. 2E), arguing against potential defective cell cycle transitions in these settings (26).
FIGURE 2.
Developmental regulation of apoptosis by AIP. A, fetal liver cells isolated from AIP+/+ or AIP−/− embryos at E12.5 were analyzed for simultaneous expression of CD71 and Ter119 by multiparametric flow cytometry. B, Ter119−/CD71− or Ter119−/CD71+ fetal liver cell populations isolated from AIP+/+ or AIP−/− embryos were analyzed for annexin V reactivity by flow cytometry. The percentage of annexin V-positive cells is indicated. C, fetal liver cells isolated from AIP+/+ or AIP−/− embryos were double-labeled for Annexin V/DAPI and analyzed by multiparametric flow cytometry. For A and C, the percentage of cells in each quadrant is indicated. D, quantification of annexin V reactivity in wild-type (AIP+/+) or AIP−/− fetal liver cells. Data are mean ± S.E. of replicates of a representative experiment of at least two independent determinations. p = 0.029 (DAPI negative); p = 0.05 (DAPI-positive). E, fetal liver populations from the indicated AIP genotypes were analyzed for CD71+/CD71− ratio by flow cytometry. F, mitochondria or cytosolic extracts were isolated from E11 AIP+/+, AIP+/−, or AIP−/− embryos and analyzed by Western blotting.
Because AIP functions as a regulator of survivin (24), which is required for erythropoiesis in vivo (30), we next looked at potential differences in survivin levels and/or subcellular distribution in AIP−/− embryos. Compared with wild-type or heterozygous embryos, homozygous deletion of AIP at E11 resulted in a nearly complete loss of survivin protein expression in both its cytosolic and mitochondrial subcellular compartments by Western blotting (Fig. 2F). Conversely, the embryonic levels of another mitochondrial protein, Smac, were not affected in the presence or absence of AIP in E11 embryos (Fig. 2F).
AIP Is Required for Mitochondrial Import of Survivin
Transfection of breast adenocarcinoma MCF-7 cells with AIP-directed siRNA (24) significantly reduced the expression of endogenous AIP in both mitochondrial and cytosolic fractions (Fig. 3A). Consistent with the results obtained with AIP−/− embryos, this was associated with a nearly complete loss of survivin expression in its mitochondrial compartment and significant reduction of cytoplasmic levels (Fig. 3A). In contrast, a control, non-targeting siRNA had no effect on AIP or survivin levels in mitochondrial or cytosolic fractions (Fig. 3A).
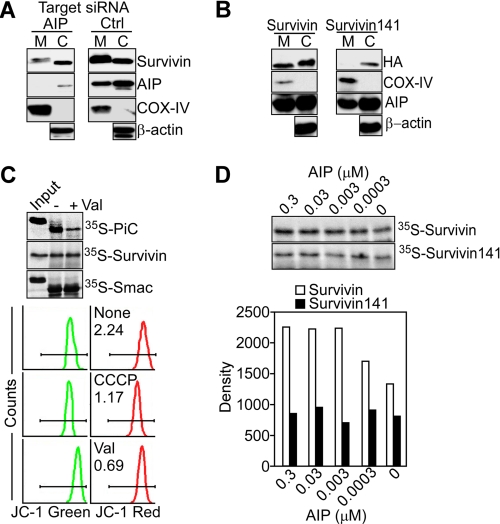
FIGURE 3.
AIP regulation of mitochondrial import of survivin. A, MCF-7 cells transfected with control non-targeting (Ctrl) or AIP-directed siRNA were fractionated in mitochondrial (M) or cytosolic (C) fractions and analyzed by Western blotting. COX-IV or β-actin was used as a mitochondrial or cytosolic marker, respectively. B, HA-tagged wild-type survivin or the survivin 1–141 mutant was transfected in MCF-7 cells, and mitochondrial or cytosolic fractions were analyzed by Western blotting. C, liver mitochondrial import of 35S-PiC, 35S-survivin, or 35S-Smac was evaluated in the presence (+) or absence (-) of the mitochondrial uncoupler valinomycin (val) and quantified by autoradiography (top panel). Bottom panel, effect of the mitochondrial uncouplers valinomycin or carboxyl cyanide 3-chlorophenyl hydrazone (CCCP) on mitochondrial membrane potential as determined by JC-1 staining and flow cytometry. The ratio between JC-1 green fluorescence/JC-1 red fluorescence is indicated per each condition tested. D, 35S-survivin or the 35S-survivin 1–141 mutant was analyzed for import in isolated mitochondria in the presence of recombinant AIP (0.3 nm-0.3 μm) (top panel). Bottom panel, densitometric quantification of radiolabeled protein bands. Representative experiment of at least three independent determinations.
Previous studies have shown that survivin binds AIP via its last COOH-terminal Asp-142 (24). Accordingly, a survivin 1–141 truncated mutant that does not bind AIP (24) was undetectable in isolated mitochondria of transfected MCF-7 cells (Fig. 3B). Conversely, the survivin 1–141 mutant accumulated in the cytosol of MCF-7 cells, albeit at lower levels than full-length survivin (Fig. 3B), consistent with a role of AIP in promoting survivin stability in vivo (24). In control experiments, full-length survivin indistinguishably accumulated in mitochondria or cytosolic fractions of transfected MCF-7 cells (Fig. 3B).
Using a mitochondrial import assay in vitro, 35S-survivin was readily imported in isolated liver mitochondria, in a reaction unaffected by disruption of the organelle transmembrane potential using the uncoupler valinomycin (Fig. 3C). Dissipation of the mitochondrial membrane potential also did not affect the import of another mitochondrial intermembrane space protein, Smac, whereas it significantly reduced the mitochondrial import of 35S-PiC, used as control for the import reaction (Fig. 3C). Under these conditions, addition of increasing concentrations of recombinant AIP to the import reaction dose-dependently enhanced the accumulation of 35S-survivin in isolated liver mitochondria (Fig. 3D). In contrast, the AIP-defective survivin 1–141 truncated mutant exhibited only background accumulation in mitochondria, in a reaction not further affected at any concentration of AIP added (Fig. 3D).
Specificity of AIP-directed Mitochondrial Import of Survivin
Survivin associates with another molecular chaperone involved in mitochondrial preprotein import, i.e. Hsp90 (23), and a role of this recognition in survivin localization to mitochondria was investigated next (19). At variance with the data obtained with AIP, preincubation of HeLa cells with the small molecule Hsp90 ATPase inhibitor GA, which inhibits preprotein import through the Tom70 mitochondrial import receptor (19), did not affect the import of survivin to mitochondria (Fig. 4A). As control, GA significantly reduced the cytosolic level of Akt, a prototype Hsp90 client protein (Fig. 4A). Similarly, GA efficiently inhibited the mitochondrial import of 35S-PiC through the Tom70 receptor (Fig. 4B), consistent with previous observations (19). Conversely, GA had no effect on the mitochondrial import of [35S]survivin (Fig. 4B).
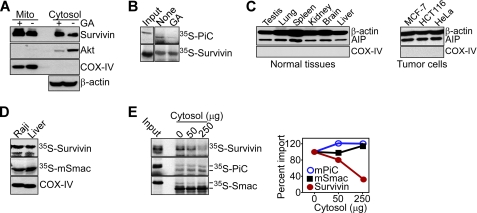
FIGURE 4.
Requirements of AIP-directed mitochondrial import of survivin. A, HeLa cells were treated with (+) or without (-) 2 μm GA for 24 h, and aliquots of isolated mitochondrial (Mito) or cytosol extracts were analyzed by Western blotting. Akt was used as a control Hsp90 client protein. B, mitochondrial import of 35S-survivin or 35S-PiC was evaluated in the absence or presence of GA. C, cytosol fractions isolated from the indicated mouse tissues (left) or human tumor cell lines (right) were analyzed by Western blotting. D, mitochondria from Raji cells or mouse liver were incubated with 35S-labeled proteins, survivin and Smac, and mitochondrial import was analyzed by autoradiography. Detection of COX-IV by Western blotting was used to confirm that comparable amount of mitochondria were applied in the import assay. E, mitochondrial import of the indicated 35S-labeled proteins was determined in the presence of mouse liver cytosol extracts and autoradiography (left). Markers, molecular size of preproteins and mature proteins. Right, densitometric quantification of protein bands. Representative experiment of at least two independent determinations.
Requirements of AIP-mediated Mitochondrial Import of Survivin
A potential basis for the differential subcellular localization of mitochondrial survivin in tumor, as opposed to normal cells (11), was investigated next. In initial experiments, AIP was found abundantly expressed and at comparable levels in all normal tissues as well as examined tumor cell lines (Fig. 4C). Similarly, 35S-survivin was imported indistinguishably in mitochondria isolated from normal mouse liver or B lymphoma Raji cells (Fig. 4D), arguing against the possibility that organelle-intrinsic properties mediated the differential localization of mitochondrial survivin selectively in tumor cells. Conversely, addition of increasing concentrations of cytosolic extracts from normal liver to the import reaction inhibited in a dose-dependent manner the accumulation of wild-type survivin in tumor, i.e. Raji, mitochondria (Fig. 4E). This response was specific, as comparable concentrations of cytosolic extracts from normal liver had no effect on the mitochondrial import of 35S-PiC or 35S-Smac (Fig. 4E).
AIP Recognition Does Not Affect Survivin-dependent Mitosis
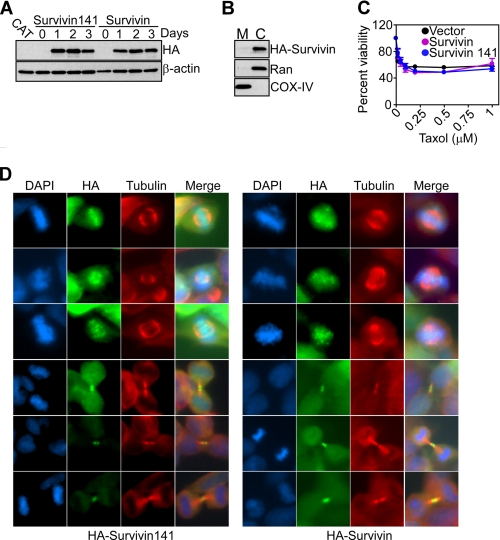
Survivin is a multifunctional protein with essential roles in cell division (31), and a potential participation of AIP in this pathway was examined next. For these experiments, we generated HEK293T cells stably transfected with Tet-regulated inducible expression of HA wild-type survivin or the AIP-defective survivin 1–141 truncated mutant (24). In control studies, wild-type or mutant survivin was comparably expressed at similar levels in HEK293T cells in response to Tet stimulation by Western blotting (Fig. 5A). Consistent with their normal phenotype (11), transfected HEK293T cells expressed survivin only in cytosol, with no localization of wild-type or the survivin 1–141 mutant to mitochondria by Western blotting of isolated subcellular fractions (Fig. 5B and not shown). Accordingly, Tet-regulated expression of wild-type survivin or the survivin 1–141 mutant did not protect HEK293T transfectants against apoptosis induced by the chemotherapeutic agent taxol (Fig. 5C). Conversely, wild-type survivin engineered to accumulate in mitochondria by conjugation with the cytochrome c organelle import sequence efficiently inhibited apoptosis in normal cells (11). Although unable to inhibit apoptosis, wild-type or mutant survivin indistinguishably associated with mitotic chromosomes and polymerized microtubules in Tet-stimulated HEK293T cultures and comparably supported mitotic transitions at metaphase, anaphase, and telophase by dual color fluorescence microscopy (Fig. 5D).
FIGURE 5.
AIP recognition of survivin does not influence mitotic transitions. A, HEK293T cells stably transfected with Tet-inducible full-length survivin or the survivin 1–141 mutant were induced with Tet and analyzed for expression of HA-reactive material at the indicated time intervals by Western blotting. Chloramphenicol acetyltransferase (CAT) was used as a control. B, mitochondrial (M) or cytosolic (C) extracts were isolated from Tet-induced HEK293T survivin transfectants and analyzed by Western blotting. Ran and COX-IV were cytosolic or mitochondrial markers, respectively. C, HEK293T cells expressing vector, wild-type survivin, or the survivin 1–141 mutant were treated with the indicated increasing concentrations of taxol and analyzed for cell viability by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide. Data are mean ± S.E. of replicates of a representative experiment of at least two independent determinations. D, stably transfected HEK293T cells conditionally expressing survivin or survivin 1–141 were analyzed for metaphase or telophase transitions by immunofluorescence microscopy with the indicated antibodies. DNA was stained with DAPI. Merge, image merging analysis.
AIP-Tom20-regulated Mitochondrial Import of Survivin
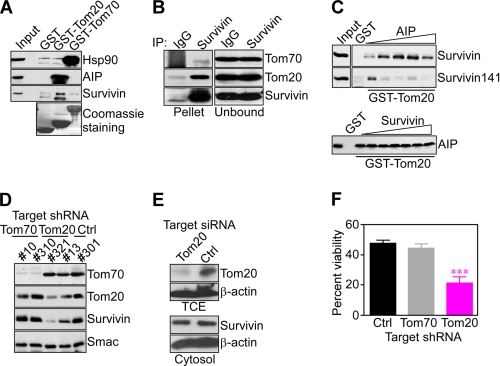
A potential participation of the mitochondrial preprotein import machinery in organelle accumulation of survivin was investigated next. Consistent with earlier reports (19), recombinant Tom70 associated with endogenous Hsp90 but not AIP in capture assays using isolated mitochondrial extracts in vivo (Fig. 6A). Reciprocally, recombinant Tom20 bound AIP (22), whereas Hsp90 was unreactive (Fig. 6A). Under these conditions, survivin strongly bound to a Tom20-AIP complex in capture assays (Fig. 6A). Similarly, survivin immune complexes precipitated from Raji cell extracts contained coassociated Tom20 in vivo (Fig. 6B). In these experiments, AIP was excluded from a survivin-Tom20 complex, suggesting that this interaction is transient and potentially reversed upon delivery of survivin to the mitochondrial import machinery in vivo. In control experiments, only a weak, albeit detectable, interaction between survivin and a Tom70-Hsp90 complex was observed in capture assays (Fig. 6A) as well as immunoprecipitation (Fig. 6B). Although recombinant Tom20 only weakly associated with isolated survivin in vitro (Fig. 6C, top panel), addition of increasing concentrations of recombinant AIP to the binding reaction resulted in dose-dependent, enhanced association of survivin to Tom20 (Fig. 6C, top panel). By contrast, the AIP-defective survivin 1–141 mutant only associated with recombinant Tom20 in the absence of AIP, whereas increasing concentrations of AIP blocked this interaction (Fig. 6C, top panel), potentially reflecting competition for the survivin-Tom20 binding site. As a control, recombinant AIP directly bound Tom20 in vitro (22), and this recognition was unaffected by increasing concentrations of recombinant survivin (Fig. 6C, bottom panel). In contrast, GST did not associate with recombinant proteins, with or without exogenously added AIP or survivin (Fig. 6C).
FIGURE 6.
Tom20 regulation of mitochondrial import of survivin. A, aliquots of GST, GST-Tom20, or GST-Tom70 recombinant proteins were mixed with Raji cell extracts, and bead-bound material was analyzed by Western blotting. Bottom panel, Coomassie Blue staining of recombinant GST fusion proteins. B, Raji mitochondrial extract was immunoprecipitated with an antibody to survivin or IgG, and proteins in pellets or supernatants (unbound) were analyzed by Western blotting. C, recombinant wild-type survivin or the survivin 1–141 mutant and GST-Tom20 beads were mixed with increasing concentration of recombinant AIP, and bead-bound proteins were analyzed by Western blotting (top panel). Bottom panel, GST or GST-Tom20 was incubated with recombinant AIP in the presence of increasing concentrations of survivin (up to 0.4 μm), and bound proteins were analyzed by Western blotting. D, independently established clones (#) of HeLa cells stably transfected with control (Ctrl) or Tom70- or Tom20-directed shRNA were analyzed by Western blotting. E, HeLa cells were transfected with control, non-targeting (Ctrl) or Tom20-directed siRNA and analyzed by Western blotting. TCE, total cell extracts. F, clones of HeLa cells with stable knockdown of Tom70 or Tom20 were treated with 0.5 μm staurosporine for 24 h and analyzed for cell viability by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide. Data are mean ± S.E. of replicates (n = 3). ***, p < 0.0001. Two different clones per conditions were tested with identical results in two independent experiments.
To further investigate a role of mitochondrial import receptors on survivin cytoprotection, we next generated HeLa cells with stable knockdown of Tom20 or Tom70 by shRNA. Tom20- or Tom70-directed shRNA efficiently silenced the expression of the intended target receptor in two independent clones analyzed, but not vice versa (Fig. 6D). Under these conditions, stable silencing of Tom20 inhibited the accumulation of endogenous survivin in HeLa cell mitochondria (Fig. 6D), further reinforcing a role of an AIP-Tom20 recognition in survivin mitochondrial import. In contrast, shRNA silencing of Tom70 did not affect the mitochondrial pool of survivin in HeLa cells (Fig. 6D). Conversely, siRNA silencing of Tom20 did not significantly affect the cytosolic pool of survivin in HeLa cells (Fig. 6E). Functionally, stable shRNA knockdown of Tom20 sensitized HeLa cells to apoptosis induced by the broad cell death stimulus staurosporine (Fig. 6F). In contrast, shRNA silencing of Tom70 or transfection with control, non-targeting shRNA had no effect on staurosporine-induced apoptosis (Fig. 6F).
DISCUSSION
In this study, we have shown that the immunophilin AIP (20, 21) is an essential gene in vivo and that its homozygous deletion in mice causes embryonic lethality at E13.5–14 (27, 28), aberrantly enhanced apoptosis during early erythropoietic differentiation, and loss of survivin expression in both its cytosolic and mitochondrial compartments in vivo. Functionally, AIP directly mediates the mitochondrial import of survivin through a Tom20- but not Tom70-dependent recognition, thus enabling its anti-apoptotic function in vivo (11).
In addition to its role as a cochaperone stabilizing the aryl hydrocarbon receptor (32, 33) in the xenobiotic response to polycyclic aromatic compounds, for instance, 2,3,7,8-tetrachlorodibenzo-p-dioxin or “dioxin” (34), AIP has recently attracted attention for its potential link to certain types of pituitary adenomas (35). Accordingly, ∼15% of patients with a rare neuroendocrine syndrome called familial isolated pituitary adenoma, but not sporadic adenomas (36), have been shown to harbor a variety of defects in the AIP locus (37), including inactivating germ line mutations as well as large genomic deletions (38) that disrupt the COOH-terminal of the AIP protein. Although it is unclear how AIP deficiency in these settings may contribute to pituitary adenomas (39), the observed loss of heterozygosity in patients carrying AIP mutant tumors (40), combined with the reported development of pituitary adenomas in AIP+/− mice (28), has prompted the model that AIP may function in an as yet unidentified pathway of neuroendocrine tumor suppression (41).
Although not ruling out this possibility, the data presented here point to a more complex scenario for AIP function(s) in vivo, which includes a novel, developmentally regulated role in cell survival. In this context, adult-type erythropoiesis becomes established in the embryo between E11.5 and E14 through rapid expansion of erythroid progenitors in the liver, essential for the survival of the embryo beyond E13.5 (42). Ter119− erythroid subsets, which are functionally CFU-e cells, are the most likely to undergo apoptosis during development, a process counteracted by the hormone erythropoietin (25). Our multiparametric flow cytometry profiling of liver cells from AIP knockout embryos identified high levels of apoptosis in these immature CD71−/Ter119− erythropoietic progenitors, which may interfere with the required rapid establishment of erythropoiesis at this stage of development and contribute, at least in part, to the observed lethal phenotype. Importantly, these cells did not exhibit changes in the CD71+/CD71− ratio, which is sensitive to defects in cell cycle transitions (26), reaffirming a pivotal role of increased apoptosis in the observed loss of erythropoietic progenitors in AIP−/− embryos. Other aspects of AIP−/− embryos described here appear in broad agreement with previous reports, reaffirming the invariable embryonic lethality of homozygous AIP deletion by E14–14.5 and the presence of extensive vascular defects, potentially because of developmental abnormalities in blood vessel formation (27, 28). At variance with a previous report (28), however, we found no significant increase in pituitary adenomas in heterozygous AIP+/− mice, compared with their wild-type littermates. The reason for this discrepancy is presently unknown, but potential differences in genetic background or penetrance of this phenotype may account for the results.
Although the basis for the essential developmental role of AIP in vivo remains to be fully elucidated, the data presented here suggest that at least some aspects of this pathway may be attributable to regulation of survivin function, especially subcellular localization to mitochondria (11), which is a molecular prerequisite for survivin cytoprotection in vivo (14). Accordingly, and reminiscent of the phenotype of AIP−/− embryos observed here and in other studies (27, 28), conditional deletion of the survivin gene in the hematopoietic compartment produces lethal defects in erythropoiesis with dramatic downstream reduction of enucleated erythrocytes (30). This pathway has been reported as highly evolutionary conserved (43), potentially reflecting exaggerated apoptosis in the absence of survivin in vivo. Similarly, survivin is essential for proper blood vessel development, and conditional ablation of the survivin gene in endothelial cells also results in embryonic lethality, hemorrhages, and cardiac malformations (44), similar to the phenotype observed for AIP−/− embryos (27).
A role for AIP in mitochondrial preprotein import through the Tom20 import receptor has been proposed previously (22). In agreement with these findings, the AIP pathway of mitochondrial import of survivin described here was specific for AIP and did not involve Hsp90, another survivin-binding chaperone (23) that contributes to mitochondrial preprotein import (19). In addition, AIP-mediated mitochondrial accumulation of survivin was not affected by the organelle transmembrane potential but required the import receptor Tom20, which bound survivin in a recognition cooperatively favored by AIP. Although it is unclear which internal sequence(s) in survivin may participate in organelle targeting (45), the data presented here suggest an overall model in which AIP binds survivin in the cytosol via its COOH terminus sequence, EQLAAMD, with the last Asp residue playing a critical role in the recognition (24). In turn, a survivin-AIP complex is protected against proteolysis in the cytosol (24) and directly delivers survivin to the Tom20 mitochondrial import machinery (this study) for translocation across the mitochondrial membrane (22), thus enabling its anti-apoptotic function (11). Consistent with this scenario, stable shRNA knockdown of Tom20, but not Tom70, abolished survivin import to mitochondria and increased the sensitivity of tumor cells to apoptosis. This is reminiscent of earlier work that also implicated Tom20 in cytoprotection by mediating the insertion of anti-apoptotic Bcl-2 into the mitochondrial outer membrane (46).
Here, experiments with conditional expression of survivin or the AIP-defective survivin 1–141 truncated mutant demonstrated that the AIP-Tom20 recognition is selectively implicated in mitochondrial import of survivin, and thus cytoprotection (11), whereas it had no effect on the localization of survivin to the mitotic apparatus or its essential roles at cell division (31). Despite considerable efforts (47), it has been difficult to unambiguously discriminate between the two main functions of survivin in apoptosis inhibition and the regulation of mitosis, and especially which of these properties is most exploited during tumorigenesis in vivo (7). As shown here, the selectivity by which AIP regulates the mitochondrial import of survivin uniquely marks the function of survivin in cytoprotection, thus providing a specific mechanism to further probe its requirements in development and cancer (7).
Consistent with this view, one of the most distinctive features of mitochondrial survivin is its nearly unique expression in tumor as opposed to normal cells (11), potentially contributing to resistance to therapy and unfavorable disease outcome (7). Here, the differential recruitment of survivin to mitochondria of tumor versus normal cell types (11) was not attributable to differences in AIP expression, which was ubiquitously present in normal or transformed cells, or mitochondrial-intrinsic properties of the tumor type. Conversely, AIP-directed accumulation of survivin in mitochondria was progressively inhibited by cytosolic extracts from normal cells in a pathway that was specific for survivin, as the import of other mitochondrial inter-membrane space molecules was not affected. In this context, it is possible that differential binding of survivin to regulatory molecules in the cytosol, including chaperones (23), may further assist in AIP-directed mitochondrial import selectively in tumor cells, potentially by affecting the availability or recognition of putative survivin organelle targeting sequence(s) (45).
In summary, these data identify a novel cytoprotective pathway centered on an AIP-Tom20 recognition that mediates the mitochondrial import of survivin and thus enables its anti-apoptotic function in vivo (11). Although this mechanism is essential to maintain productive erythropoiesis and tissue integrity during development, its exploitation in tumors likely produces an heightened anti-apoptotic threshold that contributes to disease progression (7). Conversely, because the AIP-survivin recognition is specifically associated with inhibition of apoptosis, molecular antagonists of this pathway may be beneficial to selectively disable survivin cytoprotection in tumors while leaving unscathed its essential functions at cell division in normal tissues.
Acknowledgment
We thank Dr. Takahiro Nagase (Kazusa DNA Research Institute, Japan) for providing the Tom20 and Tom70 cDNAs.
This work was supported, in whole or in part, by National Institutes of Health Grants CA78810, CA140043, HL54131, HL084168, and CA118005.
- IAP
- inhibitor of apoptosis
- TOM
- translocase of outer membrane
- AIP
- aryl hydrocarbon receptor-interacting protein
- GA
- geldanamycin
- PiC
- phosphate carrier gene
- E11
- embryonic day 11.
REFERENCES
- 1. Srinivasula S. M., Ashwell J. D. (2008) Mol. Cell 30, 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gyrd-Hansen M., Meier P. (2010) Nat. Rev. Cancer 10, 561–574 [DOI] [PubMed] [Google Scholar]
- 3. Igney F. H., Krammer P. H. (2002) Nat. Rev. Cancer 2, 277–288 [DOI] [PubMed] [Google Scholar]
- 4. Wilson T. R., Johnston P. G., Longley D. B. (2009) Curr. Cancer Drug Targets 9, 307–319 [DOI] [PubMed] [Google Scholar]
- 5. Mehrotra S., Languino L. R., Raskett C. M., Mercurio A. M., Dohi T., Altieri D. C. (2010) Cancer Cell 17, 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mita A. C., Mita M. M., Nawrocki S. T., Giles F. J. (2008) Clin. Cancer Res. 14, 5000–5005 [DOI] [PubMed] [Google Scholar]
- 7. Altieri D. C. (2008) Nat. Rev. Cancer 8, 61–70 [DOI] [PubMed] [Google Scholar]
- 8. Xia F., Altieri D. C. (2006) Cancer Res. 66, 3392–3395 [DOI] [PubMed] [Google Scholar]
- 9. Small S., Keerthivasan G., Huang Z., Gurbuxani S., Crispino J. D. (2010) Leukemia 24, 1920–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li F., Cheng Q., Ling X., Stablewski A., Tang L., Foster B. A., Johnson C. S., Rustum Y. M., Porter C. W. (2010) Am. J. Pathol. 176, 1629–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dohi T., Beltrami E., Wall N. R., Plescia J., Altieri D. C. (2004) J. Clin. Invest. 114, 1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Song Z., Yao X., Wu M. (2003) J. Biol. Chem. 278, 23130–23140 [DOI] [PubMed] [Google Scholar]
- 13. Ceballos-Cancino G., Espinosa M., Maldonado V., Melendez-Zajgla J. (2007) Oncogene 26, 7569–7575 [DOI] [PubMed] [Google Scholar]
- 14. Dohi T., Xia F., Altieri D. C. (2007) Mol. Cell 27, 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dohi T., Okada K., Xia F., Wilford C. E., Samuel T., Welsh K., Marusawa H., Zou H., Armstrong R., Matsuzawa S., Salvesen G. S., Reed J. C., Altieri D. C. (2004) J. Biol. Chem. 279, 34087–34090 [DOI] [PubMed] [Google Scholar]
- 16. Baker M. J., Frazier A. E., Gulbis J. M., Ryan M. T. (2007) Trends Cell Biol. 17, 456–464 [DOI] [PubMed] [Google Scholar]
- 17. Bolender N., Sickmann A., Wagner R., Meisinger C., Pfanner N. (2008) EMBO Rep. 9, 42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rapaport D. (2005) J. Cell Biol. 171, 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Young J. C., Hoogenraad N. J., Hartl F. U. (2003) Cell 112, 41–50 [DOI] [PubMed] [Google Scholar]
- 20. Carver L. A., Bradfield C. A. (1997) J. Biol. Chem. 272, 11452–11456 [DOI] [PubMed] [Google Scholar]
- 21. Ma Q., Whitlock J. P., Jr. (1997) J. Biol. Chem. 272, 8878–8884 [PubMed] [Google Scholar]
- 22. Yano M., Terada K., Mori M. (2003) J. Cell Biol. 163, 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fortugno P., Beltrami E., Plescia J., Fontana J., Pradhan D., Marchisio P. C., Sessa W. C., Altieri D. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13791–13796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kang B. H., Altieri D. C. (2006) J. Biol. Chem. 281, 24721–24727 [DOI] [PubMed] [Google Scholar]
- 25. Socolovsky M., Murrell M., Liu Y., Pop R., Porpiglia E., Levchenko A. (2007) PLoS Biol. 5, e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pop R., Shearstone J. R., Shen Q., Liu Y., Hallstrom K., Koulnis M., Gribnau J., Socolovsky M. (2010) PLoS Biol. 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin B. C., Sullivan R., Lee Y., Moran S., Glover E., Bradfield C. A. (2007) J. Biol. Chem. 282, 35924–35932 [DOI] [PubMed] [Google Scholar]
- 28. Raitila A., Lehtonen H. J., Arola J., Heliövaara E., Ahlsten M., Georgitsi M., Jalanko A., Paetau A., Aaltonen L. A., Karhu A. (2010) Am. J. Pathol. 177, 1969–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang J., Socolovsky M., Gross A. W., Lodish H. F. (2003) Blood 102, 3938–3946 [DOI] [PubMed] [Google Scholar]
- 30. Leung C. G., Xu Y., Mularski B., Liu H., Gurbuxani S., Crispino J. D. (2007) J. Exp. Med. 204, 1603–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Musacchio A. (2010) Science 330, 183–184 [DOI] [PubMed] [Google Scholar]
- 32. LaPres J. J., Glover E., Dunham E. E., Bunger M. K., Bradfield C. A. (2000) J. Biol. Chem. 275, 6153–6159 [DOI] [PubMed] [Google Scholar]
- 33. Meyer B. K., Perdew G. H. (1999) Biochemistry 38, 8907–8917 [DOI] [PubMed] [Google Scholar]
- 34. Nukaya M., Lin B. C., Glover E., Moran S. M., Kennedy G. D., Bradfield C. A. (2010) J. Biol. Chem. 285, 35599–35605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vierimaa O., Georgitsi M., Lehtonen R., Vahteristo P., Kokko A., Raitila A., Tuppurainen K., Ebeling T. M., Salmela P. I., Paschke R., Gündogdu S., De Menis E., Mäkinen M. J., Launonen V., Karhu A., Aaltonen L. A. (2006) Science 312, 1228–1230 [DOI] [PubMed] [Google Scholar]
- 36. DiGiovanni R., Serra S., Ezzat S., Asa S. L. (2007) Endocr. Pathol. 18, 76–78 [DOI] [PubMed] [Google Scholar]
- 37. Ozfirat Z., Korbonits M. (2010) Mol. Cell. Endocrinol. 326, 71–79 [DOI] [PubMed] [Google Scholar]
- 38. Georgitsi M., Heliövaara E., Paschke R., Kumar A. V., Tischkowitz M., Vierimaa O., Salmela P., Sane T., De Menis E., Cannavò S., Gündogdu S., Lucassen A., Izatt L., Aylwin S., Bano G., Hodgson S., Koch C. A., Karhu A., Aaltonen L. A. (2008) J. Clin. Endocrinol. Metab. 93, 4146–4151 [DOI] [PubMed] [Google Scholar]
- 39. Leontiou C. A., Gueorguiev M., van der Spuy J., Quinton R., Lolli F., Hassan S., Chahal H. S., Igreja S. C., Jordan S., Rowe J., Stolbrink M., Christian H. C., Wray J., Bishop-Bailey D., Berney D. M., Wass J. A., Popovic V., Ribeiro-Oliveira A., Jr., Gadelha M. R., Monson J. P., Akker S. A., Davis J. R., Clayton R. N., Yoshimoto K., Iwata T., Matsuno A., Eguchi K., Musat M., Flanagan D., Peters G., Bolger G. B., Chapple J. P., Frohman L. A., Grossman A. B., Korbonits M. (2008) J. Clin. Endocrinol. Metab. 93, 2390–2401 [DOI] [PubMed] [Google Scholar]
- 40. Beckers A., Daly A. F. (2007) Eur. J. Endocrinol. 157, 371–382 [DOI] [PubMed] [Google Scholar]
- 41. Heliövaara E., Raitila A., Launonen V., Paetau A., Arola J., Lehtonen H., Sane T., Weil R. J., Vierimaa O., Salmela P., Tuppurainen K., Mäkinen M., Aaltonen L. A., Karhu A. (2009) Am. J. Pathol. 175, 2501–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu H., Liu X., Jaenisch R., Lodish H. F. (1995) Cell 83, 59–67 [DOI] [PubMed] [Google Scholar]
- 43. Ma A. C., Chung M. I., Liang R., Leung A. Y. (2009) Leukemia 23, 712–720 [DOI] [PubMed] [Google Scholar]
- 44. Zwerts F., Lupu F., De Vriese A., Pollefeyt S., Moons L., Altura R. A., Jiang Y., Maxwell P. H., Hill P., Oh H., Rieker C., Collen D., Conway S. J., Conway E. M. (2007) Blood 109, 4742–4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koehler C. M. (2004) Annu. Rev. Cell Dev. Biol. 20, 309–335 [DOI] [PubMed] [Google Scholar]
- 46. Motz C., Martin H., Krimmer T., Rassow J. (2002) J. Mol. Biol. 323, 729–738 [DOI] [PubMed] [Google Scholar]
- 47. Colnaghi R., Connell C. M., Barrett R. M., Wheatley S. P. (2006) J. Biol. Chem. 281, 33450–33456 [DOI] [PubMed] [Google Scholar]